1. Introduction
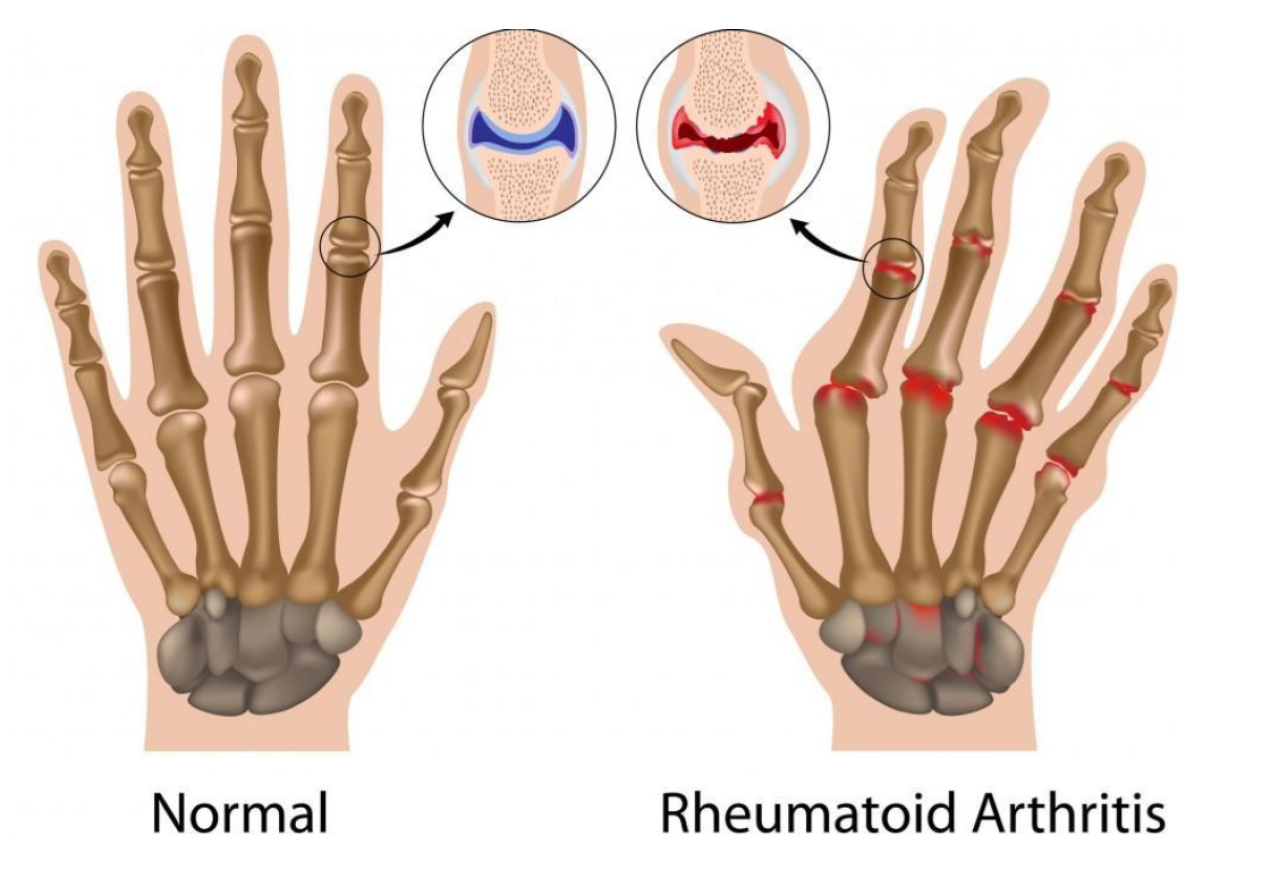
Rheumatoid arthritis (RA) is chronic disease that causes joints inflammation making them swollen, stiff and painful. It primarily affects the small joints of the hands, wrists and feet. As per
Figure 1, the arthritic hand shows visible joint swelling, deformities, and misalignment, while the normal hand maintains typical joint structure and alignment. If left untreated, RA can worsen over time and spread to more joints including knees, elbows and shoulders. Radiography is the standard imaging modality for diagnosing and monitoring the progression of joint changes. Bone erosion and joint space narrowing (JSN) are the most relevant pathological features that can be visualized with radiographs [
1]. To quantify the severity and rate the joint damage, several scoring systems have been proposed. The most commonly used includes the Sharp method [
2] and its variants [
3,
4]. They scale the joint damage with a range of 0 to 5 for the erosion and 0 to 4 for the JSN. Despite considerable efforts to enhance the speed and sensitivity of these scoring methods, they remain constrained by poor reproducibility, low sensitivity to change and the time-consuming nature of expert evaluation. These constraints underscore the potential value of deep learning as a promising solution to improve upon the sensitivity to change while increasing reproducibility and significantly reducing the time needed to score radiographs. Deep learning has evolved as a common method for medical image analysis related to several diseases. Convolutional Neural Networks (CNN) are a popular framework that has gained a crucial role in various medical image diagnosis task including bone fracture detection [
5] and cancer classification [
6].
