Submitted:
07 May 2025
Posted:
08 May 2025
Read the latest preprint version here
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Data Acquisition
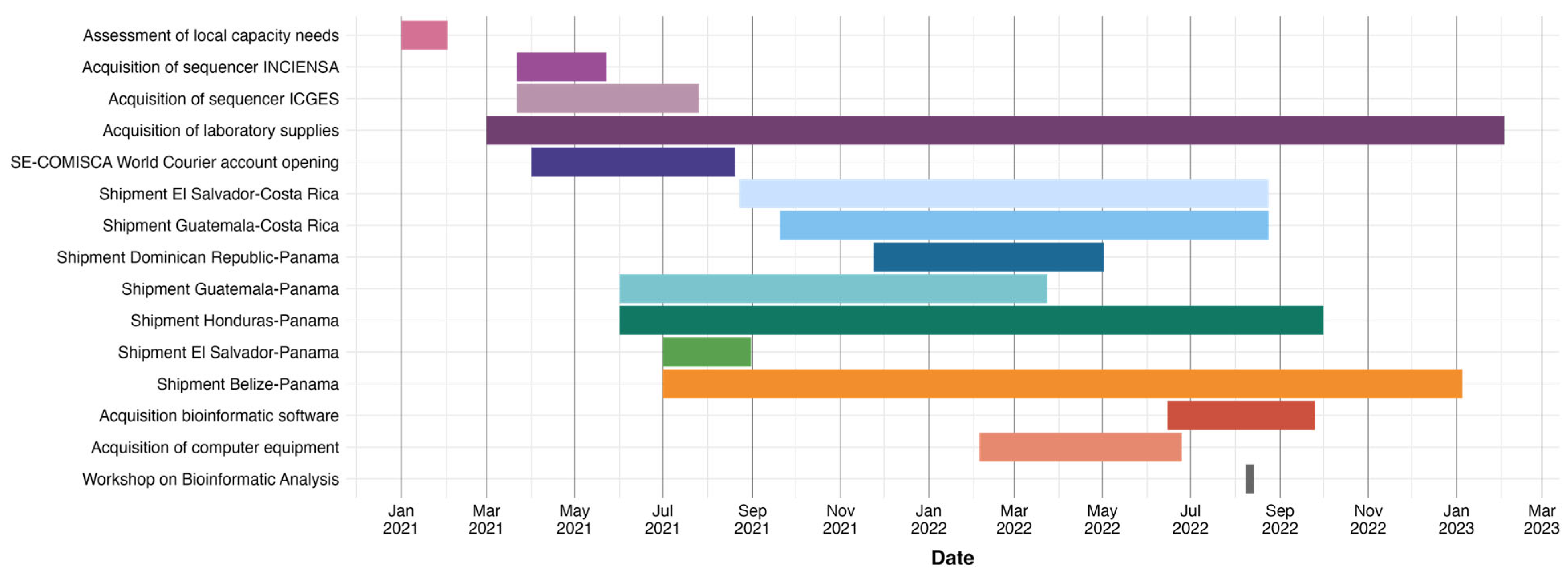
2.2. Timeline of Key Events
2.3. COVID-19 Epidemiological Data
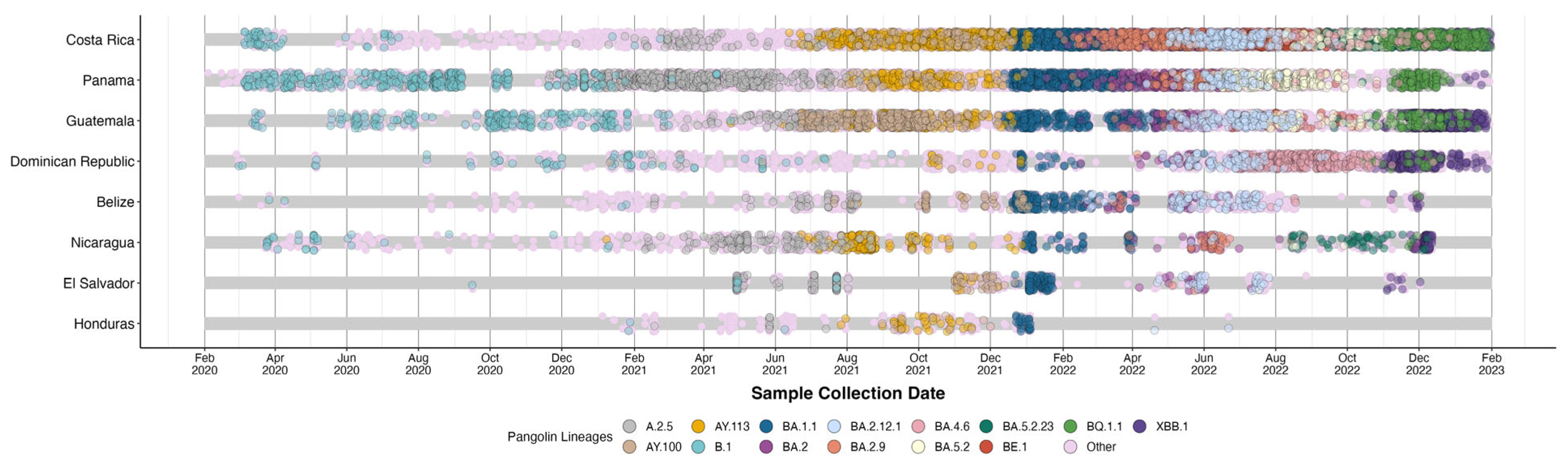
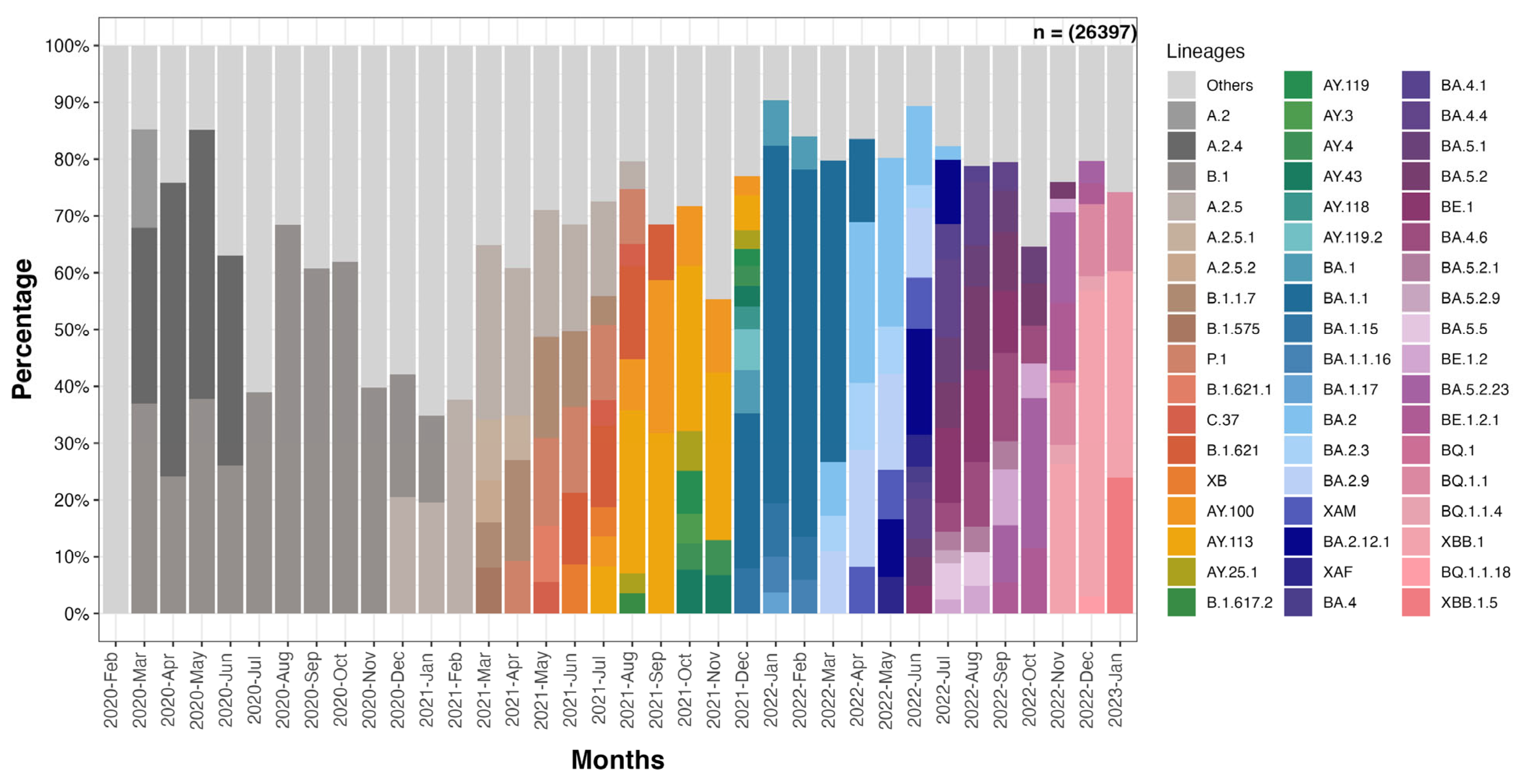
2.4. Analysis of Circulating Variants
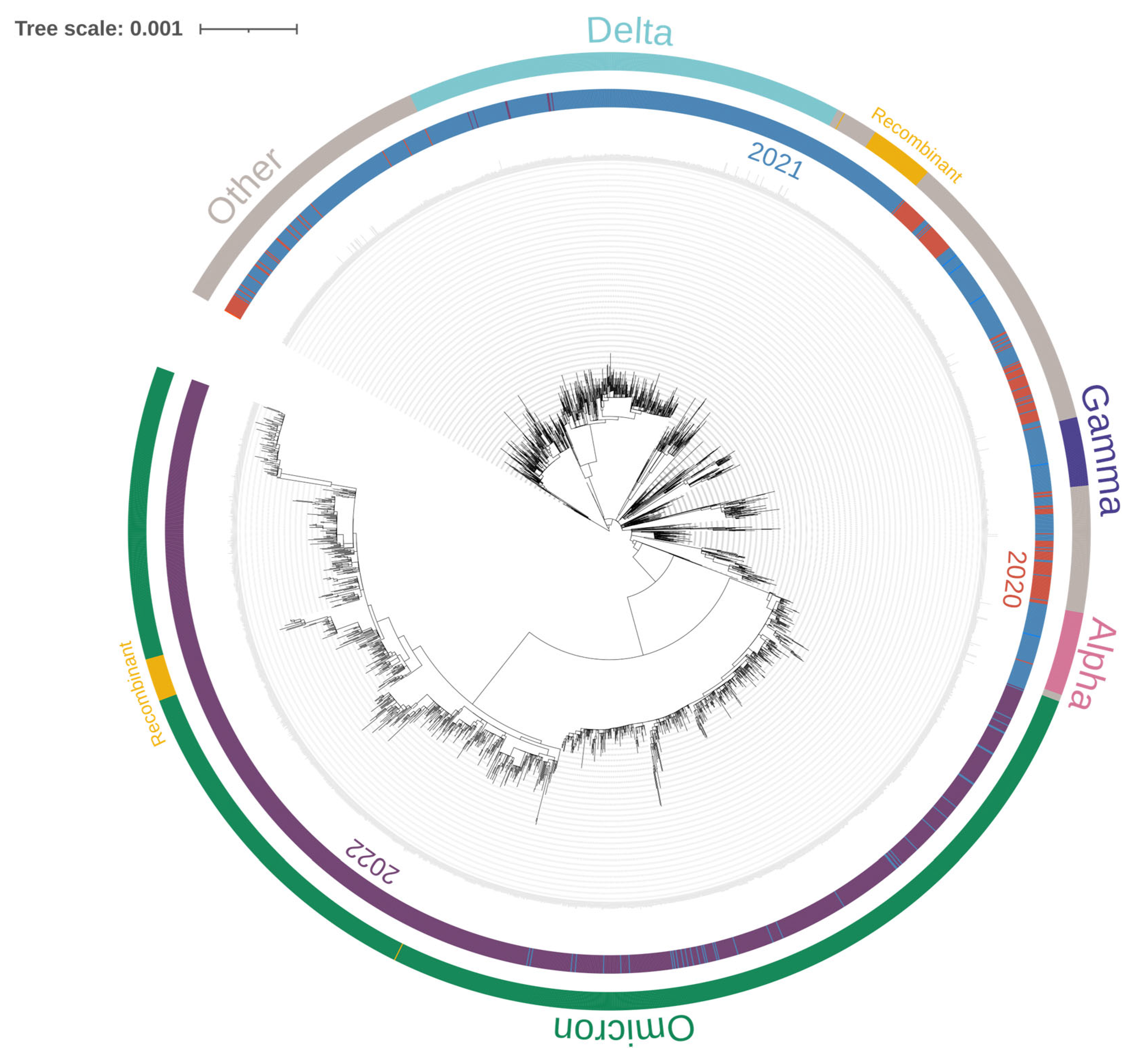
2.5. Phylogenetic Analysis
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| COMISCA | Council of Ministers of Health of Central America |
| COVID-19 | Coronavirus disease 2019 |
| GISAID | Global Initiative on Sharing All Influenza Data |
| ICGES | Instituto Conmemorativo Gorgas de Estudios de la Salud |
| INCIENSA | Instituto Costarricense de Investigación y Enseñanza en Nutrición y Salud |
| PAHO | Pan American Health Organization |
| RNA | Ribonucleic acid |
| SARS | Severe acute respiratory syndrome |
| SE-COMISCA | COMISCA Executive Secretary |
| VOC | Variants of concern |
