1. Introduction
Neuropharmacology is a branch of neuroscience investigating how drugs affect the nervous system, particularly the brain, to treat or modify neurological disorders. It integrates multiple scientific disciplines, including cellular biology, molecular biology, biochemistry, and various branches of neuroscience, to understand the mechanisms of drug action on neural pathways. Neuropharmacologists aim to identify drugs that can alleviate symptoms or modify disease progression in conditions such as Alzheimer's, Parkinson's, and epilepsy [Oyebanjo 2024]. This field also examines the role of neurotransmitters, receptors, ion channels, and intracellular signaling in health and disease states [Yang, X 2021]. Understanding these mechanisms is vital for developing targeted therapies to improve patient outcomes. In addition to its role in disease management, neuropharmacology is critical in advancing knowledge about brain function and behavior. Researchers can uncover new insights into the molecular basis of cognition, mood regulation, and motor control by studying the interaction between pharmaceutical agents and the nervous system [Sultana 2024]. This knowledge is crucial for developing treatments for existing disorders and predicting potential side effects and interactions of new drugs. The field continues to evolve rapidly, driven by technological advances like high-resolution imaging and computational modeling, which enable more precise and personalized therapeutic
AI is the simulation of human intelligence by machines, especially computer systems. It encompasses processes such as learning, reasoning, and self-correction. AI systems use algorithms to analyze data, identify patterns, and make predictions or decisions, often surpassing human capabilities in certain tasks [Mahapatra C. 2025]. This technology finds applications ranging from autonomous systems to natural language processing. AI is revolutionizing healthcare and pharmacology by enabling advanced data analysis and decision-making [Alowais, S 2023]. In healthcare, AI aids in early disease detection, personalized treatment planning, and medical imaging diagnostics [Mahapatra, C. 2025]. AI accelerates pharmacology drug discovery and development processes, optimizing candidate identification, clinical trials, and side effect prediction. For instance, AI-driven algorithms analyze vast datasets to identify molecular targets, design compounds, and predict their efficacy and safety, reducing the cost and time required for drug development [Gupta, R 2021]. AI also supports pharmacists by automating routine tasks and enabling personalized patient care. Beyond healthcare, AI is transforming the finance, transportation, and education industries [Rashid, A. B 2024]. In finance, AI enhances fraud detection, risk assessment, and algorithmic trading by analyzing large volumes of financial data in real-time [Inampudi, R. K 2023]. Autonomous vehicles and smart traffic systems in transportation rely on AI to improve safety and efficiency [Hosseinian, S. M 2024]. In education, AI-powered tools provide personalized learning experiences, adapting content and pacing to individual students’ needs [Kaswan, K. S 2024]. The intersection of AI and neuropharmacology is an emerging field with significant potential to transform the development of neurotherapeutics. AI can enhance our understanding of complex neural pathways, identify drug targets, and personalize treatments for neurological disorders [Tsuji, S 2021]. For example, machine learning models are being developed to simulate brain activity, predict epileptic seizures, and explore the interaction of drugs with neuronal networks [Raikar, A. S 2024]. These advances could lead to breakthroughs in the management of conditions like Alzheimer's disease by refining cognitive-enhancing treatments or identifying biomarkers for early diagnosis.
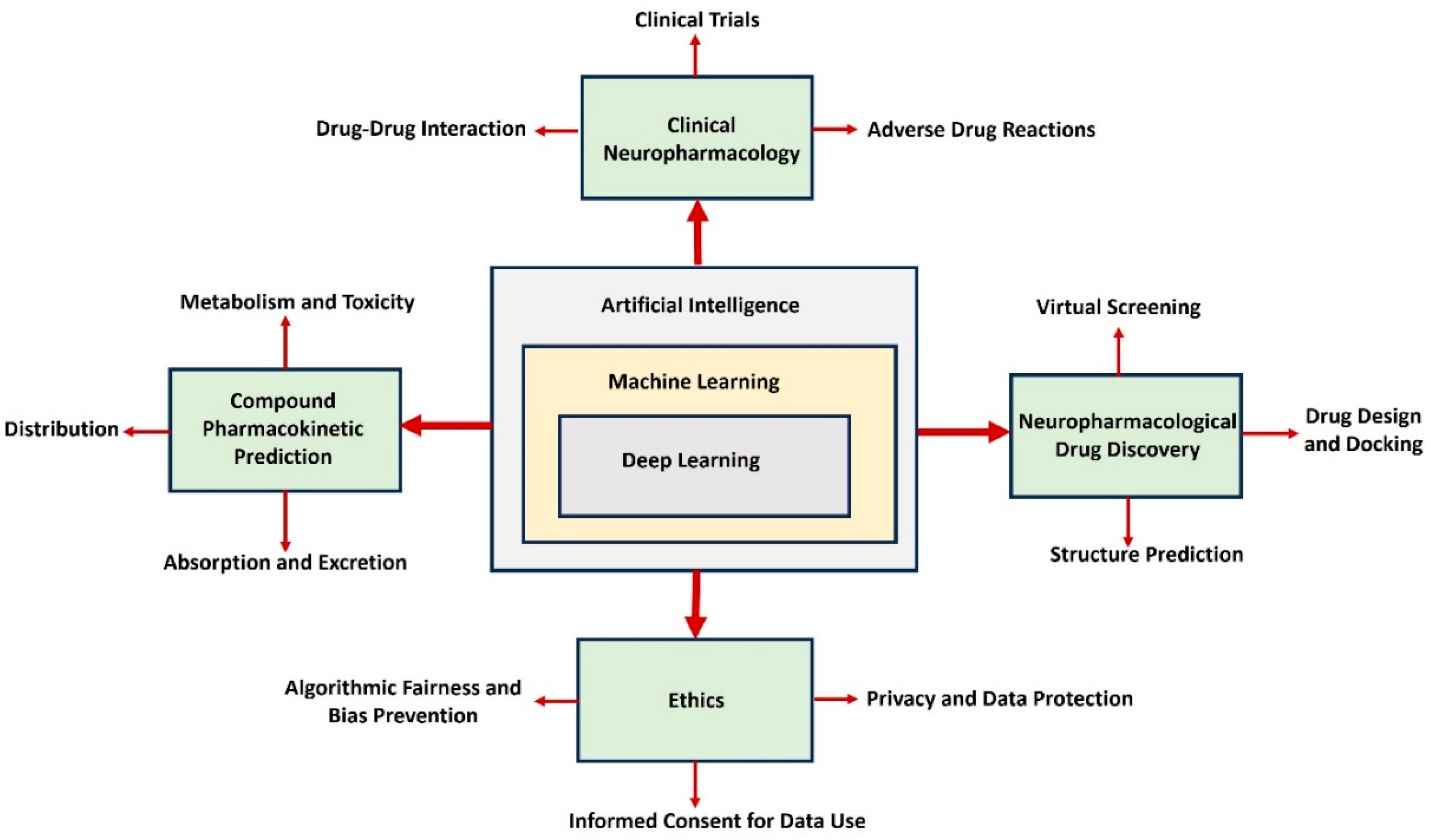
Figure 1 demonstrates the integration of AI, machine learning, and deep learning, emphasizing their transformative applications in neuropharmacological research and drug development.
AI is crucial in advancing multiple facets of drug discovery, including clinical neuropharmacology, pharmacokinetic predictions, and drug design processes [Vatansever, S 2021]. Its key functionalities encompass predicting drug-drug interactions, evaluating adverse drug reactions, and modeling pharmacokinetic parameters such as absorption, distribution, metabolism, and excretion. Furthermore, AI facilitates drug discovery through virtual screening, structure prediction, and the development of innovative drug molecules [Jiménez-Luna 2021]. The diagram also highlights the collaborative relationship between these domains to enhance drug development efficiency and precision. For instance, clinical neuropharmacology leverages AI for more accurate detection of drug-drug interactions, reducing risks in clinical trials. Similarly, pharmacokinetic modeling uses AI to predict how compounds behave within the body, streamlining the selection of safer and more effective candidates [Paliwal, A2024]. AI-based tools in neuropharmacological drug discovery accelerate the design and refinement of therapeutic compounds, significantly optimizing drug docking and structure prediction [Schauperl, M.,2022]. The inclusion of ethical considerations, such as addressing biases, ensuring fairness, protecting privacy, and obtaining informed consent for data use, underscores the importance of responsible AI implementation [Díaz-Rodríguez 2023]. Collectively, these advancements depict how AI is revolutionizing the drug discovery process by making it faster, more accurate, and ethically aligned. By integrating AI into neuropharmacology, researchers can accelerate drug discovery, reduce costs, and improve patient outcomes, marking a new era in neuroscience. Despite its promise, challenges such as data privacy, ethical considerations, and the integration of AI tools into clinical practice remain [Karimian, G., 2022]. Additionally, the reliance on high-quality, diverse datasets to train AI systems underscores the need for collaboration between researchers, clinicians, and tech developers. Transparency and explainability in AI-driven decisions are crucial to gaining trust among practitioners and patients [Wysocki, O 2023]. Equally important is establishing robust regulatory frameworks to ensure that AI applications adhere to ethical standards while safeguarding patient rights. This involves addressing potential algorithm biases, ensuring equitable access to AI-driven healthcare solutions, and maintaining patient confidentiality through stringent data protection measures. By fostering interdisciplinary cooperation and prioritizing fairness and accountability, the healthcare industry can maximize the benefits of AI while minimizing risks.