In the context of RA, deep learning-based method have been explored primarily for automatic joint assessment. The proposed methods fall into two primary categories. The first one, known as single stage methods, includes research works that evaluate all joints simultaneously [
7]. As for those belonging to the second category, called two-stage methods, they first identify the joints and then conduct an individual assessment [
8,
9,
10]. In this context, the elaborated efforts boast results comparable to expert annotations. However, several limiting factors persist preventing their wide adoption either in academic or industrial domain. Most of existing works showcase results on different private datasets, which are inaccessible [
11,
12,
13,
14,
15]. Furthermore, there is a lack of rigorous evaluations of several proposed methods’ performance [
8,
16,
17]. Consequently, the comparison of these approaches to identify a definitive state-of-the-art solution still challenging task.
Starting from this report, this paper introduces a novel framework to enhance the classification of RA severity through the combination of Wavelet Transform and CNN. By leveraging the complementary strengths of both techniques, our approach aims to mitigate the limitations of conventional classification methodologies and improve the accuracy, sensitivity, and specificity of RA diagnosis.
In the following sections, We will present the theoretical foundation of the core technique underlying our proposed method, outline the key components that form the basis of our approach, present the experiments conducted to demonstrate the feasibility and effectiveness of our proposed method.
2. Background
This section provides an overview of the key techniques and models relevant to the problem domain, offering the necessary context for understanding the rationale behind and design of our proposal.
2.1. Wavelet Transform
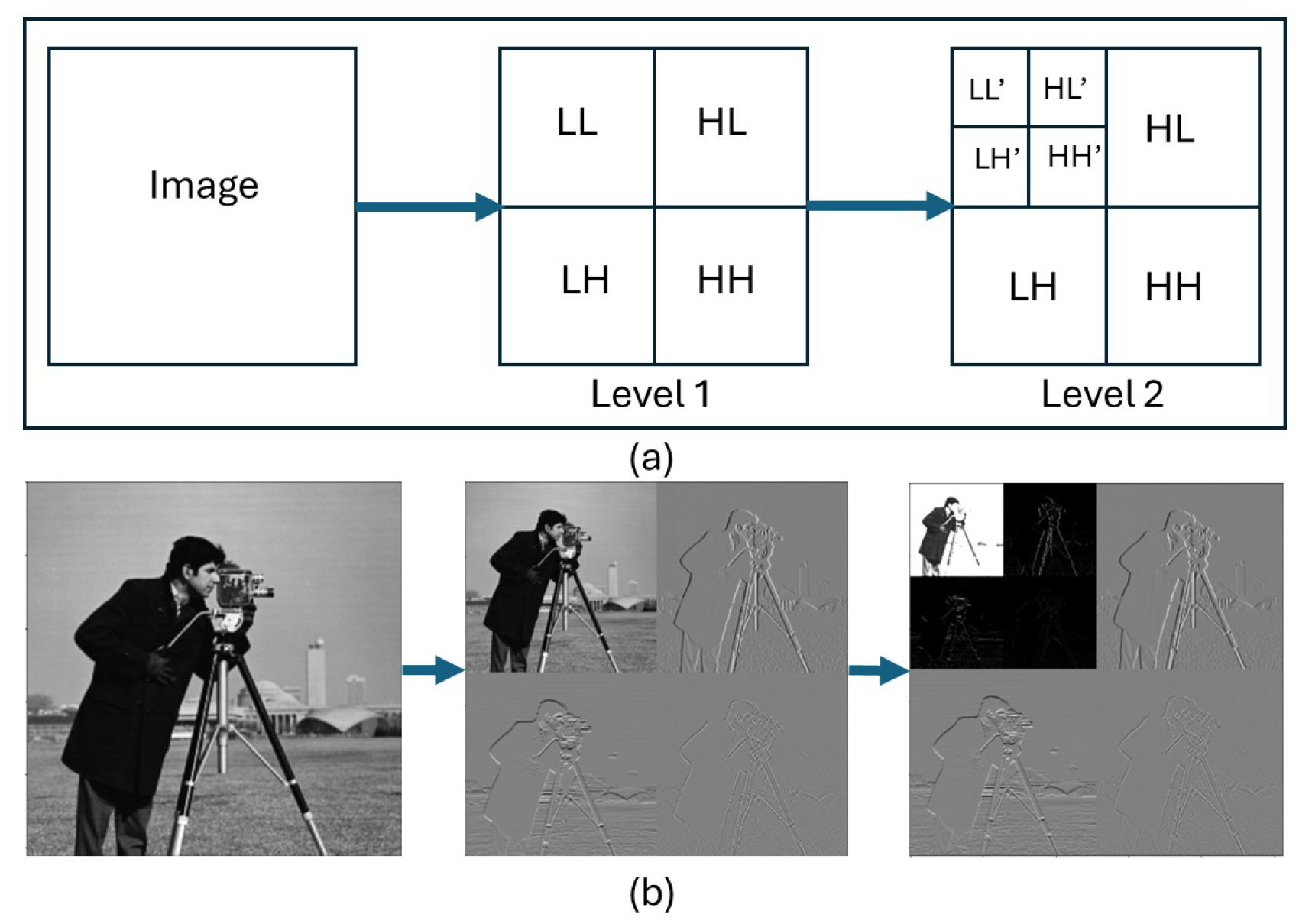
Wavelet Transform is a highly effective technique used in image processing allowing the examination of image data in various level of details. Discrete Wavelet Transform (DWT) is the most commonly used in image processing to decompose an image into set of
approximation (low-frequency) and
detail (high-frequency) sub-bands as shown in
Figure 2. The sub-band LL apply a low-pass filter to the image retained only low-frequency components. It provides a "rough outline" of the image which explains its name
approximation coefficients. The sub-bands (LH,HL, HH), called
detail coefficients, represent the high-frequency details of the image, including sharp edges, textures, and noise. A further decomposition of the LL sub-band is required to obtain the next level of wavelet coefficients.