| VOI | Variants of interest |
| VUM | Variants under monitoring |
| WGS | Whole genome sequencing |
| WHO | World Health Organization |
References
- Zhou, P.; Yang, X. Lou; Wang, X. G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H. R.; Zhu, Y.; Li, B.; Huang, C. L.; Chen, H. D.; Chen, J.; Luo, Y.; Guo, H.; Jiang, R. Di; Liu, M. Q.; Chen, Y.; Shen, X. R.; Wang, X.; Zheng, X. S.; Zhao, K.; Chen, Q. J.; Deng, F.; Liu, L. L.; Yan, B.; Zhan, F. X.; Wang, Y. Y.; Xiao, G. F.; Shi, Z. L. A Pneumonia Outbreak Associated with a New Coronavirus of Probable Bat Origin. Nature 2020, 579 (7798), 270–273. [CrossRef]
- Cucinotta, D.; Vanelli, M. WHO Declares COVID-19 a Pandemic. Acta Biomedica 2020, 91 (1), 157–160. [CrossRef]
- Li, J.; Lai, S.; Gao, G. F.; Shi, W. The Emergence, Genomic Diversity and Global Spread of SARS-CoV-2. Nature 2021, 600 (7889), 408–418. [CrossRef]
- World Health Organization. Tracking SARS-CoV-2 variants. https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/ (accessed 2024-12-08).