A comprehensive review is essential to summarize the state-of-the-art advancements, highlight challenges, and propose directions for future research in this niche field [Alfarano, A 2024]. Our objective is to create a detailed review paper that provides an in-depth analysis of the advanced applications of AI in neuropharmacology. This includes evaluating current AI-driven methodologies for drug discovery, understanding neural networks, and predicting drug responses. The review aims to identify knowledge gaps, inspire further research, and propose frameworks for integrating AI innovations into neuropharmacological practices. We hope to contribute to more effective and personalized treatments for neurological disorders by bridging the knowledge between AI advancements and neuropharmacology. Furthermore, this review will explore the potential of AI to revolutionize patient-specific treatment plans by leveraging its ability to process vast datasets and identify patterns in neurological disease progression. It will also examine ethical considerations, including data privacy and algorithmic transparency, to ensure the responsible application of AI technologies in healthcare. The paper seeks to guide researchers and practitioners toward implementing AI-based strategies to address unmet clinical needs, reduce treatment costs, and enhance the quality of life for patients with complex neurological conditions by providing a critical synthesis of existing studies.
2. Applications of AI For Neurotransmitter Analysis
Neuropharmacology examines neurotransmitters, which perform diverse functions in the nervous system. These chemical messengers are common targets for drugs aimed at treating neurological and psychiatric conditions [Nimgampalle 2023].
Table 1 lists primary neurotransmitters, their agonists and antagonists, and their functional roles in neurological disorders.
Accurate assessment of neurotransmitter systems is essential for understanding neurological and psychiatric disorders, as well as for developing targeted therapies [Mahapatra C. et al., 2025]. AI has emerged as a transformative tool in this area, enhancing the precision, efficiency, and depth of neurotransmitter analysis. Neurotransmitter systems are complex, involving interactions between synthesis, release, receptor binding, reuptake, and degradation processes [Nimgampalle 2023]. Traditional methods for studying neurotransmitters, such as high-performance liquid chromatography (HPLC), mass spectrometry, and positron emission tomography (PET), have provided valuable insights [Varrone, A. 2022]. However, these techniques often require extensive time, expertise, and resources, and they may struggle to capture the dynamic nature of neurotransmitter activity in real-time or across large populations. AI has brought significant advancements to neurotransmitter analysis by enabling the processing of large, complex datasets and the identification of subtle patterns that traditional methods may miss [Mahapatra C. et al.,2024]. Machine learning algorithms can analyze data from a variety of sources, including imaging studies, electrophysiological recordings, and biochemical assays, to map neurotransmitter activity and its relationship to neural circuits and behavior [RothRichard 2020]. This capability is particularly valuable in understanding disorders such as depression, schizophrenia, Parkinson’s disease, and addiction, which often involve dysregulation of neurotransmitter systems. In imaging-based neurotransmitter analysis, AI enhances the interpretation of PET and functional MRI (fMRI) data [Zuo, C 2024]. PET imaging is commonly used to quantify neurotransmitter receptor availability, transporters, and overall neurotransmitter dynamics. AI algorithms can automate the analysis of PET scans, improving accuracy and reducing human error [Varrone, A. 2022]. Additionally, AI models can integrate imaging data with other modalities, such as genetic and proteomic data, to provide a more comprehensive understanding of neurotransmitter systems [Mahapatra C. et al.,2024]. AI also supports dynamic modeling of neurotransmitter systems, capturing temporal changes that are critical for understanding brain function. For example, reinforcement learning and recurrent neural networks (RNNs) have been employed to model neurotransmitter release and reuptake processes in response to stimuli [Schmidgall, S 2024]. These models help researchers predict how neurotransmitter systems adapt to various conditions, such as stress, drug exposure, or therapeutic interventions [Mahapatra C. et al.,2018]. In drug development, AI has become a valuable tool for analyzing the effects of pharmacological agents on neurotransmitter systems. By simulating the interaction between drugs and neurotransmitter receptors, AI models can predict efficacy, side effects, and optimal dosages [Vatansever, S 2021]. This predictive capability reduces the reliance on time-consuming and expensive experimental testing. For example, in studying treatments for dopamine-related disorders such as Parkinson’s disease, AI algorithms can identify drug candidates that effectively modulate dopaminergic activity while minimizing adverse effects [Mahapatra C. et al.,2017]. Moreover, AI facilitates personalized approaches to neurotransmitter analysis by leveraging individual-level data. Using machine learning, researchers can analyze patient-specific profiles, including genetic variants, brain imaging results, and clinical symptoms, to tailor interventions that address unique neurotransmitter imbalances [Oyebanjo, O. T 2024]. This personalized approach is particularly promising in neuropsychiatric conditions like depression and anxiety, where responses to treatments targeting serotonin or norepinephrine systems can vary widely [Jones, C 2021]. Despite these advancements, challenges remain in applying AI to neurotransmitter analysis. The quality and consistency of input data are critical, as AI models are only as reliable as the datasets they are trained on. Standardizing data collection methods and ensuring diverse representation in datasets are ongoing priorities [Mahapatra C. et al.,2015]. Additionally, the interpretability of AI models is an important consideration, particularly in clinical applications where transparency and validation are essential. Looking ahead, the integration of AI with emerging technologies, such as single-cell sequencing, advanced neuroimaging, and wearable biosensors, holds immense potential for neurotransmitter analysis [Meng, Z 2024]. These combined approaches will provide deeper insights into the role of neurotransmitters in health and disease, ultimately leading to more effective and personalized therapies. By harnessing AI’s capabilities, neuropharmacology is poised to advance our understanding of brain chemistry and revolutionize the treatment of neurological and psychiatric disorders.
3. Applications of AI For Drug Discovery in Neuropharmacology
AI is revolutionizing neuropharmacology by enhancing the understanding and treatment of neurological disorders. AI addresses long-standing challenges and drives innovative solutions, from accelerating drug discovery to modeling disease mechanisms.
Drug Discovery and Development
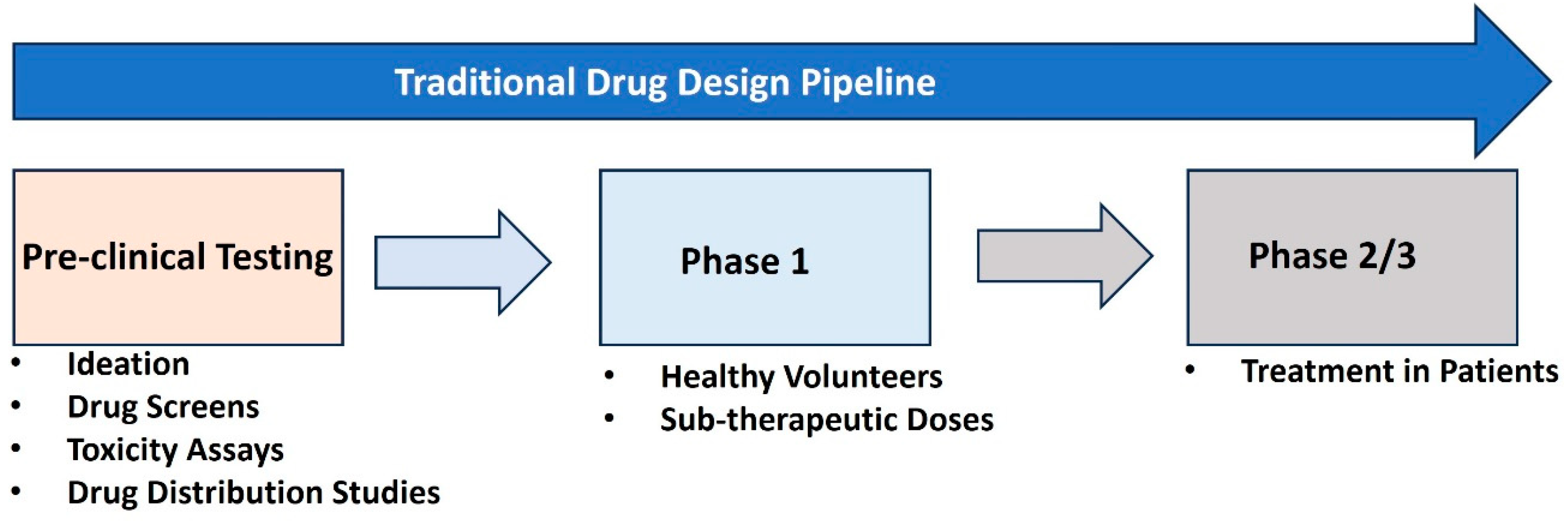
Figure 2 illustrates the Traditional Drug Design Pipeline, which consists of three key stages in drug development [Sadybekov, A 2023]. The first stage, Pre-clinical Testing, involves ideation, drug screening, toxicity assays, and drug distribution studies to assess the safety and biological activity of the drug candidates in vitro (test tubes) or in vivo (animal models). After passing this stage, the drug enters Phase 1, where it is tested on healthy volunteers at sub-therapeutic doses to evaluate safety, dosage, and pharmacokinetics. Finally, in Phase 2/3, the drug is administered to patient populations to assess its therapeutic effectiveness and further monitor safety, leading to potential approval for widespread use if successful. This pipeline represents a systematic and iterative process to ensure drugs are safe and effective before clinical application.