In image processing, the most common application of DWT are image denoising or edge detection. In both application, the coefficients obtained in the first step undergoes a thresholding step to selectively filter out noise and enhance true edges. Finding an optimum threshold that balance noise removal and feature preservation is the most challenging task in this context. The threshold determines the wavelet coefficient to be retained and those to be discarded or shrinked, thereby it influences the quality of the processed image.
2.2. Convolutional Neural Network CNN
CNN is a class of Artificial Neural Network (ANN) primarily designed to extract features form a grid-like data structures, such as image. CNNs are widely used in computer vision applications due to their effectiveness in processing visual data. They consist of multiple layers containing an input layer, one or more hidden layers, and an output layer. The input layer is the entry point that receives the raw data (e.g. image data) before it passes through the network’s subsequent layers. The output layer is the final layer that produces the predicted result of the model, based on the learned features from all previous layers. Its structure and activation function depend on the type of task the CNN is solving. Hidden layers are the intermediate layers between the input and the output layer. They extract and learn features from the input data, transforming raw pixel values into increasingly abstract representations. Three main types can be identified in the hidden layers:
-
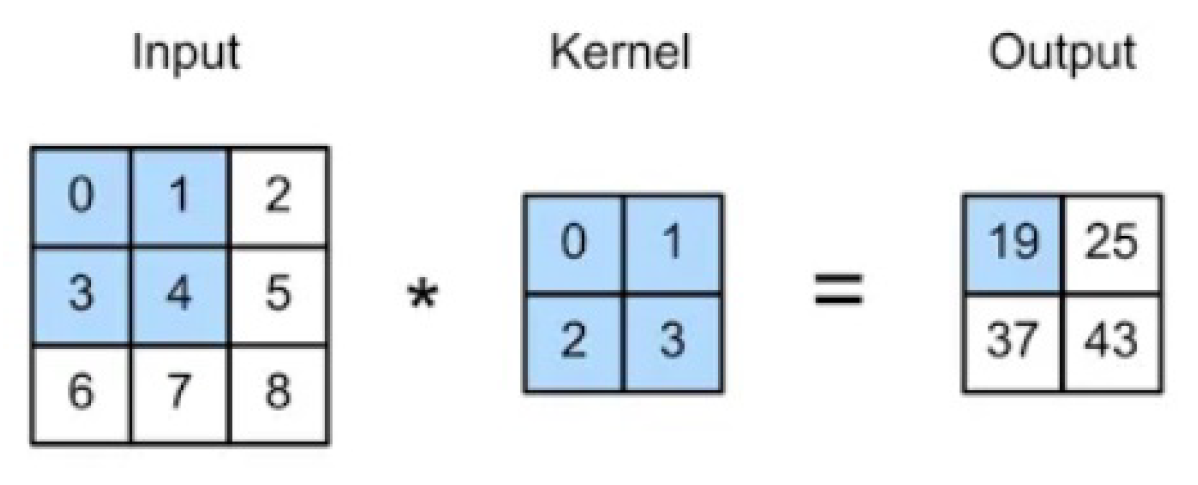
Convolutional layer: is the core building block of a CNN, used to extract the feature from the input images. A set of learnable filters known as the kernels slides over the input image data and computes the dot product between kernel weight and the corresponding input image patch. The output of this layer is a feature map highlighting where the feature appears in the input.
Figure 3.
The convolution operation.
Figure 3.
The convolution operation.
Pooling layer: is used to progressively reduce the size of feature maps while retaining the most important information. It helps to downsample the input, making the model more computationally efficient and less prone to overfitting. The most common types of pooling layers are max pooling and average pooling.
Fully-connected (FC) layer: also known as Dense layer, is a layer in the network where each node in the output layer connects directly to a node in the previous layer. The main function of this layer is to make the classification based on the features extracted through the previous layers and their different filters.
Depending on the architecture that a CNN is made of, several popular CNN models can be identified. AlexNet, ResNet and DenseNet are among the most common ones.
3. Methodology
3.1. Overview of the Wavelet CNN Model
This section presents the details of our proposal to combine the power of Wavelet transform and CNN for the classification of RA severity.