- Pan American Health Organization. Update on the emergence of SARS-CoV-2 Omicron sublineages and recombination events (18 November 2022). https://iris.paho.org/handle/10665.2/56767 (accessed 2024-04-21).
- Pan American Health Organization. Epidemiological Update: SARS-CoV-2 variants in the Region of the Americas (1 December 2021). 2021. https://iris.paho.org/handle/10665.2/55321 (accessed 2024-04-21).
- Singh, P.; Sharma, K.; Shaw, D.; Bhargava, A.; Negi, S. S. Mosaic Recombination Inflicted Various SARS-CoV-2 Lineages to Emerge into Novel Virus Variants: A Review Update. Indian Journal of Clinical Biochemistry 2023, 38 (4), 418–425. [CrossRef]
- Molina-Mora, J. A.; Cordero-Laurent, E.; Godínez, A.; Calderón-Osorno, M.; Brenes, H.; Soto-Garita, C.; Pérez-Corrales, C.; Drexler, J. F.; Moreira-Soto, A.; Corrales-Aguilar, E.; Duarte-Martínez, F. SARS-CoV-2 Genomic Surveillance in Costa Rica: Evidence of a Divergent Population and an Increased Detection of a Spike T1117I Mutation. Infection, Genetics and Evolution 2021, 92, 104872. [CrossRef]
- Molina-Mora, J. A. Insights into the Mutation T1117I in the Spike and the Lineage B.1.1.389 of SARS-CoV-2 Circulating in Costa Rica. Gene Rep 2022, 27, 101554. [CrossRef]
- Rambaut, A.; Holmes, E. C.; O’Toole, Á.; Hill, V.; McCrone, J. T.; Ruis, C.; du Plessis, L.; Pybus, O. G. A Dynamic Nomenclature Proposal for SARS-CoV-2 Lineages to Assist Genomic Epidemiology. Nat Microbiol 2020, 5 (11), 1403–1407. [CrossRef]
- Oude Munnink, B. B.; Worp, N.; Nieuwenhuijse, D. F.; Sikkema, R. S.; Haagmans, B.; Fouchier, R. A. M.; Koopmans, M. The next Phase of SARS-CoV-2 Surveillance: Real-Time Molecular Epidemiology. Nat Med 2021, 27 (9), 1518–1524. [CrossRef]
- Stockdale, J. E.; Liu, P.; Colijn, C. The Potential of Genomics for Infectious Disease Forecasting. Nat Microbiol 2022, 7 (11), 1736–1743. [CrossRef]
- Mathieu, E.; Ritchie, H.; Rodés-Guirao, L.; Appel, C.; Giattino, C.; Hasell, J.; Macdonald, B.; Dattani, S.; Beltekian, D.; Ortiz-Ospina, E.; Roser, M. Coronavirus Pandemic (COVID-19). https://ourworldindata.org/covid-cases.
- O’Toole, Á.; Scher, E.; Underwood, A.; Jackson, B.; Hill, V.; McCrone, J. T.; Colquhoun, R.; Ruis, C.; Abu-Dahab, K.; Taylor, B.; Yeats, C.; du Plessis, L.; Maloney, D.; Medd, N.; Attwood, S. W.; Aanensen, D. M.; Holmes, E. C.; Pybus, O. G.; Rambaut, A. Assignment of Epidemiological Lineages in an Emerging Pandemic Using the Pangolin Tool. Virus Evol 2021, 7 (2). [CrossRef]
- Alpert, T.; Brito, A. F.; Lasek-Nesselquist, E.; Rothman, J.; Valesano, A. L.; MacKay, M. J.; Petrone, M. E.; Breban, M. I.; Watkins, A. E.; Vogels, C. B. F.; Kalinich, C. C.; Dellicour, S.; Russell, A.; Kelly, J. P.; Shudt, M.; Plitnick, J.; Schneider, E.; Fitzsimmons, W. J.; Khullar, G.; Metti, J.; Dudley, J. T.; Nash, M.; Beaubier, N.; Wang, J.; Liu, C.; Hui, P.; Muyombwe, A.; Downing, R.; Razeq, J.; Bart, S. M.; Grills, A.; Morrison, S. M.; Murphy, S.; Neal, C.; Laszlo, E.; Rennert, H.; Cushing, M.; Westblade, L.; Velu, P.; Craney, A.; Cong, L.; Peaper, D. R.; Landry, M. L.; Cook, P. W.; Fauver, J. R.; Mason, C. E.; Lauring, A. S.; St. George, K.; MacCannell, D. R.; Grubaugh, N. D. Early Introductions and Transmission of SARS-CoV-2 Variant B.1.1.7 in the United States. Cell 2021, 184 (10), 2595-2604.e13. [CrossRef]
- Katoh, K. MAFFT: A Novel Method for Rapid Multiple Sequence Alignment Based on Fast Fourier Transform. Nucleic Acids Res 2002, 30 (14), 3059–3066. [CrossRef]
- Minh, B. Q.; Schmidt, H. A.; Chernomor, O.; Schrempf, D.; Woodhams, M. D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol Biol Evol 2020, 37 (5), 1530–1534. [CrossRef]
- Kalyaanamoorthy, S.; Minh, B. Q.; Wong, T. K. F.; von Haeseler, A.; Jermiin, L. S. ModelFinder: Fast Model Selection for Accurate Phylogenetic Estimates. Nat Methods 2017, 14 (6), 587–589. [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (ITOL) v4: Recent Updates and New Developments. Nucleic Acids Res 2019, 47 (W1), W256–W259. [CrossRef]
- Pan American Health Organization. Evaluation of the Pan American Health Organization Response to COVID-19 2020–2022. Volume I. Final Report; Pan American Health Organization, 2023. [CrossRef]
- COMISCA. Acuerdo Cooperativo Salud Global COMISCA/CDC apoya el fortalecimiento al Laboratorio INCIENSA. https://www.sica.int/noticias/acuerdo-cooperativo-salud-global-comisca-cdc-apoya-el-fortalecimiento-al-laboratorio-inciensa_1_127875.html (accessed 2024-10-01).