AI has significantly enhanced the traditional drug discovery pipeline by streamlining and optimizing each of its stages. During the pre-clinical testing phase, AI-driven algorithms excel in ideation and drug screening by predicting molecular interactions and identifying promising compounds from vast chemical libraries [Pu, Z 2024]. Machine learning models can analyze in vitro and in vivo data to predict a compound's toxicity and pharmacokinetic properties with higher accuracy, reducing the reliance on labor-intensive and time-consuming experimental assays [Catacutan, D. B 2024]. Additionally, AI tools help prioritize compounds with the greatest potential for success, expediting the initial evaluation process [Tiwari, P. C 2023]. In Phase 1 and Phase 2/3 of clinical trials, AI facilitates the identification of optimal dosing regimens and patient stratification, ensuring that trials are conducted more efficiently and safely [Poweleit 2023]. By analyzing vast datasets from previous clinical studies and real-world evidence, AI systems can predict adverse reactions, refine therapeutic doses, and suggest suitable biomarkers for assessing efficacy [Yang, S 2023]. This accelerates decision-making and enhances the reliability of trial outcomes. Overall, AI's integration into the drug discovery process reduces costs and timelines while maintaining the rigor needed to ensure new therapies are safe and effective for widespread use. AI plays a pivotal role in transforming the traditional drug discovery pipeline, which is often expensive and time-consuming. By utilizing machine learning algorithms, researchers can analyze vast datasets to predict the interactions of molecules with biological targets [Mahapatra C. et al.,2024]. These predictions streamline the identification of promising drug candidates, significantly reducing the need for exhaustive experimental testing. For example, AI-driven molecular docking techniques allow scientists to virtually screen thousands of compounds for their potential efficacy against neurological targets [Vicidomini, 2024]. This capability not only speeds up the discovery process but also optimizes resource allocation by focusing on the most promising candidates. Additionally, AI can predict safety profiles and potential side effects of new compounds, helping to mitigate risks during early development phases. These advancements have shortened timelines and reduced costs, making the development of treatments for complex neurological disorders more feasible and efficient.
Virtual Screening
Virtual screening (VS) has become a cornerstone of drug discovery and pharmacological research, leveraging computational techniques to evaluate vast libraries of chemical compounds [Singh, N. K 2025]. The primary goal is to identify candidates with strong potential to bind effectively to a biological target, such as a protein implicated in disease.
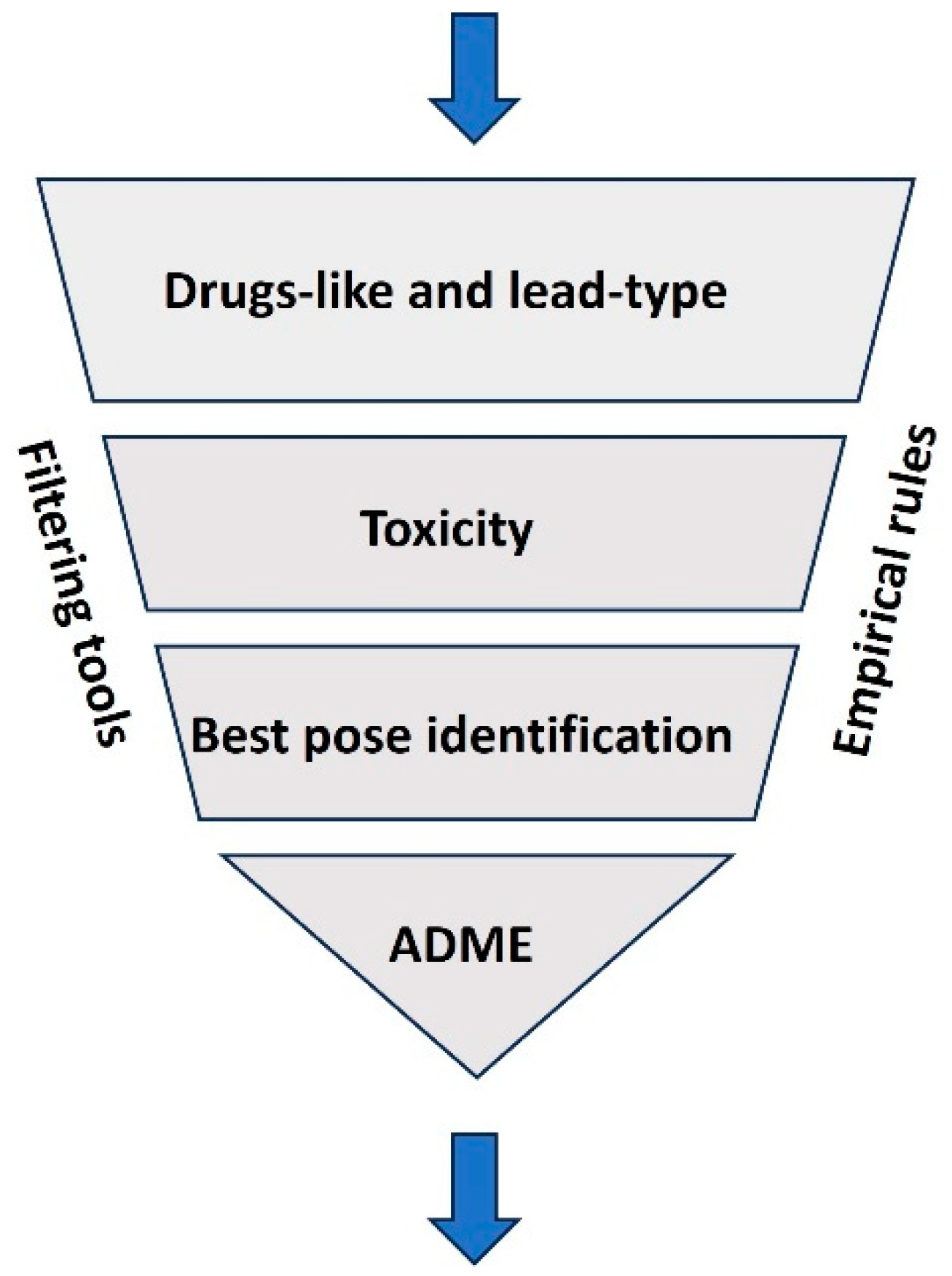
Figure 3 illustrates the VS process, which involves filtering compounds based on various criteria, such as drug-likeness, lead-likeness, and adherence to empirical rules like Lipinski's Rule of Five [Ononamadu, C 2021]. Following this, advanced algorithms assess the toxicity of the selected compounds to eliminate those with undesirable effects. The next stage involves identifying the best molecular pose, where potential drugs are evaluated for their ability to bind effectively to a target, typically a protein or receptor [Mahapatra C. et al.,2019]. Finally, ADME (Absorption, Distribution, Metabolism, and Excretion) properties are predicted to ensure the compounds are suitable for further development [Sucharitha 2022]
Virtual screening streamlines drug discovery by prioritizing the most promising candidates, reducing experimental workload, and accelerating the development process. VS methodologies are typically categorized into two main approaches: ligand-based and structure-based [Bhunia 2021]. Ligand-based screening utilizes information from known active compounds to infer the activity of similar molecules. In contrast, structure-based screening relies on the three-dimensional configuration of the target protein to pinpoint compatible compounds [Mahapatra C. et al.,2019]. This approach greatly expedites the drug discovery process by focusing efforts on promising candidates before experimental validation. AI has significantly enhanced the efficiency and precision of virtual screening [Kasaraneni 2021]. AI-powered algorithms, especially those employing machine learning and deep learning techniques, are adept at handling large datasets and uncovering intricate patterns. These technologies facilitate the prediction of molecular properties, simulate interactions between ligands and targets, and optimize potential drug candidates. For instance, AI tools can estimate binding affinities, assess toxicity risks, and predict pharmacokinetic profiles with remarkable accuracy, reducing the dependence on labor-intensive traditional methods [Abbas 2024]. Deep learning architectures such as convolutional neural networks and graph neural networks have proven instrumental in analyzing molecular data. By transforming molecular structures into computational representations, these models can predict biological activities and rank compounds by their therapeutic potential [Mahapatra C. et al.,2017]. Moreover, generative models like variational autoencoders and generative adversarial networks are increasingly used to design novel molecules tailored to specific pharmacological requirements [Martinelli 2022]. AI also enhances the efficiency of molecular docking, a key process in structure-based virtual screening. Through predictive models, AI can determine the optimal binding poses and energies of compounds with greater speed and reliability compared to traditional docking methods [Choudhury, C 2022]. Reinforcement learning algorithms further refine the optimization of drug candidates, ensuring higher specificity and efficacy [Kalusivalingam 2021]. The integration of AI into virtual screening offers multiple advantages. It accelerates the screening process, minimizes costs, and delivers results with improved accuracy. Additionally, AI systems can be customized for specific drug discovery projects, incorporating parameters such as toxicity thresholds and desired molecular properties [Serrano 2024]. Combining AI with other technologies like high-performance computing and quantum mechanics further amplifies the precision and efficiency of the drug design process [Lappala 2024]. As AI continues to advance, its impact on virtual screening is poised to revolutionize drug discovery pipelines. These innovations promise not only to speed up the development of new therapies but also to enhance their quality and success rates, transforming the landscape of pharmacological research.