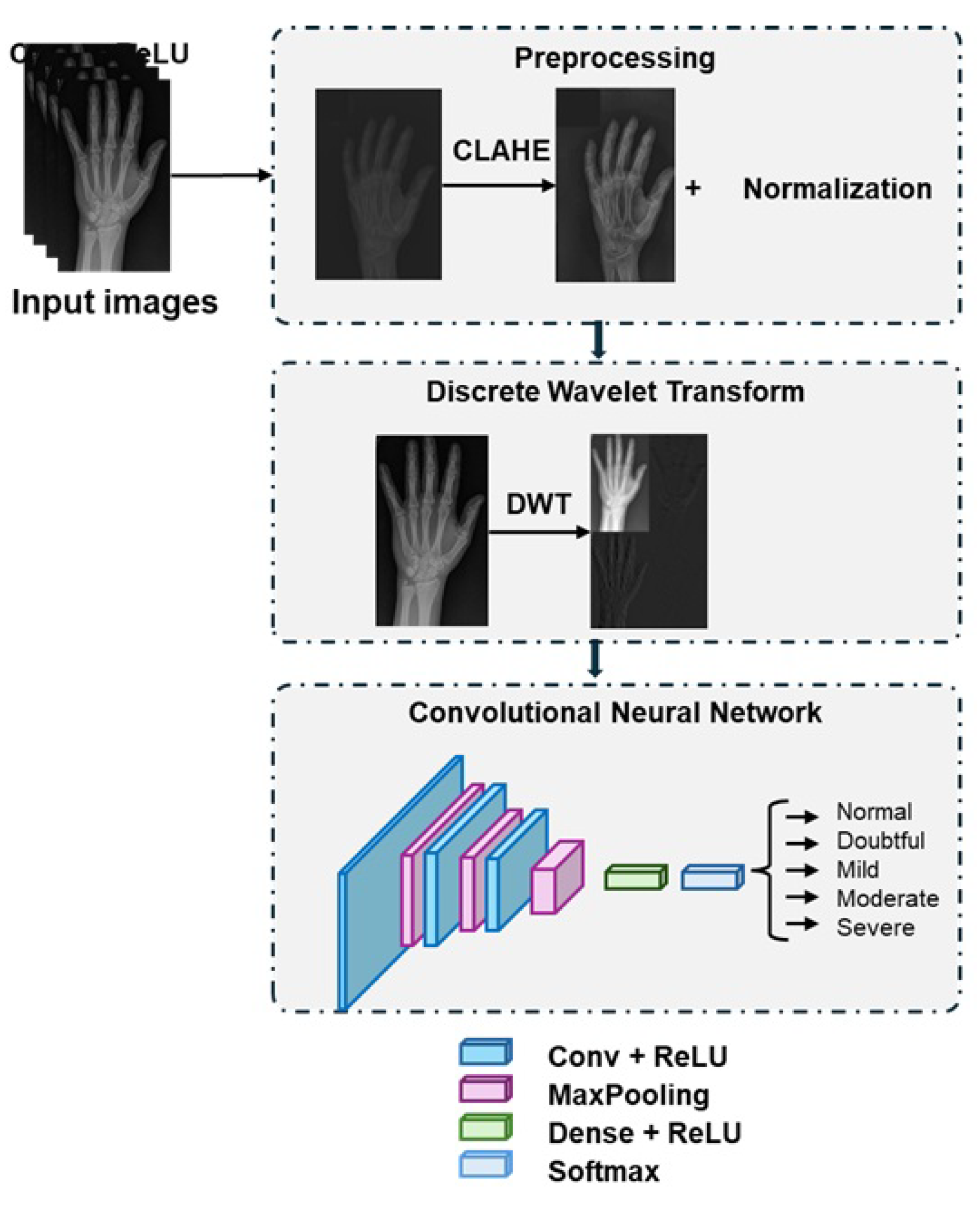
Figure 4 depicts the building blocks of the proposed method.
The proposed method carried out the classification of RA x-ray images into 5 severity level: normal, doubtful, mild, moderate, and severe. The process start by applying two preprocessing steps to enhance the image quality: CLAHE and normalization. Then, the DWT is applied to decompose the image into approximation and details coefficients capturing fine-grained texture and edge information required to identify early stage joint abnormalities. These coefficients are then processed through a CNN to predict the severity of RA.
3.2. Preprocessing
Image preprocessing is a critical step in the analysis and interpretation of medical images data. It refers to the set of operations applied to raw images with the aim of improving their quality and preparing them in the suitable format for further analysis. In this paper, two main operations are considered: contrast adjustment and resizing.