- Salazar Mather, T. P.; Gallo Marin, B.; Medina Perez, G.; Christophers, B.; Paiva, M. L.; Oliva, R.; Hijaz, B. A.; Prado, A. M.; Jarquín, M. C.; Moretti, K.; González Marqués, C.; Murillo, A.; Tobin-Tyler, E. Love in the Time of COVID-19: Negligence in the Nicaraguan Response. Lancet Glob Health 2020, 8 (6), e773. [CrossRef]
- U.S. Census Bureau. International Database (IDB). https://www.census.gov/data-tools/demo/idb/#/dashboard?COUNTRY_YEAR=2023&COUNTRY_YR_ANIM=2023&CCODE_SINGLE=**&CCODE=**&menu=countryViz (accessed 2024-07-24).
- Taboada, B.; Zárate, S.; García-López, R.; Muñoz-Medina, J. E.; Sanchez-Flores, A.; Herrera-Estrella, A.; Boukadida, C.; Gómez-Gil, B.; Mojica, N. S.; Rosales-Rivera, M.; Salas-Lais, A. G.; Gutiérrez-Ríos, R. M.; Loza, A.; Rivera-Gutierrez, X.; Vazquez-Perez, J. A.; Matías-Florentino, M.; Pérez-García, M.; Ávila-Ríos, S.; Hurtado, J. M.; Herrera-Nájera, C. I.; Núñez-Contreras, J. de J.; Sarquiz-Martínez, B.; García-Arias, V. E.; Santiago-Mauricio, M. G.; Martínez-Miguel, B.; Enciso-Ibarra, J.; Cháidez-Quiróz, C.; Iša, P.; Wong-Chew, R. M.; Jiménez-Corona, M. E.; López, S.; Arias, C. F. Dominance of Three Sublineages of the SARS-CoV-2 Delta Variant in Mexico. Viruses 2022, 14 (6). [CrossRef]
- Kumar, S.; Karuppanan, K.; Subramaniam, G. Omicron (BA.1) and Sub-variants (BA.1.1, BA.2, and BA.3) of SARS-CoV-2 Spike Infectivity and Pathogenicity: A Comparative Sequence and Structural-based Computational Assessment. J Med Virol 2022, 94 (10), 4780–4791. [CrossRef]
- Tamura, T.; Ito, J.; Uriu, K.; Zahradnik, J.; Kida, I.; Anraku, Y.; Nasser, H.; Shofa, M.; Oda, Y.; Lytras, S.; Nao, N.; Itakura, Y.; Deguchi, S.; Suzuki, R.; Wang, L.; Begum, M. M.; Kita, S.; Yajima, H.; Sasaki, J.; Sasaki-Tabata, K.; Shimizu, R.; Tsuda, M.; Kosugi, Y.; Fujita, S.; Pan, L.; Sauter, D.; Yoshimatsu, K.; Suzuki, S.; Asakura, H.; Nagashima, M.; Sadamasu, K.; Yoshimura, K.; Yamamoto, Y.; Nagamoto, T.; Schreiber, G.; Maenaka, K.; Ito, H.; Misawa, N.; Kimura, I.; Suganami, M.; Chiba, M.; Yoshimura, R.; Yasuda, K.; Iida, K.; Ohsumi, N.; Strange, A. P.; Takahashi, O.; Ichihara, K.; Shibatani, Y.; Nishiuchi, T.; Kato, M.; Ferdous, Z.; Mouri, H.; Shishido, K.; Sawa, H.; Hashimoto, R.; Watanabe, Y.; Sakamoto, A.; Yasuhara, N.; Suzuki, T.; Kimura, K.; Nakajima, Y.; Nakagawa, S.; Wu, J.; Shirakawa, K.; Takaori-Kondo, A.; Nagata, K.; Kazuma, Y.; Nomura, R.; Horisawa, Y.; Tashiro, Y.; Kawai, Y.; Irie, T.; Kawabata, R.; Motozono, C.; Toyoda, M.; Ueno, T.; Hashiguchi, T.; Ikeda, T.; Fukuhara, T.; Saito, A.; Tanaka, S.; Matsuno, K.; Takayama, K.; Sato, K. Virological Characteristics of the SARS-CoV-2 XBB Variant Derived from Recombination of Two Omicron Subvariants; 2023; Vol. 14. [CrossRef]
- Gardy, J. L.; Loman, N. J. Towards a Genomics-Informed, Real-Time, Global Pathogen Surveillance System. Nat Rev Genet 2018, 19 (1), 9–20. [CrossRef]
- Saravanan, K. A.; Panigrahi, M.; Kumar, H.; Rajawat, D.; Nayak, S. S.; Bhushan, B.; Dutt, T. Role of Genomics in Combating COVID-19 Pandemic. Gene. Elsevier B.V. May 20, 2022. [CrossRef]
- Durón, R. M.; Sánchez, E.; Choi, J. N.; Peralta, G.; Ventura, S. G.; Soto, R. J.; Rodríguez, G.; Ahrens, C.; Farach, E.; Figueroa, J.; Pineda, G.; Romero, A.; Rodríguez, O.; Discua, D.; Salgado, J. Honduras: Two Hurricanes, COVID-19, Dengue and the Need for a New Digital Health Surveillance System. J Public Health (Bangkok) 2021, 43 (2), e297–e298. [CrossRef]
- Vavrek, D.; Speroni, L.; Curnow, K. J.; Oberholzer, M.; Moeder, V.; Febbo, P. G. Genomic Surveillance at Scale Is Required to Detect Newly Emerging Strains at an Early Timepoint. 2021. [CrossRef]