AI-Assisted Docking Algorithms
AI-assisted docking algorithms represent a significant advancement in computational drug discovery, leveraging machine learning to predict binding affinities between proteins and potential ligands with high accuracy [Singh, S 2024]. These algorithms are trained on extensive datasets of experimental and simulated binding interactions, uncovering patterns that traditional methods might overlook. By integrating experimental data—such as high-throughput screening results and structural insights from X-ray crystallography or cryo-electron microscopy—with predictive algorithms, these tools refine scoring functions to estimate binding affinities more precisely [Maveyraud 2020]. Techniques like neural networks, random forests, and support vector machines enhance the evaluation of potential drug candidates, addressing the limitations of conventional approaches. One notable benefit of AI-driven docking is its ability to streamline the docking process, reducing computational demands while effectively prioritizing promising compounds [Bettanti 2024]. By narrowing large chemical libraries to a focused set of highly active candidates, these algorithms accelerate the early stages of drug discovery and increase the likelihood of identifying hits and leads with strong target interactions [Mahapatra C. et al.,2025]. Machine learning models' adaptability further ensures continuous improvement as new data becomes available, enhancing predictions and improving the overall drug discovery pipeline. Additionally, graph neural networks bring further innovation to molecular docking by accurately predicting the binding affinity and orientation of small molecules (ligands) with target proteins. Graph neural networks (GNNs) analyze experimental and computational data to determine optimal ligand conformations and binding energies within the protein's active site [Knutson 2022]. This detailed understanding helps identify promising therapeutic compounds early, expediting drug development timelines and increasing efficiency. While challenges such as data quality and interpretability persist, ongoing advancements in AI promise to revolutionize the pharmaceutical landscape, making drug development faster, more cost-effective, and highly precise.
4. AI Techniques Used in Neuropharmacological Research
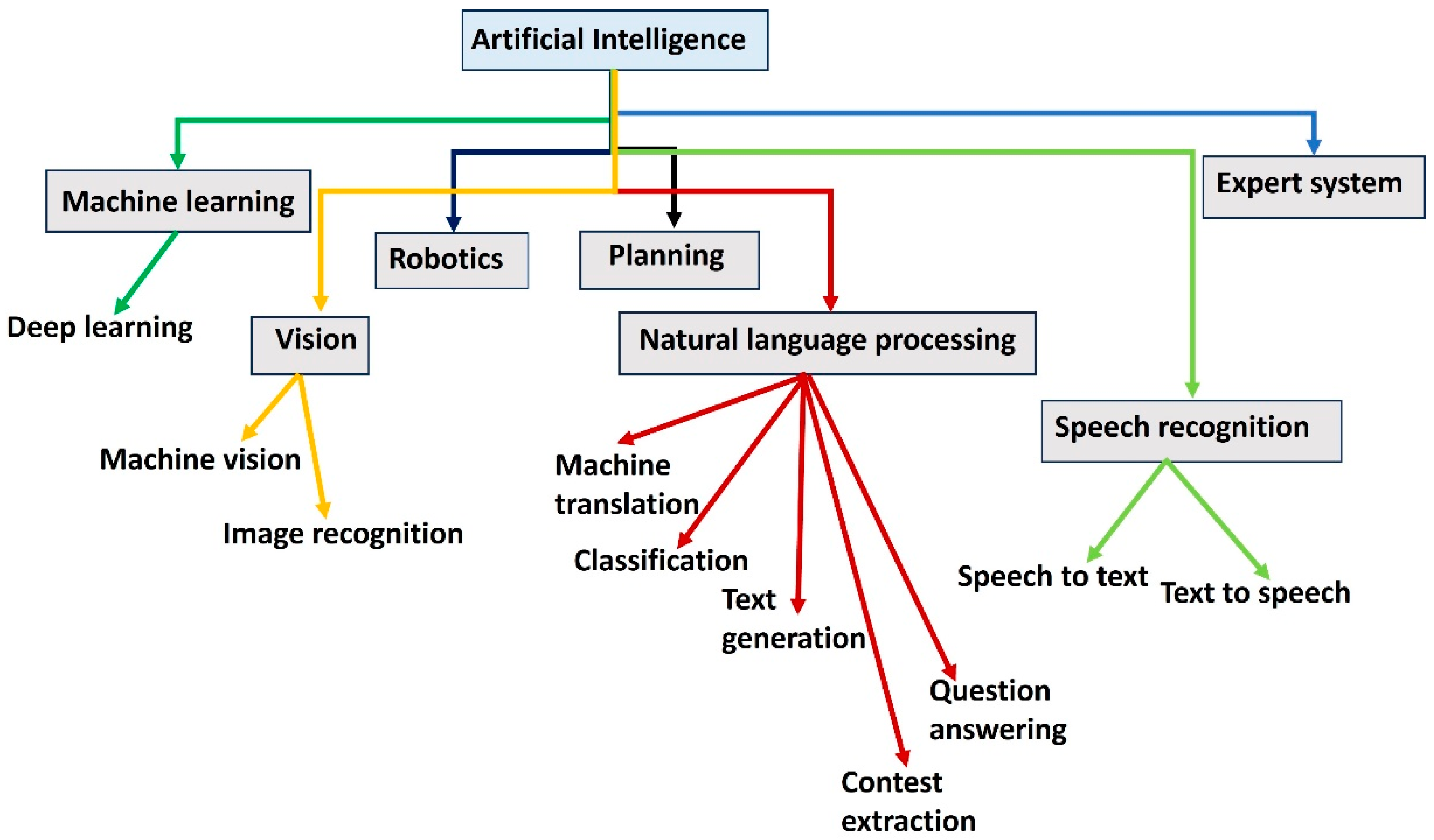
AI has emerged as a transformative tool in neuropharmacological research, aiding scientists in uncovering complex relationships within biological systems and accelerating drug discovery. The block diagram in
Figure 4 provides an overview of AI and its key domains, illustrating their interconnections and applications. At the core, AI encompasses diverse fields like machine learning, robotics, natural language processing (NLP), vision, and expert systems. Machine learning, a subset of AI, includes deep learning and focuses on enabling systems to learn from data [Janiesch 2021]. Robotics involves planning and vision, with machine vision and image recognition enabling systems to perceive and interact with their environment [Mahapatra C 2025]. NLP addresses tasks such as machine translation, classification, text generation, question answering, and context extraction, showcasing its role in understanding and generating human language [Zhang 2021]. Speech recognition, a branch of NLP, enables bidirectional conversion between speech and text [Prasanna 2022]. Expert systems represent a specialized application of AI that uses knowledge-based rules to make decisions [Yang 2024]. This diagram demonstrates how AI integrates various technologies to achieve versatile functionalities.
Machine Learning in Neuropharmacology
Machine learning (ML) involves algorithms that learn from data to make predictions or decisions without explicit programming. In neuropharmacology, ML is widely applied in various stages of drug discovery and development. For example, ML models can analyze high-dimensional datasets, such as genomic, proteomic, and metabolomic data, to identify potential drug targets [Mahapatra C. et al.,2024]. These algorithms help researchers uncover patterns that may not be evident through traditional statistical methods. Supervised learning, a subset of ML, is often used for classification and regression tasks in neuropharmacology [Obaido 2024]. For instance, ML algorithms can classify compounds based on their biological activity or predict their toxicity levels. Random forests, support vector machines (SVMs), and gradient boosting machines are commonly employed for such purposes. These algorithms are particularly effective in identifying lead compounds that exhibit desired pharmacological effects, streamlining the drug development process [Nayarisseri 2021]. Furthermore, supervised learning models are instrumental in predicting the side effects of drugs, ensuring better safety profiles before clinical trials [Mak, K 2024]. Unsupervised learning, another ML approach, is used to cluster data and uncover hidden structures within it [Glielmo 2021]. In neuropharmacological research, this method can group patients based on their response to specific treatments, paving the way for personalized medicine. For example, clustering algorithms such as k-means and hierarchical clustering can segment patients into subgroups with shared genetic or molecular characteristics, which may influence drug efficacy and safety [Trezza 2024]. Reinforcement learning, yet another ML paradigm is gaining traction in optimizing treatment protocols by dynamically adapting to individual patient needs. This adaptive approach allows for more precise interventions, particularly in managing chronic neurological disorders like epilepsy or Parkinson’s disease [van Nuland 2020].