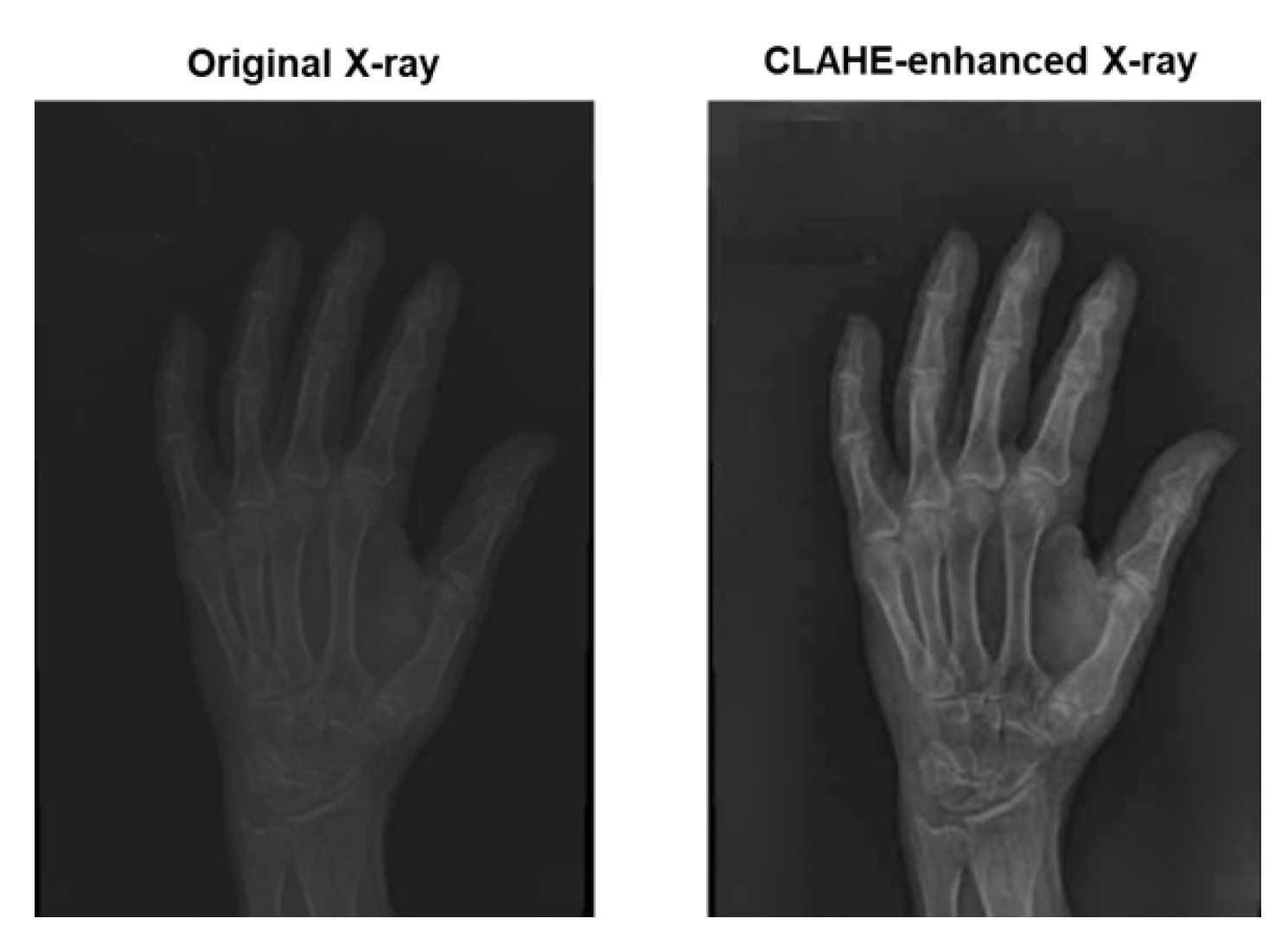
Contrast adjustment aims to remaps the original image intensities to produce a higher contrast one and consequently enhance the quality of that image. X-ray images usually suffer from differences in exposure settings, patient positioning, and scanner types. Hence, pathological features such as bone erosion and joint narrow spacing often subtle. To accurately assess disease severity and progression, we resort to the Contrast Limited Adaptive Histogram Equalization (CLAHE) [
18] to improve the image’s quality. CLAHE operates on small contextual regions called
tiles to perform an histogram equalization with a pre-defined clip limit. Should any histogram bin values be higher than the clip limit, it will be cut (clipped) and distributed uniformly to other bins. CLAHE has proven its ability to effectively enable finer enhancement of localized details while avoiding over-amplification of noise and preserving image details.
Figure 5 illustrate the impact of CLAHE to the quality of an image.
Next, we apply a min-max normalization step to scale pixel values to a common range, typically between 0 and 1. The aim was to ensure consistency in image intensities which allow the model to focus on anatomical features rather than differences in brightness or contrast. X-ray images are then resized to ensure consistent training and inference. In addition, resizing may reduce memory usage and increases training speed, while preserving enough detail for diagnosis.
3.3. Feature Extraction Using DWT
This step aims to identify and represent key patterns or characteristics from x-ray images that are relevant to assessing disease severity. This study employs the DWT as a feature extraction technique prior to the classification. It decomposes an image into multiple sub-bands, enabling the representation of both coarse and fine details across various spatial resolutions. We opted for the Daubechies wavelet to perform such a decomposition due to its computational efficiency and effectiveness in preserving medical image features. A 2-level decomposition is carried out to capture both high- and low-frequency components across multiple scales. The resulting sub-bands, specifically those generated at the second level (LL’,LH’, HL’, and HH’), are selected to be fed into the CNN. The choice of these sub-bands can be justified by the following:
First-level detail coefficients (LH, HL, HH) represent the high-frequency details of the image, including noise and may pick up on irrelevant anatomical or imaging artifacts.
Second-level coefficients filter out much of this noise, providing cleaner features for the CNN to learn from.
Selecting only the second-level sub-bands reduces the size of the input tensor, leading to faster training and lower memory use, while retaining diagnostic value.
The selected wavelet coefficients are then normalized and reshaped into a multi-channel tensor to serve as input for the CNN. By integrating multi-resolution analysis from DWT into the learning process, the model benefits from both spatial and frequency-domain features, ultimately improving its ability to discriminate between different RA severity levels over models trained on raw image data alone.