- Brito, A. F.; Semenova, E.; Dudas, G.; Hassler, G. W.; Kalinich, C. C.; Kraemer, M. U. G.; Ho, J.; Tegally, H.; Githinji, G.; Agoti, C. N.; Matkin, L. E.; Whittaker, C.; Kantardjiev, T.; Korsun, N.; Stoitsova, S.; Dimitrova, R.; Trifonova, I.; Dobrinov, V.; Grigorova, L.; Stoykov, I.; Grigorova, I.; Gancheva, A.; Jennison, A.; Leong, L.; Speers, D.; Baird, R.; Cooley, L.; Kennedy, K.; de Ligt, J.; Rawlinson, W.; van Hal, S.; Williamson, D.; Singh, R.; Nathaniel-Girdharrie, S. M.; Edghill, L.; Indar, L.; St. John, J.; Gonzalez-Escobar, G.; Ramkisoon, V.; Brown-Jordan, A.; Ramjag, A.; Mohammed, N.; Foster, J. E.; Potter, I.; Greenaway-Duberry, S.; George, K.; Belmar-George, S.; Lee, J.; Bisasor-McKenzie, J.; Astwood, N.; Sealey-Thomas, R.; Laws, H.; Singh, N.; Oyinloye, A.; McMillan, P.; Hinds, A.; Nandram, N.; Parasram, R.; Khan-Mohammed, Z.; Charles, S.; Andrewin, A.; Johnson, D.; Keizer-Beache, S.; Oura, C.; Pybus, O. G.; Faria, N. R.; Stegger, M.; Albertsen, M.; Fomsgaard, A.; Rasmussen, M.; Khouri, R.; Naveca, F.; Graf, T.; Miyajima, F.; Wallau, G.; Motta, F.; Khare, S.; Freitas, L.; Schiavina, C.; Bach, G.; Schultz, M. B.; Chew, Y. H.; Makheja, M.; Born, P.; Calegario, G.; Romano, S.; Finello, J.; Diallo, A.; Lee, R. T. C.; Xu, Y. N.; Yeo, W.; Tiruvayipati, S.; Yadahalli, S.; Wilkinson, E.; Iranzadeh, A.; Giandhari, J.; Doolabh, D.; Pillay, S.; Ramphal, U.; San, J. E.; Msomi, N.; Mlisana, K.; von Gottberg, A.; Walaza, S.; Ismail, A.; Mohale, T.; Engelbrecht, S.; Van Zyl, G.; Preiser, W.; Sigal, A.; Hardie, D.; Marais, G.; Hsiao, M.; Korsman, S.; Davies, M. A.; Tyers, L.; Mudau, I.; York, D.; Maslo, C.; Goedhals, D.; Abrahams, S.; Laguda-Akingba, O.; Alisoltani-Dehkordi, A.; Godzik, A.; Wibmer, C. K.; Martin, D.; Lessells, R. J.; Bhiman, J. N.; Williamson, C.; de Oliveira, T.; Chen, C.; Nadeau, S.; du Plessis, L.; Beckmann, C.; Redondo, M.; Kobel, O.; Noppen, C.; Seidel, S.; de Souza, N. S.; Beerenwinkel, N.; Topolsky, I.; Jablonski, P.; Fuhrmann, L.; Dreifuss, D.; Jahn, K.; Ferreira, P.; Posada-Céspedes, S.; Beisel, C.; Denes, R.; Feldkamp, M.; Nissen, I.; Santacroce, N.; Burcklen, E.; Aquino, C.; de Gouvea, A. C.; Moccia, M. D.; Grüter, S.; Sykes, T.; Opitz, L.; White, G.; Neff, L.; Popovic, D.; Patrignani, A.; Tracy, J.; Schlapbach, R.; Dermitzakis, E.; Harshman, K.; Xenarios, I.; Pegeot, H.; Cerutti, L.; Penet, D.; Stadler, T.; Howden, B. P.; Sintchenko, V.; Zuckerman, N. S.; Mor, O.; Blankenship, H. M.; de Oliveira, T.; Lin, R. T. P.; Siqueira, M. M.; Resende, P. C.; Vasconcelos, A. T. R.; Spilki, F. R.; Aguiar, R. S.; Alexiev, I.; Ivanov, I. N.; Philipova, I.; Carrington, C. V. F.; Sahadeo, N. S. D.; Branda, B.; Gurry, C.; Maurer-Stroh, S.; Naidoo, D.; von Eije, K. J.; Perkins, M. D.; van Kerkhove, M.; Hill, S. C.; Sabino, E. C.; Pybus, O. G.; Dye, C.; Bhatt, S.; Flaxman, S.; Suchard, M. A.; Grubaugh, N. D.; Baele, G.; Faria, N. R. Global Disparities in SARS-CoV-2 Genomic Surveillance. Nat Commun 2022, 13 (1). [CrossRef]
- Mir, D.; Rego, N.; Resende, P. C.; Tort, F.; Fernández-Calero, T.; Noya, V.; Brandes, M.; Possi, T.; Arleo, M.; Reyes, N.; Victoria, M.; Lizasoain, A.; Castells, M.; Maya, L.; Salvo, M.; Schäffer Gregianini, T.; Mar da Rosa, M. T.; Garay Martins, L.; Alonso, C.; Vega, Y.; Salazar, C.; Ferrés, I.; Smircich, P.; Sotelo Silveira, J.; Fort, R. S.; Mathó, C.; Arantes, I.; Appolinario, L.; Mendonça, A. C.; Benítez-Galeano, M. J.; Simoes, C.; Graña, M.; Motta, F.; Siqueira, M. M.; Bello, G.; Colina, R.; Spangenberg, L. Recurrent Dissemination of SARS-CoV-2 Through the Uruguayan–Brazilian Border. Front Microbiol 2021, 12. [CrossRef]
- Paniz-Mondolfi, A.; Muñoz, M.; Florez, C.; Gomez, S.; Rico, A.; Pardo, L.; Barros, E. C.; Hernández, C.; Delgado, L.; Jaimes, J. E.; Pérez, L.; Teherán, A. A.; Alshammary, H. A.; Obla, A.; Khan, Z.; Dutta, J.; van de Guchte, A.; Gonzalez-Reiche, A. S.; Hernandez, M. M.; Sordillo, E. M.; Simon, V.; van Bakel, H.; Llewellyn, M. S.; Ramírez, J. D. SARS-CoV-2 Spread across the Colombian-Venezuelan Border. Infection, Genetics and Evolution 2020, 86. [CrossRef]
- Patiño, L. H.; Ballesteros, N.; Muñoz, M.; Ramírez, A. L.; Luna, N.; Castañeda, S.; Gutierrez-Marin, R.; Mendoza-Ibarra, J. A.; Rodriguez, R.; Bohada, D. P.; Ramírez, J. D.; Paniz-Mondolfi, A. Mu SARS-CoV-2 (B.1.621) Variant: A Genomic Snapshot across the Colombian-Venezuelan Border. J Med Virol 2023, 95 (4). [CrossRef]