Deep Learning in Neuropharmacology
Deep learning (DL), a subset of ML, is distinguished by its use of neural networks with multiple layers to model complex data relationships. DL has proven particularly useful in neuropharmacology due to its ability to process unstructured data such as images, videos, and sequences [Askr, H 2023]. Convolutional neural networks (CNNs), for example, are frequently employed to analyze brain imaging data, aiding in the identification of biomarkers for neurological disorders like Alzheimer’s and Parkinson’s disease [Mahapatra C. et al.,2024]. These biomarkers can then serve as targets for therapeutic interventions, enabling the development of more effective drugs. In drug discovery, DL algorithms can predict the activity of drug molecules by analyzing their chemical structure. This process involves the use of GNNs that represent molecular structures as graphs, capturing intricate chemical relationships [Xiong 2021]. RNNs and transformers are used to model sequential data, such as the interactions between drugs and proteins. These techniques have significantly accelerated the identification of lead compounds, reducing the time and cost of developing new neuro pharmaceuticals [Jiang 2024]. DL also facilitates the analysis of large-scale datasets from high-throughput screening experiments, helping researchers identify promising candidates for further investigation. Moreover, DL is instrumental in analyzing electroencephalography (EEG) and functional magnetic resonance imaging (fMRI) data. These analyses help in understanding the neural mechanisms underlying drug effects and disease progression [Ebrahimzadeh 2022]. For instance, DL models can detect subtle patterns in EEG signals that correlate with the efficacy of anti-epileptic drugs, enabling better treatment strategies [Mahapatra C. et al.,2024]. Generative adversarial networks (GANs) are also being used to create synthetic data, which can augment limited datasets and improve model performance. GANs are particularly valuable in scenarios where real-world data is scarce, such as rare neurological diseases [Ibrahim 2024].
Natural Language Processing in Neuropharmacology
NLP is another critical AI technique applied in neuropharmacological research. NLP focuses on enabling machines to understand and process human language [Chowdhary 2020]. In the context of neuropharmacology, NLP is primarily used for mining scientific literature, clinical trial reports, and electronic health records (EHRs) to extract relevant information [Mahapatra C. et al.,2021]. This capability is essential given the vast and ever-growing body of biomedical research that must be synthesized to make informed decisions. NLP algorithms can identify relationships between drugs, diseases, and molecular pathways by analyzing vast amounts of text data. Named entity recognition (NER) models are used to extract entities such as drug names, gene symbols, and disease terms from unstructured text [Perera 2020]. Relation extraction techniques then establish connections between these entities, facilitating hypothesis generation. For instance, NLP systems can highlight potential interactions between drug candidates and molecular targets, providing insights that might otherwise be overlooked [Chen 2024]. In addition, NLP-based tools are employed to predict potential drug repurposing opportunities by analyzing existing drug descriptions and clinical outcomes. By leveraging data from previous trials and observational studies, NLP algorithms can identify drugs with the potential to treat conditions beyond their original indications. Sentiment analysis and topic modeling are other NLP applications that can assess patient-reported outcomes and identify emerging trends in neuropharmacology research [Wang Y 2023]. These tools provide a comprehensive view of patient experiences, enabling researchers to prioritize drug development efforts accordingly. With the integration of pre-trained language models like BERT (Bidirectional Encoder Representations from Transformers), NLP has become even more efficient in handling complex biomedical texts [Mahapatra C. et al.,2019]. These advanced models excel at understanding context and nuances, making them invaluable for tasks such as summarizing research findings or generating new hypotheses. These methods allow for the efficient analysis of complex datasets, the identification of novel drug targets, and the extraction of valuable insights from unstructured data. NLPs bridge the gap between unstructured biomedical literature and actionable insights, enhancing researchers’ ability to navigate the vast landscape of neuropharmacology. As AI continues to evolve, its integration into neuropharmacology is likely to enhance our understanding of neurological disorders and accelerate the development of effective treatments. Ultimately, these advancements promise to improve patient outcomes and contribute to the emergence of precision medicine in the field of neuropharmacology.
5. AI in Drug Target Identification
Drug target identification refers to the process of pinpointing specific biomolecules, such as proteins or genes, that play a crucial role in disease progression and can be modulated by therapeutic agents [Zou, M 2024]. It is a foundational step in drug discovery, enabling the development of targeted treatments. AI is revolutionizing the field of drug target identification by offering sophisticated methods to analyze complex biological data. Two key areas where AI is making significant contributions include predicting drug-target interactions and validating neurochemical pathways [Mahapatra C. et al.,2019]. These advancements not only accelerate the drug discovery process but also improve the precision and efficacy of therapeutics for various diseases, particularly neurological disorders.
Predicting Drug-Target Interactions
Predicting drug-target interactions (DTIs) is a critical step in the drug discovery process. AI techniques have proven to be invaluable in this domain, enabling researchers to efficiently identify potential interactions between drugs and their biological targets. Traditional methods for DTI prediction often involve labor-intensive experimental assays, but AI provides computational approaches that are faster and more cost-effective [Duo 2024]. ML and DL algorithms such as SVMs, CNNs, and GNNs are widely employed for DTI prediction. These algorithms can analyze large-scale datasets comprising chemical structures, protein sequences, and interaction profiles to uncover patterns that suggest potential drug-target pairs [Liu 2021]. One notable application of AI in DTI prediction involves the use of transfer learning. Pre-trained models on large chemical and biological datasets can be fine-tuned for specific drug discovery tasks, improving their performance on limited data [Lian 2024]. Additionally, techniques such as matrix factorization and collaborative filtering are used to predict DTIs by leveraging the similarity between drugs and targets within a given dataset [Jamali 2021]. These approaches are particularly useful in identifying repurposing opportunities for existing drugs, as they can highlight previously unrecognized interactions with novel targets. AI models can also incorporate multi-omics data, including genomic, transcriptomic, and proteomic information, to provide a more comprehensive view of drug-target interactions [Mahapatra C. et al.,2019]. This integration allows researchers to identify biomarkers associated with drug response, paving the way for precision medicine. Moreover, NLP tools are used to extract relevant information from scientific literature, patents, and clinical trial data, further enriching the knowledge base for DTI prediction [Vatansever 2021].
Validating Neurochemical Pathways
Validating neurochemical pathways is another crucial aspect of drug target identification, especially for neuropharmacological research. AI techniques enable researchers to model and analyze the intricate networks of biochemical interactions that regulate neural function [Oyebanjo 2024]. By doing so, AI helps in pinpointing key nodes within these pathways that could serve as potential drug targets. Systems biology approaches, powered by AI, are often used to construct and analyze models of neurochemical pathways. These models incorporate data from various sources, including gene expression profiles, protein-protein interaction networks, and metabolomic studies. AI algorithms can identify critical nodes within these networks that are associated with disease states or therapeutic responses. For example, network-based machine learning methods can predict how modulating specific proteins or enzymes within a pathway might affect overall neural function, guiding the selection of promising drug targets [Vatansever 2021]. DL methods have also been applied to simulate neurochemical processes and predict the effects of pharmacological interventions. For instance, RNNs and transformer models can be used to analyze time-series data from experiments involving neural activity or drug administration [Chiranjeevi 2024]. These models can uncover dynamic patterns within neurochemical pathways, offering insights into the temporal effects of drugs on neural systems. Another significant application of AI in validating neurochemical pathways involves the use of GANs. GANs can create synthetic biological data that mimics real-world observations, providing researchers with additional datasets to validate their findings [Georges-Filteau 2020]. This capability is particularly valuable in cases where experimental data is scarce or difficult to obtain. AI is also instrumental in integrating data from animal models and human studies to validate neurochemical pathways. By analyzing cross-species datasets, AI can identify conserved mechanisms that are likely to be relevant for human diseases. This approach not only enhances the reliability of preclinical findings but also reduces the risk of failures in subsequent clinical trials. Furthermore, AI-driven tools are being developed to predict the downstream effects of modulating specific nodes within neurochemical pathways [González-González 2024]. These predictions help researchers anticipate potential side effects and off-target interactions, ensuring the safety and efficacy of new therapeutic interventions. AI-based validation methods thus play a critical role in refining the drug discovery pipeline, particularly for complex and poorly understood neurological conditions.
Expanding the Role of AI in Drug Target Identification
The role of AI in drug target identification is continually expanding, driven by advancements in computational power and the availability of increasingly comprehensive datasets. Beyond the current applications, AI is being leveraged to address emerging challenges in the field. For example, hybrid models that combine multiple AI techniques are being developed to improve the accuracy of predictions. These models integrate machine learning with deep learning, enhancing their ability to capture both linear and non-linear relationships in biological data. AI is also being applied to study polypharmacology, which involves understanding how a single drug interacts with multiple targets. This is particularly important in neuropharmacology, where complex diseases like Alzheimer’s and Parkinson’s often involve multiple pathways [Mahapatra C. et al.,2018]. By modeling these interactions, AI can aid in the design of multi-target drugs that address various aspects of a disease simultaneously. Another promising area is the use of AI for de novo drug design. Generative models, such as variational autoencoders (VAEs) and reinforcement learning frameworks, are being used to create novel chemical compounds that are optimized for specific targets. These techniques not only identify potential drug candidates but also predict their pharmacokinetic and pharmacodynamic properties, streamlining the early stages of drug development. Collaboration between AI tools and high-throughput experimental techniques is further enhancing the drug discovery process. For example, AI-guided screening platforms can prioritize compounds for experimental testing, significantly reducing the time and resources required. Moreover, the integration of AI with robotics and automated laboratory systems is enabling real-time optimization of experiments, accelerating the pace of discovery [Mahapatra C. et al.,2017]. The ethical implications of AI in drug target identification are also gaining attention. Ensuring transparency and interpretability of AI models is critical for gaining regulatory approval and fostering trust among stakeholders. Researchers are increasingly focusing on developing explainable AI (XAI) methods that provide insights into how models make predictions, ensuring that their outputs can be validated and understood by human experts.