3.4. RA Severity Classification Using CNN
The features extracted using the DWT are fed into a CNN for classification. The CNN is designed to automatically learn discriminative spatial patterns and hierarchical representations from the wavelet-transformed input, enabling accurate assessment of Rheumatoid Arthritis severity. The proposed CNN structure consists of 4 blocks. The first 3 blocks holds convolution, Rectified Linear Unit (ReLU) activation function and pooling layers. The convolution layers apply multiple filters (3x3) to detect localized patterns within the wavelet sub-bands, such as bone erosion and narrowing of joint spaces. The ReLU activation function (Eq. 1) aims to introduce non-linearity into the network allowing the CNN to learn complex, non-linear patterns that are essential for recognizing pathological features in X-ray images.
The pooling layers follow each convolution stage to perform a downsampling operation that reduces the spatial dimensions of the feature maps. It select the maximum value within non-overlapping regions, typically 2×2 windows. The succession of pooling layers gradually reduce the feature maps size allowing the model to accurately recognize pathological patterns. The fourth block holds a Fully Connected (FC) layer including a Dense layer followed by a ReLU activation function and a Dropout regularization. The Dropout regularization tends to enhance both learning efficiency and generalization performance. The output layer contain a Softmax activation to convert the raw output into interpretable class probabilities.
4. Experiments
4.1. Dataset
We conducted extensive experiments using the dataset provided by [
19]. The dataset includes 185 images separated in 3 files for train, test and validation. The dataset provide a csv file where anatomical regions that are commonly examined in X-rays to assess joint damage are binary classified according to the absence/presence of RA symptom in each of the 12 joint in the hand and wrist (ulna, radius, 5 metacarpal bones, 3 phalanges, soft tissue calcination, artefact). To calculate a severity score for RA from the CSV file, we assign weight to each feature as some features are more clinically important than others and contribute differently to the severity. The total score range from 0 to 23 and data is skewed toward high severity. Hence, we opted for a data-driven quantile-based approach to appropriately classify the severity score. All images were resized to 224×224 pixels and normalized for uniform input to the deep learning models.
4.2. Experimental Setup and Evaluation Metrics
To evaluate the performance of our proposal, we resort to conventional measures used to evaluate an image classification task: accuracy, precision, recall, and f1-score metrics. Their mathematical expressions are presented in what follow:
where TP stands for true positives, FP for false positives, TN for true negatives and FN for false negatives.
The experiment was conducted using the Adam optimizer to minimize the loss function with a batch size of 8 as the dataset is relatively small. The output classes are encoded as integer 1-5, hence we opted for the Sparse Categorical Cross Entropy as a loss function. The learning rate was fixed to 1e-4 and the epochs number was 50. The dataset was split into 3 subsets: training (70%), validation (15%), and testing (15%).
4.3. Performance Comparison
In this section, we compare the performance of our proposed Wavelet CNN framework for RA severity classification with several state-of-the-art deep learning models commonly employed in medical image analysis. The models used for comparison include traditional CNN architectures such as VGG16, ResNet50 and DenseNet121.
We start our experiment by applying the proposed wavelet CNN model and calculate its performance metrics.
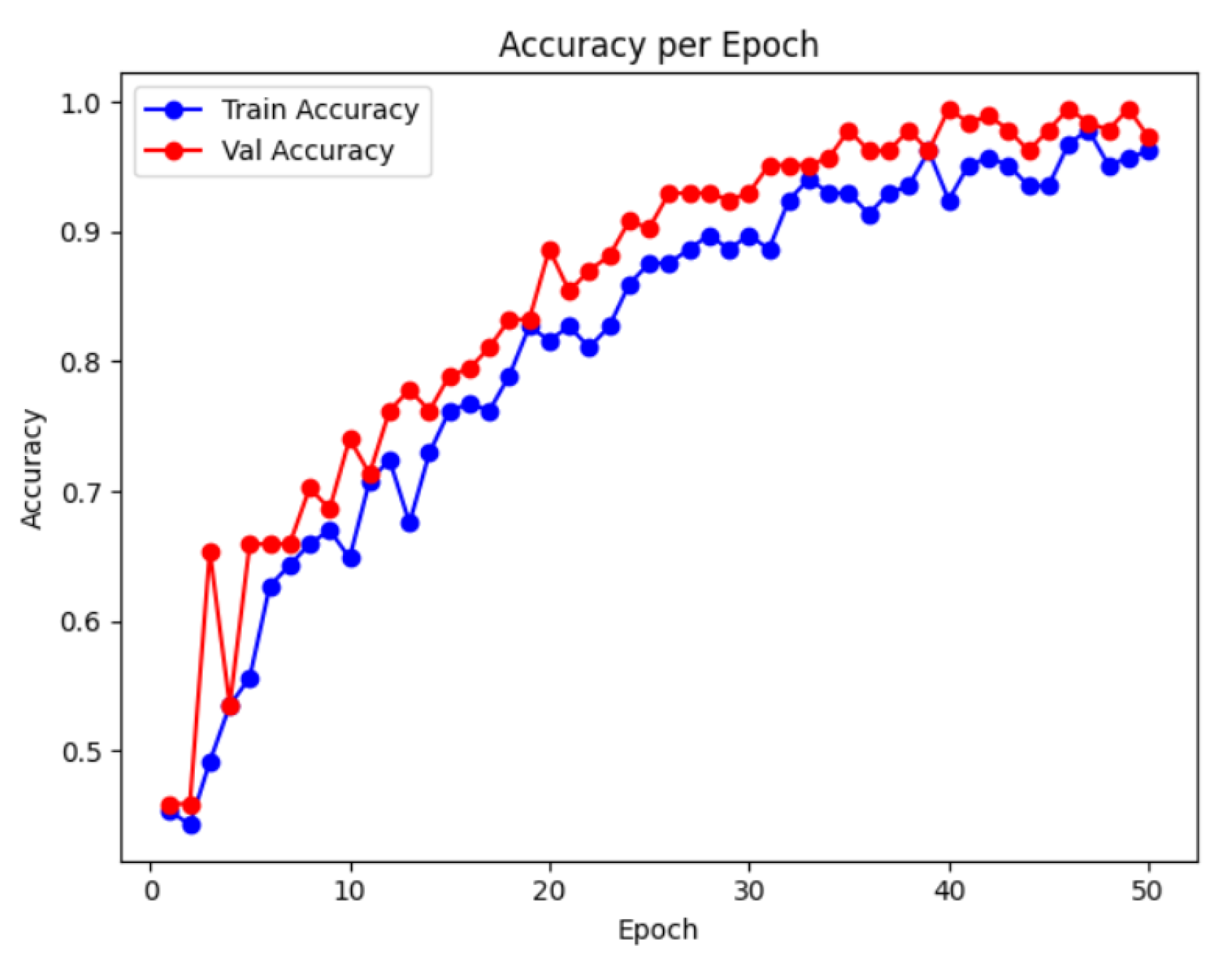
Figure 6 illustrate the training and validation accuracy over successive epochs.
As observed, the training accuracy exhibits a consistent rising trend, indicating that the model effectively learns discriminative features related to RA severity over time. Additionally, the accuracy gradually increases in the early epochs, stabilizing around epoch 40, which suggests good generalization performance and minimal overfitting. The convergence of training and validation accuracy curves demonstrates that the model is not only fitting the training data but is also capable of accurately classifying unseen samples. This performance trend supports the effectiveness of combining wavelet-based feature extraction with deep convolutional learning for RA image classification.