- Hodcroft, E. B.; Zuber, M.; Nadeau, S.; Vaughan, T. G.; Crawford, K. H. D.; Althaus, C. L.; Reichmuth, M. L.; Bowen, J. E.; Walls, A. C.; Corti, D.; Bloom, J. D.; Veesler, D.; Mateo, D.; Hernando, A.; Comas, I.; González-Candelas, F.; González-Candelas, F.; Goig, G. A.; Chiner-Oms, Á.; Cancino-Muñoz, I.; López, M. G.; Torres-Puente, M.; Gomez-Navarro, I.; Jiménez-Serrano, S.; Ruiz-Roldán, L.; Bracho, M. A.; García-González, N.; Martínez-Priego, L.; Galán-Vendrell, I.; Ruiz-Hueso, P.; De Marco, G.; Ferrús, M. L.; Carbó-Ramírez, S.; D’Auria, G.; Coscollá, M.; Ruiz-Rodríguez, P.; Roig-Sena, F. J.; Sanmartín, I.; Garcia-Souto, D.; Pequeno-Valtierra, A.; Tubio, J. M. C.; Rodríguez-Castro, J.; Rabella, N.; Navarro, F.; Miró, E.; Rodríguez-Iglesias, M.; Galán-Sanchez, F.; Rodriguez-Pallares, S.; de Toro, M.; Escudero, M. B.; Azcona-Gutiérrez, J. M.; Alberdi, M. B.; Mayor, A.; García-Basteiro, A. L.; Moncunill, G.; Dobaño, C.; Cisteró, P.; García-de-Viedma, D.; Pérez-Lago, L.; Herranz, M.; Sicilia, J.; Catalán-Alonso, P.; Muñoz, P.; Muñoz-Cuevas, C.; Rodríguez-Rodríguez, G.; Alberola-Enguidanos, J.; Nogueira, J. M.; Camarena, J. J.; Rezusta, A.; Tristancho-Baró, A.; Milagro, A.; Martínez-Cameo, N. F.; Gracia-Grataloup, Y.; Martró, E.; Bordoy, A. E.; Not, A.; Antuori-Torres, A.; Benito, R.; Algarate, S.; Bueno, J.; del Pozo, J. L.; Boga, J. A.; Castelló-Abietar, C.; Rojo-Alba, S.; Alvarez-Argüelles, M. E.; Melon, S.; Aranzamendi-Zaldumbide, M.; Vergara-Gómez, A.; Fernández-Pinero, J.; Martínez, M. J.; Vila, J.; Rubio, E.; Peiró-Mestres, A.; Navero-Castillejos, J.; Posada, D.; Valverde, D.; Estévez-Gómez, N.; Fernandez-Silva, I.; de Chiara, L.; Gallego-García, P.; Varela, N.; Moreno, R.; Tirado, M. D.; Gomez-Pinedo, U.; Gozalo-Margüello, M.; Eliecer-Cano, M.; Méndez-Legaza, J. M.; Rodríguez-Lozano, J.; Siller, M.; Pablo-Marcos, D.; Oliver, A.; Reina, J.; López-Causapé, C.; Canut-Blasco, A.; Hernáez-Crespo, S.; Cordón, M. L. A.; Lecároz-Agara, M. C.; Gómez-González, C.; Aguirre-Quiñonero, A.; López-Mirones, J. I.; Fernández-Torres, M.; Almela-Ferrer, M. R.; Gonzalo-Jiménez, N.; Ruiz-García, M. M.; Galiana, A.; Sanchez-Almendro, J.; Cilla, G.; Montes, M.; Piñeiro, L.; Sorarrain, A.; Marimón, J. M.; Gomez-Ruiz, M. D.; López-Hontangas, J. L.; González Barberá, E. M.; Navarro-Marí, J. M.; Pedrosa-Corral, I.; Sanbonmatsu-Gámez, S.; Pérez-González, C.; Chamizo-López, F.; Bordes-Benítez, A.; Navarro, D.; Albert, E.; Torres, I.; Gascón, I.; Torregrosa-Hetland, C. J.; Pastor-Boix, E.; Cascales-Ramos, P.; Fuster-Escrivá, B.; Gimeno-Cardona, C.; Ocete, M. D.; Medina-Gonzalez, R.; González-Cantó, J.; Martínez-Macias, O.; Palop-Borrás, B.; de Toro, I.; Mediavilla-Gradolph, M. C.; Pérez-Ruiz, M.; González-Recio, Ó.; Gutiérrez-Rivas, M.; Simarro-Córdoba, E.; Lozano-Serra, J.; Robles-Fonseca, L.; de Salazar, A.; Viñuela-González, L.; Chueca, N.; García, F.; Gómez-Camarasa, C.; Carvajal, A.; de la Puente, R.; Martín-Sánchez, V.; Fregeneda-Grandes, J. M.; Molina, A. J.; Argüello, H.; Fernández-Villa, T.; Farga-Martí, M. A.; Domínguez-Márquez, V.; Costa-Alcalde, J. J.; Trastoy, R.; Barbeito-Castiñeiras, G.; Coira, A.; Pérez-del-Molino, M. L.; Aguilera, A.; Planas, A. M.; Soriano, A.; Fernandez-Cádenas, I.; Pérez-Tur, J.; Marcos, M. Á.; Moreno-Docón, A.; Viedma, E.; Mingorance, J.; Galán-Montemayor, J. C.; Parra-Grande, M.; Stadler, T.; Neher, R. A. Spread of a SARS-CoV-2 Variant through Europe in the Summer of 2020. Nature 2021, 595 (7869), 707–712. [CrossRef]
- Serwin, K.; Aksak-Was˛, B.; Parczewski, M. Phylodynamic Dispersal of SARS-CoV-2 Lineages Circulating across Polish–German Border Provinces. Viruses 2022, 14 (5). [CrossRef]
- Arora, P.; Mrig, S.; Goldust, Y.; Kroumpouzos, G.; Karadağ, A. S.; Rudnicka, L.; Galadari, H.; Szepietowski, J. C.; Di Lernia, V.; Goren, A.; Kassir, M.; Goldust, M. New Coronavirus (Sars-Cov-2) Crossing Borders beyond Cities, Nations, and Continents: Impact of International Travel. Balkan Med J 2021, 38 (4), 205–211. [CrossRef]
- Choi, J. Y.; Smith, D. M. SARS-CoV-2 Variants of Concern. Yonsei Med J 2021, 62 (11), 961–968. [CrossRef]
- Zárate, S.; Taboada, B.; Muñoz-Medina, J. E.; Iša, P.; Sanchez-Flores, A.; Boukadida, C.; Herrera-Estrella, A.; Selem Mojica, N.; Rosales-Rivera, M.; Gómez-Gil, B.; Salas-Lais, A. G.; Santacruz-Tinoco, C. E.; Montoya-Fuentes, H.; Alvarado-Yaah, J. E.; Molina-Salinas, G. M.; Espinoza-Ayala, G. E.; Enciso-Moreno, J. A.; Gutiérrez-Ríos, R. M.; Loza, A.; Moreno-Contreras, J.; García-López, R.; Rivera-Gutierrez, X.; Comas-García, A.; Wong-Chew, R. M.; Jiménez-Corona, M.-E.; del Angel, R. M.; Vazquez-Perez, J. A.; Matías-Florentino, M.; Pérez-García, M.; Ávila-Ríos, S.; Castelán-Sánchez, H. G.; Delaye, L.; Martínez-Castilla, L. P.; Escalera-Zamudio, M.; López, S.; Arias, C. F. The Alpha Variant (B.1.1.7) of SARS-CoV-2 Failed to Become Dominant in Mexico. Microbiol Spectr 2022, 10 (2). [CrossRef]