Compound pharmacokinetic prediction
AI demonstrates remarkable potential in predicting the pharmacokinetic properties of compounds, including metabolism, toxicity, distribution, absorption, and excretion. These facets are integral to understanding how drugs interact within the human body, ensuring safety and efficacy in therapeutic interventions. One of the foremost applications of AI in neuropharmacology is the prediction of a compound's pharmacokinetics. Machine learning models, trained on extensive datasets, enable the prediction of key parameters such as absorption rate and bioavailability [Yingngam 2024]. By leveraging neural networks and DL, researchers can simulate how drugs traverse biological membranes and identify factors influencing absorption. This expedites the evaluation of candidate molecules, reducing reliance on time-consuming and costly in vivo studies. Metabolism is another critical area where AI contributes significantly. Understanding how drugs are metabolized—primarily by liver enzymes—is essential to predict their efficacy and potential side effects. AI models use enzyme-substrate interaction data to predict metabolic pathways and identify potential metabolites [Salas 2024]. Advanced computational tools can even simulate enzyme kinetics, providing insights into how genetic polymorphisms may affect drug metabolism in different populations. Toxicity prediction remains a cornerstone of drug safety. AI algorithms utilize data from preclinical toxicity studies to predict potential adverse effects of compounds. Techniques such as NLP and graph neural networks allow researchers to analyze chemical structures and biological interaction data to flag molecules with high toxicity risks early in the drug development pipeline [Mahapatra C. et al.,2017]. This proactive approach mitigates the risk of costly failures during clinical trials. AI also plays a pivotal role in understanding drug distribution. Models informed by physicochemical properties of compounds and physiological parameters predict how drugs distribute across tissues, including crossing the blood-brain barrier—a major challenge in neuropharmacology. Predictive tools help optimize molecular structures to enhance CNS penetration, a crucial factor in developing treatments for neurological disorders. Finally, the prediction of excretion pathways is streamlined using AI. Excretion, which determines the duration a drug remains active in the body, is influenced by renal and hepatic functions [Paliwal 2024]. AI-driven models assess renal clearance and biliary excretion by correlating compound properties with historical excretion data [Oyanna 2024]. This accelerates the development of drugs with favorable elimination profiles, minimizing risks of bioaccumulation and toxicity. In summary, AI provides a multifaceted toolkit for addressing complex pharmacokinetic challenges in neuropharmacology. By predicting absorption, distribution, metabolism, toxicity, and excretion, AI accelerates drug discovery while improving safety and efficacy. As computational methodologies advance, the integration of AI into neuropharmacological research will continue to transform how therapeutic compounds are developed and optimized for clinical use.
6. Personalized Medicine in Neuropharmacology
Personalized medicine, also known as precision medicine, is transforming the landscape of neuropharmacology by providing tailored treatments based on an individual’s genetic, molecular, and clinical profile [Griñán-Ferré 2024]. This approach leverages advanced technologies, particularly AI, to optimize therapeutic strategies for neurological disorders. Two pivotal areas of application include AI-driven biomarker discovery and the development of therapies specifically tailored to individual patients’ needs.
AI-Driven Biomarker Discovery
Biomarkers are measurable indicators of biological processes, diseases, or responses to therapy. In neuropharmacology, identifying reliable biomarkers is crucial for understanding the mechanisms underlying neurological disorders and predicting patient responses to treatment. AI has become an indispensable tool in biomarker discovery due to its ability to process and analyze complex datasets from diverse sources, such as genomics, proteomics, metabolomics, and neuroimaging. ML algorithms are at the forefront of biomarker discovery. These algorithms can sift through vast datasets to identify patterns and correlations that might be overlooked using traditional statistical methods. For instance, ML techniques such as random forests, SVMs, and neural networks have been employed to pinpoint genetic variants associated with susceptibility to disorders like Alzheimer’s disease and schizophrenia [Ahmed 2021]. These insights help researchers identify potential therapeutic targets and stratify patient populations based on their genetic risk profiles. DL has shown particular promise in analyzing unstructured data such as imaging studies and electrophysiological recordings. CNNs, for example, have been used to analyze brain scans, enabling the identification of subtle structural or functional abnormalities associated with conditions such as multiple sclerosis and Parkinson’s disease [Mahapatra C. et al.,2016]. By integrating imaging biomarkers with molecular and clinical data, AI provides a comprehensive view of disease mechanisms, paving the way for more targeted interventions. NLP, another AI technology, facilitates biomarker discovery by extracting relevant information from scientific literature, electronic health records, and clinical trial reports. NLP tools can identify emerging biomarkers and their associated pathways, ensuring that researchers remain updated on the latest developments in the field. Moreover, AI-driven integrative analyses that combine data from multiple modalities—such as gene expression profiles, protein interactions, and metabolomic pathways—enhance the robustness and reliability of biomarker discovery efforts. AI-driven biomarker discovery also plays a vital role in monitoring disease progression and evaluating therapeutic efficacy. For example, biomarkers identified through AI analysis of cerebrospinal fluid or blood samples can serve as indicators of neuroinflammation or neuronal damage, offering real-time insights into a patient’s response to treatment [Arrambide 2024]. These advancements are critical for developing precision therapies and improving clinical outcomes in patients with complex neurological conditions.
Tailoring Therapies for Neurological Disorders
The ultimate goal of personalized medicine in neuropharmacology is to tailor therapies that address the unique needs of individual patients. Neurological disorders such as epilepsy, depression, and neurodegenerative diseases exhibit significant heterogeneity in their etiology, symptoms, and progression [Maristany 2024]. AI is enabling clinicians to account for this variability by developing customized treatment plans based on each patient’s specific profile. AI models are increasingly used to predict patient responses to various therapeutic interventions. These models analyze data from genetic tests, neuroimaging, and patient histories to recommend treatments that are most likely to be effective. For example, pharmacogenomic studies, powered by AI, identify genetic variants that influence how patients metabolize or respond to specific drugs [Taherdoost 2024]. This information allows clinicians to select medications and dosages that minimize side effects and maximize efficacy. For instance, in treating depression, pharmacogenomic insights can guide the choice of antidepressants based on the patient’s cytochrome P450 enzyme profile, reducing the trial-and-error process often associated with finding the right medication [Eap 2021]. DL algorithms have also been applied to optimize neuromodulation therapies, such as transcranial magnetic stimulation (TMS) and deep brain stimulation (DBS) [Soleimani 2023]. By analyzing brain activity patterns, these algorithms can personalize stimulation parameters to achieve optimal therapeutic effects for conditions like treatment-resistant depression or Parkinson’s disease. AI-driven models can further adapt these parameters in real-time, ensuring that the therapy remains effective as the patient’s condition evolves. In the realm of neurodegenerative diseases, AI has facilitated the development of disease-modifying therapies that target specific pathological mechanisms. For example, ML models have been used to identify subtypes of Alzheimer’s disease based on biomarkers such as amyloid-beta and tau protein levels [Mahapatra C. et al.,2016]. These subtypes help researchers design therapies tailored to the underlying disease mechanisms, improving treatment outcomes. Similarly, AI-driven approaches are aiding in the discovery of combination therapies that address multiple pathways involved in complex diseases. AI is also revolutionizing the design of clinical trials for neurological disorders. Adaptive trial designs, guided by AI algorithms, allow researchers to modify trial parameters based on interim results, improving the likelihood of success [Fountzilas 2022]. Additionally, patient stratification based on AI-identified biomarkers ensures that trials enroll participants who are most likely to benefit from the intervention, reducing variability and enhancing the reliability of results. These advancements accelerate the development of new therapies and ensure that patients receive treatments aligned with their individual needs.
7. Case Studies
AI in Neuropharmacology for Alzheimer’s Disease Research
Alzheimer’s disease (AD), a progressive neurodegenerative disorder, poses a significant challenge to researchers due to its complex etiology and lack of effective treatments. AI has emerged as a transformative tool in advancing neuropharmacological research for Alzheimer’s disease by facilitating the identification of biomarkers, drug target discovery, and patient stratification. One notable application of AI in Alzheimer’s research is the analysis of neuroimaging data. Techniques such as DL, particularly CNNs, have been utilized to analyze brain scans for detecting early signs of AD. For example, AI models can identify subtle structural changes, such as hippocampal atrophy, that may not be apparent to human observers [Aditya 2024]. These insights are critical for early diagnosis and timely intervention. In one study, a CNN-based model achieved over 90% accuracy in distinguishing between mild cognitive impairment (MCI) and early-stage Alzheimer’s, highlighting its potential as a diagnostic aid [El-Assy 2024]. AI has also been instrumental in identifying molecular biomarkers associated with Alzheimer’s disease. ML algorithms have analyzed vast datasets from genomics, proteomics, and transcriptomics to uncover genes and proteins implicated in the disease’s progression. For instance, AI-driven studies have identified key biomarkers such as amyloid-beta and tau protein aggregates, which are central to Alzheimer’s pathology [Kale 2024]. Additionally, emerging biomarkers like neurofilament light chain (NfL) and synaptic density markers have been discovered through integrative AI analyses, providing new targets for drug development [Krishnamurthy 2024]. Another significant contribution of AI in Alzheimer’s research is drug repurposing. By analyzing large-scale databases of existing drugs and their molecular targets, AI algorithms can predict which compounds might be effective against Alzheimer’s. For example, a DL model successfully identified potential candidates such as antihypertensive and anti-inflammatory drugs that may mitigate neuroinflammation and oxidative stress, key contributors to AD progression [Mir 2021]. Clinical trials for Alzheimer’s therapies also benefit from AI technologies. AI can optimize patient recruitment by identifying individuals most likely to benefit from experimental treatments based on their biomarker profiles and genetic predispositions. Furthermore, AI-driven analysis of trial data helps detect subtle treatment effects, ensuring that promising therapies are not prematurely discarded. For instance, predictive modeling has been used to identify responders and non-responders in trials for anti-amyloid drugs, thereby refining therapeutic strategies and improving trial outcomes [Argueta 2022]. AI is also aiding in the development of precision medicine approaches for Alzheimer’s. By integrating data from genetic, neuroimaging, and clinical studies, AI models can personalize treatment plans. This is particularly useful for identifying subgroups of patients who may benefit from targeted therapies, such as those aimed at reducing amyloid-beta plaques. Additionally, AI is helping to predict disease progression, allowing for better planning of interventions and care strategies.