Table 1 shows the performance comparison of the proposed Wavelet CNN over traditional CNN models. As observed, our proposed model outperforms all baseline models across every evaluation metric. The integration of Discrete Wavelet Transform (DWT) enables the network to focus on multi-scale texture and frequency features such as joint space narrowing and bone erosion, which are critical indicators of RA progression. The CNN layers subsequently learn spatial hierarchies of these features more effectively than conventional architectures that rely solely on raw image inputs. These results demonstrate the superiority of the proposed model in capturing both global and local pathological patterns, providing a more accurate and clinically reliable RA severity classification system.
4.4. Discussion
The experimental results clearly demonstrate the key role that the integration of wavelet transforms into the CNN pipeline plays to improve the performance of RA classification. This integration enables more effective extraction of spatial-frequency information from medical images. Decomposing the input image into multiple frequency subbands provides two key advantages. First, it improves the model’s ability to identify the subtle structural deformities and erosions in joints. Second, by leveraging wavelet decomposition, the model retains more information about the original image structure while reducing redundancy.
The Wavelet CNN demonstrated consistently better precision and recall. This indicates that the model is not only more accurate overall but is also more reliable in minimizing false positives and false negatives, which is critical in clinical decision-making. Another important aspect is the model’s robustness across image variations. Rheumatoid Arthritis imaging datasets often suffer from variability in contrast, noise levels, and joint positioning. The multiscale analysis provided by wavelet transforms inherently addresses these issues by capturing invariant features across different frequency bands, improving classification performance under diverse imaging conditions.
In clinical contexts, early and accurate detection of RA is crucial for initiating timely treatment and preventing irreversible joint damage. The promising results of the Wavelet CNN suggest that such architectures could serve as reliable tools in computer-aided diagnosis (CAD) systems, providing rheumatologists with valuable second opinions and improving diagnostic throughput. We argue that the Wavelet CNN represents a meaningful advancement in the application of deep learning to RA classification by successfully bridging the gap between conventional image processing techniques and modern neural network design.
5. Conclusions
This study presented a Wavelet Convolutional Neural Network (Wavelet CNN) approach for the classification of Rheumatoid Arthritis (RA) severity using x-ray imaging. By integrating multi-resolution wavelet transforms with deep convolutional architectures, the model effectively captured both global and local features critical for distinguishing RA severity levels. The experimental results demonstrated that the Wavelet CNN outperformed conventional CNN models in terms of classification accuracy and robustness, particularly in handling subtle joint deformities and varying image quality. These findings suggest that wavelet-based deep learning can offer a promising direction for automated RA diagnosis and severity assessment, supporting more objective and early clinical decision-making. Future work may explore the integration of clinical metadata and the application of the model to other joint-related disorders.
Author Contributions
Conceptualization, methodology, validation, formal analysis, resources, data curation, writing—original draft preparation, and funding acquisition were all performed by the author.
Funding
The authors extend their appreciation to Prince Sattam bin Abdulaziz University for funding this research work through the project number (PSAU/2025/03/33627).