- Stefanelli, P.; Trentini, F.; Guzzetta, G.; Marziano, V.; Mammone, A.; Schepisi, M. S.; Poletti, P.; Grané, C. M.; Manica, M.; del Manso, M.; Andrianou, X.; Ajelli, M.; Rezza, G.; Brusaferro, S.; Merler, S.; Di Martino, A.; Ambrosio, L.; Lo Presti, A.; Fiore, S.; Fabiani, C.; Benedetti, E.; Di Mario, G.; Facchini, M.; Puzelli, S.; Calzoletti, L.; Fontana, S.; Venturi, G.; Fortuna, C.; Marsili, G.; Amendola, A.; Stuppia, L.; Savini, G.; Picerno, A.; Lopizzo, T.; Dell’Edera, D.; Minchella, P.; Greco, F.; Viglietto, G.; Atripaldi, L.; Limone, A.; D’Agaro, P.; Licastro, D.; Pongolini, S.; Sambri, V.; Dirani, G.; Zannoli, S.; Affanni, P.; Colucci, M. E.; Capobianchi, M. R.; Icardi, G.; Bruzzone, B.; Lillo, F.; Orsi, A.; Pariani, E.; Baldanti, F.; Molecolare, U. V.; Gismondo, M. R.; Maggi, F.; Caruso, A.; Ceriotti, F.; Boniotti, M. B.; Barbieri, I.; Bagnarelli, P.; Menzo, S.; Garofalo, S.; Scutellà, M.; Pagani, E.; Collini, L.; Ghisetti, V.; Brossa, S.; Ru, G.; Bozzetta, E.; Chironna, M.; Parisi, A.; Rubino, S.; Serra, C.; Piras, G.; Coghe, F.; Vitale, F.; Tramuto, F.; Scalia, G.; Palermo, C. I.; Mancuso, G.; Pollicino, T.; Di Gaudio, F.; Vullo, S.; Reale, S.; Cusi, M. G.; Rossolini, G. M.; Pistello, M.; Mencacci, A.; Camilloni, B.; Severini, S.; Di Benedetto, M.; Terregino, C.; Monne, I.; Biscaro, V. Co-Circulation of SARS-CoV-2 Alpha and Gamma Variants in Italy, February and March 2021. Eurosurveillance 2022, 27 (5). [CrossRef]
- Oróstica, K. Y.; Mohr, S. B.; Dehning, J.; Bauer, S.; Medina-Ortiz, D.; Iftekhar, E. N.; Mujica, K.; Covarrubias, P. C.; Ulloa, S.; Castillo, A. E.; Daza-Sánchez, A.; Verdugo, R. A.; Fernández, J.; Olivera-Nappa, Á.; Priesemann, V.; Contreras, S. Early Mutational Signatures and Transmissibility of SARS-CoV-2 Gamma and Lambda Variants in Chile. Sci Rep 2024, 14 (1). [CrossRef]
- Rahimi, F.; Kamali, N.; Bezmin Abadi, A. T. The Mu Strain: The Last but Not Least Circulating Variant of Interest’ Potentially Affecting the COVID-19 Pandemic. Future Virology. Future Medicine Ltd. January 1, 2022, pp 5–8. [CrossRef]
- Giovanetti, M.; Fonseca, V.; Wilkinson, E.; Tegally, H.; San, E. J.; Althaus, C. L.; Xavier, J.; Nanev Slavov, S.; Viala, V. L.; Ranieri Jerônimo Lima, A.; Ribeiro, G.; Souza-Neto, J. A.; Fukumasu, H.; Lehmann Coutinho, L.; Venancio Da Cunha, R.; Freitas, C.; Campelo De A E Melo, C. F.; Navegantes De Araújo, W.; Do Carmo Said, R. F.; Almiron, M.; De Oliveira, T.; Coccuzzo Sampaio, S.; Elias, M. C.; Covas, D. T.; Holmes, E. C.; Lourenço, J.; Kashima, S.; De Alcantara, L. C. J. Replacement of the Gamma by the Delta Variant in Brazil: Impact of Lineage Displacement on the Ongoing Pandemic. Virus Evol 2022, 8 (1). [CrossRef]
- Fall, A.; Eldesouki, R. E.; Sachithanandham, J.; Morris, C. P.; Norton, J. M.; Gaston, D. C.; Forman, M.; Abdullah, O.; Gallagher, N.; Li, M.; Swanson, N. J.; Pekosz, A.; Klein, E. Y.; Mostafa, H. H.; Feinstone, W. H. The Displacement of the SARS-CoV-2 Variant Delta with Omicron: An Investigation of Hospital Admissions and Upper Respiratory Viral Loads. 2022. [CrossRef]
- Wang, Y.; Long, Y.; Wang, F.; Li, C.; Liu, W. Characterization of SARS-CoV-2 Recombinants and Emerging Omicron Sublineages. Int J Med Sci 2023, 20 (1), 151–162. [CrossRef]
- Lee, W. L.; Armas, F.; Guarneri, F.; Gu, X.; Formenti, N.; Wu, F.; Chandra, F.; Parisio, G.; Chen, H.; Xiao, A.; Romeo, C.; Scali, F.; Tonni, M.; Leifels, M.; Chua, F. J. D.; Kwok, G. W.; Tay, J. Y.; Pasquali, P.; Thompson, J.; Alborali, G. L.; Alm, E. J. Rapid Displacement of SARS-CoV-2 Variant Delta by Omicron Revealed by Allele-Specific PCR in Wastewater. Water Res 2022, 221. [CrossRef]
- Andre, M.; Lau, L. S.; Pokharel, M. D.; Ramelow, J.; Owens, F.; Souchak, J.; Akkaoui, J.; Ales, E.; Brown, H.; Shil, R.; Nazaire, V.; Manevski, M.; Paul, N. P.; Esteban-Lopez, M.; Ceyhan, Y.; El-Hage, N. From Alpha to Omicron: How Different Variants of Concern of the SARS-Coronavirus-2 Impacted the World. Biology (Basel) 2023, 12 (9). [CrossRef]
- Shiraz, R.; Tripathi, S. Enhanced Recombination Among SARS-CoV-2 Omicron Variants Contributes to Viral Immune Escape. 2022. [CrossRef]