AI in Neuropharmacology for Parkinson’s Disease Drug Development
Parkinson’s disease (PD) is a neurodegenerative disorder characterized by motor symptoms, such as tremors and rigidity, as well as non-motor symptoms, including cognitive decline and mood disturbances. AI is playing an increasingly vital role in advancing drug development for Parkinson’s by enabling the identification of novel therapeutic targets, optimizing clinical trials, and improving disease monitoring. A major challenge in Parkinson’s drug development is understanding the molecular mechanisms underlying the disease. AI-driven approaches have been used to analyze multi-omics data to identify potential drug targets. For instance, ML algorithms have uncovered critical pathways involving alpha-synuclein aggregation and mitochondrial dysfunction [Srinivasan 2021]. These discoveries have paved the way for the development of disease-modifying therapies aimed at halting or slowing disease progression. DL techniques have also been employed to predict DTIs for Parkinson’s. By analyzing large datasets of chemical structures and biological targets, AI models can predict which compounds may interact with specific proteins implicated in Parkinson’s pathology. For example, computational models have identified small molecules that inhibit alpha-synuclein aggregation, offering promising leads for therapeutic development [Ghosh 2025]. In addition to target discovery, AI has enhanced clinical trial design for Parkinson’s therapies. AI algorithms analyze patient data to stratify participants based on disease stage, genetic mutations (e.g., LRRK2 or GBA variants), and biomarker profiles [Courtman 2024]. This stratification ensures that trials test therapies on the most relevant patient subgroups, increasing the likelihood of success. For example, AI has been used to design trials targeting individuals with early-stage Parkinson’s, where neuroprotective treatments may have the greatest impact. Monitoring disease progression is another area where AI has demonstrated immense potential. Wearable devices and mobile health technologies generate continuous data on motor symptoms, such as tremor frequency and gait abnormalities [Lobo 2024]. AI models analyze these data streams to provide real-time insights into disease progression and treatment efficacy. For instance, DL algorithms applied to wearable sensor data have been shown to predict disease progression with high accuracy, aiding in the evaluation of new therapies. Drug repurposing efforts for Parkinson’s have also benefited from AI. By mining existing drug libraries, AI models have identified compounds with potential neuroprotective effects. For example, antihypertensive drugs and GLP-1 receptor agonists, originally developed for diabetes, have been flagged as potential candidates for slowing Parkinson’s progression [Colin 2023]. These findings have accelerated the drug discovery timeline and reduced associated costs. AI is further enabling personalized treatment approaches for Parkinson’s. By integrating data from genetic testing, neuroimaging, and clinical assessments, AI algorithms recommend tailored therapeutic regimens. For instance, patients with specific genetic mutations may benefit from targeted therapies, such as LRRK2 inhibitors [Tolosa 2020]. This precision approach ensures that treatments address the underlying causes of the disease in individual patients, improving outcomes and quality of life. Additionally, AI technologies are fostering collaboration among researchers and clinicians by creating shared platforms for analyzing and interpreting data. These platforms enable real-time data sharing and analysis, which is crucial for rapidly evolving research areas like Parkinson’s. Predictive models are also being employed to anticipate adverse effects of experimental treatments, ensuring safer and more efficient development processes. The application of AI in neuropharmacology has significantly advanced research and drug development for Alzheimer’s and Parkinson’s diseases. In Alzheimer’s research, AI has facilitated early diagnosis, biomarker discovery, and drug repurposing, while optimizing clinical trials. For Parkinson’s disease, AI has enhanced target identification, trial design, disease monitoring, and personalized treatment strategies. The integration of AI into these areas has not only accelerated the drug discovery pipeline but also improved the precision and effectiveness of therapeutic approaches. As AI technologies continue to evolve, they hold immense promise for transforming the landscape of neuropharmacological research and improving the lives of individuals affected by these devastating neurodegenerative disorders.
8. Ethical and Practical Challenges in AI of Neuropharmacology
The integration of AI into neuropharmacology offers tremendous opportunities for advancing drug discovery, improving treatment outcomes, and personalizing therapies for neurological disorders. However, the application of AI also presents a range of ethical and practical challenges that need to be addressed to maximize its potential while safeguarding against potential risks. Two critical areas of concern are data privacy and the interpretability of AI models.
Data Privacy Concerns
AI-driven neuropharmacological research relies heavily on access to large datasets, including genomic, clinical, and neuroimaging data. These datasets are often derived from individual patients, making privacy and security a significant concern. Protecting the sensitive information of patients is paramount, as breaches in data privacy can lead to misuse, discrimination, or stigmatization. One of the main challenges is ensuring compliance with data protection regulations, such as the General Data Protection Regulation (GDPR) in the European Union or the Health Insurance Portability and Accountability Act (HIPAA) in the United States [Bakare 2024]. These laws mandate stringent requirements for obtaining patient consent, anonymizing data, and limiting access to authorized parties. However, the complexity of neuropharmacological datasets, which often require cross-institutional and cross-border collaborations, complicates adherence to these regulations. Another concern is the risk of re-identification. Even anonymized datasets can sometimes be reverse-engineered to reveal personal information, particularly when combined with other publicly available data. For instance, neuroimaging or genomic data can be cross-referenced with demographic information to re-identify individuals, undermining privacy protections [Oliver 2022]. To mitigate this, advanced techniques such as differential privacy and federated learning are being explored. Differential privacy involves adding noise to the data to obscure individual identities, while federated learning enables AI models to train on decentralized data without transferring it to a central repository [Zhu 2020]. Moreover, there is an ethical obligation to ensure informed consent. Patients must fully understand how their data will be used, shared, and stored. In many cases, the technical nature of AI-driven research can make it difficult for patients to grasp the implications of consenting to data use. Researchers and institutions must therefore develop clear, accessible communication strategies to educate participants about data privacy and usage [Jones 2020]. Finally, the proprietary nature of some AI algorithms poses additional privacy challenges. Many AI systems used in neuropharmacology are developed by private companies that may not fully disclose their data handling practices [Syed 2024]. This lack of transparency raises questions about accountability and the potential for unethical data usage. Ensuring data privacy also requires robust cybersecurity measures. As cyberattacks become increasingly sophisticated, the risk of sensitive medical data being compromised grows. Institutions must invest in state-of-the-art encryption methods, regular vulnerability assessments, and staff training to prevent breaches [Javaid 2023]. Collaboration between stakeholders, including governments, research institutions, and technology companies, is essential to establish standardized practices for secure data sharing.
Interpretability of AI Models
AI models, particularly those based on deep learning, are often described as “black boxes” due to their complexity and lack of transparency [Hassija 2024]. This poses a significant challenge in neuropharmacology, where understanding the rationale behind AI-driven predictions is essential for clinical decision-making and drug development. One key issue is the difficulty in explaining how AI models arrive at specific conclusions. For example, a deep learning model might predict that a certain drug will interact with a target molecule implicated in a neurological disorder. However, without a clear understanding of the underlying mechanisms, researchers and clinicians may be hesitant to act on these predictions. This lack of interpretability can hinder the adoption of AI tools in neuropharmacology, as regulatory bodies and stakeholders demand transparency and accountability in scientific research. To address this, the field is increasingly focusing on the development of explainable AI (XAI). XAI techniques aim to make AI models more transparent by providing insights into their decision-making processes [Patidar 2024]. For instance, feature attribution methods can highlight which inputs (e.g., specific genes, proteins, or imaging features) were most influential in a model’s prediction. Visualization tools, such as saliency maps, can also help researchers interpret results by showing which areas of a brain scan contributed to a diagnostic decision. Another practical challenge is ensuring that AI models generalize well across diverse datasets. Many AI models are trained on specific datasets that may not fully represent the broader patient population. This can result in biased predictions that disproportionately affect certain groups, such as underrepresented ethnicities or age groups [Ladin 2024]. Ensuring interpretability requires not only understanding a model’s predictions but also assessing its fairness and robustness. In addition, interpretability is crucial for building trust among clinicians and patients [ElShawi 2021]. Clinicians need to understand how AI models work to confidently integrate them into their practice, while patients must feel assured that AI-driven recommendations are grounded in scientifically sound reasoning. The lack of interpretability could lead to resistance from both groups, slowing the adoption of AI in neuropharmacology. Finally, the dynamic nature of AI models presents another challenge. Many AI systems continuously learn and adapt as they are exposed to new data. While this capability can improve accuracy over time, it also complicates interpretability, as the reasoning behind a model’s predictions may change. Establishing frameworks for monitoring and validating AI models throughout their lifecycle is essential to maintain transparency and reliability [Almagrabi 2024]. The ethical implications of interpretability also extend to accountability. If an AI model makes an erroneous prediction that results in harm, determining responsibility becomes difficult. Clear guidelines on liability and accountability are needed to ensure that both developers and users of AI systems understand their roles and obligations.