Data Availability Statement
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Bird, A.; Oakden-Rayner, L.; McMaster, C.; Smith, L.A.; Zeng, M.; Wechalekar, M.D.; Ray, S.; Proudman, S.; Palmer, L.J. Artificial intelligence and the future of radiographic scoring in rheumatoid arthritis: a viewpoint. Arthritis Research & Therapy 2022, 24, 268. [Google Scholar]
- Sharp, J.T. Scoring radiographic abnormalities in rheumatoid arthritis. Radiologic Clinics of North America 1996, 34, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Genant, H.K.; Peterfy, C.G.; Westhovens, R.; Becker, J.C.; Aranda, R.; Vratsanos, G.; Teng, J.; Kremer, J.M. Abatacept inhibits progression of structural damage in rheumatoid arthritis: results from the long-term extension of the AIM trial. Annals of the rheumatic diseases 2008, 67, 1084–1089. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.J.; Gherghe, A.M.; Van Der Heijde, D. Radiographic progression in clinical trials in rheumatoid arthritis: a systemic literature review of trials performed by industry. RMD open 2020, 6, e001277. [Google Scholar] [CrossRef] [PubMed]
- Kalmet, P.H.; Sanduleanu, S.; Primakov, S.; Wu, G.; Jochems, A.; Refaee, T.; Ibrahim, A.; Hulst, L.v.; Lambin, P.; Poeze, M. Deep learning in fracture detection: a narrative review. Acta orthopaedica 2020, 91, 215–220. [Google Scholar] [CrossRef]
- Fakoor, R.; Ladhak, F.; Nazi, A.; Huber, M. Using deep learning to enhance cancer diagnosis and classification. In Proceedings of the Proceedings of the international conference on machine learning. ACM New York, NY, USA, 2013, Vol. 28, pp. 3937–3949.
- Wang, Z.; Liu, J.; Gu, Z.; Li, C. An Efficient CNN for Hand X-Ray Overall Scoring of Rheumatoid Arthritis. Complexity 2022, 2022, 5485606. [Google Scholar] [CrossRef]
- Dang, S.; Allison, L. Using deep learning to assign rheumatoid arthritis scores. In2020 IEEE 21st International Conference on Information Reuse and Integration for Data Science (IRI), 2020.
- Chaturvedi, N. DeepRA: predicting joint damage from radiographs using CNN with attention. arXiv 2021, arXiv:2102.06982. [Google Scholar]
- Hargreaves, C.A.; Tan, Y.M.; Quek, R.H.C. Rheumatoid Arthritis: Automated Scoring of Radiographic Joint Damage 2021.
- Sun, D.; Nguyen, T.M.; Allaway, R.J.; Wang, J.; Chung, V.; Yu, T.V.; Mason, M.; Dimitrovsky, I.; Ericson, L.; Li, H.; et al. A crowdsourcing approach to develop machine learning models to quantify radiographic joint damage in rheumatoid arthritis. JAMA network open 2022, 5, e2227423. [Google Scholar] [CrossRef]
- Murakami, S.; Hatano, K.; Tan, J.; Kim, H.; Aoki, T. Automatic identification of bone erosions in rheumatoid arthritis from hand radiographs based on deep convolutional neural network. Multimedia tools and applications 2018, 77, 10921–10937. [Google Scholar] [CrossRef]
- Üreten, K.; Erbay, H.; Maraş, H.H. Detection of rheumatoid arthritis from hand radiographs using a convolutional neural network. Clinical rheumatology 2020, 39, 969–974. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.J.; Su, C.P.; Lai, C.C.; Chen, W.R.; Chen, C.; Ho, L.Y.; Chu, W.C.; Lien, C.Y. Deep learning-based computer-aided diagnosis of rheumatoid arthritis with hand X-ray images conforming to modified total sharp/van der Heijde score. Biomedicines 2022, 10, 1355. [Google Scholar] [CrossRef] [PubMed]
- Radke, K.L.; Kors, M.; Müller-Lutz, A.; Frenken, M.; Wilms, L.M.; Baraliakos, X.; Wittsack, H.J.; Distler, J.H.; Abrar, D.B.; Antoch, G.; et al. Adaptive IoU thresholding for improving small object detection: a proof-of-concept study of hand erosions classification of patients with rheumatic arthritis on X-ray images. Diagnostics 2022, 13, 104. [Google Scholar] [CrossRef] [PubMed]
- Genant, H.K. Methods of assessing radiographic change in rheumatoid arthritis. The American journal of medicine 1983, 75, 35–47. [Google Scholar] [CrossRef] [PubMed]
- Hirano, T.; Nishide, M.; Nonaka, N.; Seita, J.; Ebina, K.; Sakurada, K.; Kumanogoh, A. Development and validation of a deep-learning model for scoring of radiographic finger joint destruction in rheumatoid arthritis. Rheumatology advances in practice 2019, 3, rkz047. [Google Scholar] [CrossRef] [PubMed]
- Zuiderveld, K. Contrast limited adaptive histogram equalization. Graphics gems 1994, 474–485. [Google Scholar]
- x-ray rheumatology. https://universe.roboflow.com/roboflow-100/x-ray-rheumatology/dataset/2. Generated on Aug 30, 2022.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).