- Paulino-Ramírez, R.; Pham, K.; Breban, M. I.; Peguero, A.; Jabier, M.; Sánchez, N.; Eustate, I.; Ruiz, I.; Grubaugh, N. D.; Hahn, A. M. Genome Sequence of a Recombinant SARS-CoV-2 Lineage XAM (BA.1.1/BA.2.9) Strain from a Clinical Sample in Santo Domingo, Dominican Republic. Microbiol Resour Announc 2023, 12 (1). [CrossRef]
- Wang, Q.; Iketani, S.; Li, Z.; Liu, L.; Guo, Y.; Huang, Y.; Bowen, A. D.; Liu, M.; Wang, M.; Yu, J.; Valdez, R.; Lauring, A. S.; Sheng, Z.; Wang, H. H.; Gordon, A.; Liu, L.; Ho, D. D. Alarming Antibody Evasion Properties of Rising SARS-CoV-2 BQ and XBB Subvariants. Cell 2023, 186 (2), 279-286.e8. [CrossRef]
- Parums, D. V. Editorial: The XBB.1.5 (‘Kraken’) Subvariant of Omicron SARS-CoV-2 and Its Rapid Global Spread. Medical Science Monitor. International Scientific Information, Inc. 2023. [CrossRef]
- Vogel, L. What to Know about Omicron XBB.1.5. CMAJ. Canadian Medical Association Journal 2023, 195 (3), E127–E128. [CrossRef]
- Brito, A. F.; Semenova, E.; Dudas, G.; Hassler, G. W.; Kalinich, C. C.; Kraemer, M. U. G.; Ho, J.; Tegally, H.; Githinji, G.; Agoti, C. N.; Matkin, L. E.; Whittaker, C.; Howden, B. P.; Sintchenko, V.; Zuckerman, N. S.; Mor, O.; Blankenship, H. M.; Oliveira, T. de; Lin, R. T. P.; Siqueira, M. M.; Resende, P. C.; Vasconcelos, A. T. R.; Spilki, F. R.; Aguiar, R. S.; Alexiev, I.; Ivanov, I. N.; Philipova, I.; Carrington, C. V. F.; Sahadeo, N. S. D.; Gurry, C.; Maurer-Stroh, S.; Naidoo, D.; von Eije, K. J.; Perkins, M. D.; Kerkhove, M. van; Hill, S. C.; Sabino, E. C.; Pybus, O. G.; Dye, C.; Bhatt, S.; Flaxman, S.; Suchard, M. A.; Grubaugh, N. D.; Baele, G.; Faria, N. R. Global Disparities in SARS-CoV-2 Genomic Surveillance. August 26, 2021. [CrossRef]
- Wheeler, N. E.; Price, V.; Cunningham-Oakes, E.; Tsang, K. K.; Nunn, J. G.; Midega, J. T.; Anjum, M. F.; Wade, M. J.; Feasey, N. A.; Peacock, S. J.; Jauneikaite, E.; Baker, K. S. Innovations in Genomic Antimicrobial Resistance Surveillance. The Lancet Microbe. Elsevier Ltd December 1, 2023, pp e1063–e1070. [CrossRef]
- Molina-Mora, J. A.; Reales-González, J.; Camacho, E.; Duarte-Martínez, F.; Tsukayama, P.; Soto-Garita, C.; Brenes, H.; Cordero-Laurent, E.; Ribeiro dos Santos, A.; Guedes Salgado, C.; Santos Silva, C.; Santana de Souza, J.; Nunes, G.; Negri, T.; Vidal, A.; Oliveira, R.; Oliveira, G.; Muñoz-Medina, J. E.; Salas-Lais, A. G.; Mireles-Rivera, G.; Sosa, E.; Turjanski, A.; Monzani, M. C.; Carobene, M. G.; Remes Lenicov, F.; Schottlender, G.; Fernández Do Porto, D. A.; Kreuze, J. F.; Sacristán, L.; Guevara-Suarez, M.; Cristancho, M.; Campos-Sánchez, R.; Herrera-Estrella, A. Overview of the SARS-CoV-2 Genotypes Circulating in Latin America during 2021. Front Public Health 2023, 11. [CrossRef]
- Concha-Toloza, M.; González, L. C.; Estrella, A. H. H.; Porto, D. F. Do; Campos-Sánchez, R.; Molina-Mora, J. A. Genomic, Socio-Environmental, and Sequencing Capability Patterns in the Surveillance of SARS-CoV-2 in Latin America and the Caribbean up to 2023. November 15, 2024. [CrossRef]
- Lira-Morales, J. D.; López-Cuevas, O.; Medrano-Félix, J. A.; González-Gómez, J. P.; González-López, I.; Castro-Del Campo, N.; Gomez-Gil, B.; Chaidez, C. Genomic Surveillance of SARS-CoV-2 in México: Three Years since Wuhan, China’s First Reported Case. Viruses 2023, 15 (11), 2223. [CrossRef]
- Ribeiro Dias, M. F.; Andriolo, B. V.; Silvestre, D. H.; Cascabulho, P. L.; Leal da Silva, M. Genomic Surveillance and Sequencing of SARS-CoV-2 across South America. Revista Panamericana de Salud Pública 2023, 47, 1. [CrossRef]
| Country | COVID-19 Cases per Million | Total deaths | Total reported | COVID-19 Cases per Million | Total deaths |
|---|---|---|---|---|---|
| Belize | 174223.1 | 688 | 70610 | 1183 | 1.68 |
| Costa Rica | 228954.6 | 9158 | 1186176 | 9960 | 0.84 |
| Dominican Republic | 58793.97 | 4384 | 660187 | 2628 | 0.40 |
| El Salvador | 31845.41 | 4230 | 201785 | 914 | 0.45 |
| Guatemala | 68736.55 | 20092 | 1226529 | 4623 | 0.38 |
| Honduras | 45103.27 | 11104 | 470556 | 287 | 0.06 |
| Nicaragua | 2240.661 | 245 | 15569 | 1065 | 6.84 |
| Panama | 233567.4 | 8596 | 1029701 | 6613 | 0.64 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