The ethical and practical challenges of AI in neuropharmacology—particularly data privacy concerns and the interpretability of AI models—highlight the need for careful governance and innovation. Addressing data privacy issues requires robust security measures, adherence to regulations, and transparent communication with patients. At the same time, enhancing the interpretability of AI models is essential for fostering trust, ensuring fairness, and enabling informed decision-making. By addressing these challenges, the field of neuropharmacology can harness the full potential of AI to improve research and clinical outcomes while upholding ethical standards. Collaborative efforts among researchers, policymakers, and industry leaders will be crucial in navigating these challenges and ensuring that AI serves as a responsible and effective tool in advancing neuropharmacological science.
9. Future Prospects and Innovations in the Application of AI in Neuropharmacology
AI has revolutionized numerous fields, and its integration into neuropharmacology holds immense potential for advancing the discovery and development of new therapies. Two key areas of AI application in neuropharmacology that are expected to experience significant innovation are Integrative Neuropharmacology and Real-Time AI Applications in Neurology. These fields, which deal with the complex interplay between the brain, drugs, and neurological conditions, stand to benefit immensely from AI-driven advancements.
Integrative Neuropharmacology
Integrative Neuropharmacology is a subfield of pharmacology that focuses on the interactions between drugs and the nervous system, aiming to identify how pharmacological agents can alter brain function and behavior. One of the biggest challenges in neuropharmacology is understanding the intricate molecular and neural pathways that govern brain functions. AI is poised to transform this area by offering advanced tools for simulating, modeling, and predicting these interactions with unprecedented precision. AI’s ability to process vast amounts of data has already proven essential in neuropharmacology. ML algorithms, researchers can analyze large-scale datasets from genomic, proteomic, and transcriptomic studies to uncover new molecular targets for drug development. AI tools can help identify patterns in data that may be too subtle or complex for traditional analytical methods. For instance, DL models can predict how certain compounds might affect brain activity by analyzing data from imaging techniques like fMRI and EEGs. These insights can be used to discover novel drugs that can modulate brain functions, providing new therapeutic options for conditions such as schizophrenia, depression, and Alzheimer's disease. Another area where AI will have a profound impact is in the development of personalized medicine. By integrating data from multiple sources, including patient-specific genetic information and previous treatment responses, AI can help tailor treatments that are more likely to succeed for individual patients. This is particularly valuable in neuropharmacology, where the same drug may have very different effects on patients due to genetic variations or differences in the underlying causes of neurological diseases. AI-driven predictive models can assist in selecting the right drug, dosage, and treatment regimen, thereby improving clinical outcomes and minimizing adverse effects. In addition to drug discovery, AI can also play a critical role in drug repurposing. Many existing drugs, originally developed for one condition, can be effective in treating other diseases. By using AI algorithms to screen large libraries of FDA-approved drugs, researchers can identify potential candidates for conditions that lack effective treatments [Gangwal 2024]. This can significantly shorten the time and cost required to bring new therapies to market.
Real-Time AI Applications in Neurology
Neurology is another area where AI is making significant strides, especially when it comes to real-time applications. Neurological disorders, such as epilepsy, stroke, Parkinson’s disease, and multiple sclerosis, often present with complex, rapidly changing symptoms that require real-time intervention. AI technologies have the potential to revolutionize the way these conditions are monitored, diagnosed, and treated in real time, leading to improved patient outcomes. One promising application is in the real-time monitoring of patients with neurological conditions. Wearable devices, combined with AI-powered analytics, can continuously track neurological parameters such as brain wave patterns, motor function, and speech patterns [Gutman 2024]. For example, AI algorithms can analyze EEG data in real time to detect early signs of seizures in patients with epilepsy. By identifying abnormal brain activity before a full-blown seizure occurs, AI can trigger alerts to caregivers and healthcare providers, allowing for timely interventions that can prevent injury or reduce the severity of the event. This predictive capability could greatly improve the quality of life for patients with chronic neurological conditions. Similarly, in the context of Parkinson’s disease, AI-powered systems can track a patient’s motor symptoms in real-time, such as tremors, gait disturbances, and rigidity. This data can be used to adjust medication regimens dynamically, optimizing therapeutic efficacy and minimizing side effects. ML models can also identify patterns in a patient’s movement data, allowing for early detection of disease progression or changes in treatment response. In the realm of stroke, real-time AI applications are making a significant impact on both diagnosis and treatment. AI algorithms can quickly analyze medical imaging, such as CT scans or MRIs, to detect signs of a stroke, even in its early stages. In cases where rapid intervention is crucial, such as ischemic strokes, AI can help doctors make faster, more accurate decisions about treatment options, including thrombolytic therapy or endovascular procedures [Parvathy 2024]. AI can also assist in predicting patient outcomes, helping clinicians develop more personalized treatment plans. Another area where AI is transforming neurology is in the development of personalized treatment strategies based on real-time data. By integrating data from multiple sources, such as genetic information, clinical observations, and ongoing monitoring, AI can continuously adapt treatment protocols to suit the specific needs of each patient. This dynamic approach is particularly valuable in conditions like multiple sclerosis, where disease activity can vary significantly between patients and even in the same patient over time. Furthermore, AI is facilitating the development of real-time brain-computer interfaces (BCIs), which allow for direct communication between the brain and external devices [Karikari 2023]. These systems can help patients with severe neurological impairments, such as those with locked-in syndrome or spinal cord injuries, regain some degree of control over their environment. By analyzing neural signals in real-time, AI can translate brain activity into control commands for prosthetic limbs, communication devices, or even exoskeletons, enhancing the autonomy and quality of life for individuals with neurological disabilities.
10. Conclusion
The integration of AI into neuropharmacology represents a transformative era in neuroscience and drug discovery. This comprehensive review has highlighted how AI technologies such as machine learning, deep learning, and natural language processing are revolutionizing the field by addressing critical challenges and introducing innovative solutions. From drug discovery to personalized medicine, AI has demonstrated its potential to accelerate research, enhance precision, and reduce costs. AI-driven advancements have streamlined processes such as predicting drug-target interactions, optimizing virtual screening, and modeling pharmacokinetics. These improvements not only expedite the identification of effective therapies but also contribute to a deeper understanding of complex neurological disorders like Alzheimer’s and Parkinson’s diseases. By integrating multimodal datasets, AI has enabled the discovery of novel biomarkers, refined clinical trial designs, and fostered precision medicine approaches, ensuring that treatments are tailored to individual patient needs. Moreover, AI's capacity to bridge diverse data sources and methodologies has opened new avenues for research in previously challenging areas. Complex neural pathways and interactions between pharmacological agents and the brain are being demystified through computational models that combine genetic, proteomic, and neuroimaging data. These integrative approaches are critical for tackling multifaceted conditions such as epilepsy, schizophrenia, and neurodegenerative diseases, where traditional methods often fall short. By providing a unified platform for analysis and interpretation, AI enhances the reproducibility and scalability of neuropharmacological research, making breakthroughs more accessible and impactful. Despite its promise, challenges such as ethical considerations, data privacy concerns, and the interpretability of AI models remain. Addressing these issues requires a multidisciplinary effort involving researchers, clinicians, policymakers, and technologists. Transparency, accountability, and adherence to robust regulatory frameworks are essential to harness AI's full potential while safeguarding ethical standards. Looking ahead, the future of AI in neuropharmacology lies in advancing integrative neuropharmacology and real-time applications. AI's role in simulating neural pathways, personalizing treatments, and monitoring patients in real-time offers unprecedented opportunities to improve clinical outcomes. Emerging technologies like brain-computer interfaces and wearable devices further extend AI's reach, providing innovative solutions for managing neurological conditions. In conclusion, the convergence of AI and neuropharmacology is not merely a technological advancement but a paradigm shift that redefines how we approach the treatment of neurological disorders. By fostering collaboration and innovation, the field can continue to break new ground, ultimately improving the lives of patients and shaping the future of neuroscience. As AI technologies evolve, their integration into neuropharmacology will not only enhance therapeutic precision but also deepen our understanding of the brain's complex mechanisms, paving the way for a new era in medical science.
References
- Abbas, M. K. G., Rassam, A., Karamshahi, F., Abunora, R., & Abouseada, M. (2024). The Role of AI in Drug Discovery. Chembiochem, 25(14), e202300816. [CrossRef]
- Aditya Shastry, K., & Sanjay, H. A. (2024). Artificial intelligence techniques for the effective diagnosis of Alzheimer’s disease: a review. Multimedia Tools and Applications, 83(13), 40057-40092. [CrossRef]
- Ahmed, H., Alarabi, L., El-Sappagh, S., Soliman, H., & Elmogy, M. (2021). Genetic variations analysis for complex brain disease diagnosis using machine learning techniques: opportunities and hurdles. PeerJ Computer Science, 7, e697. [CrossRef]
- Alfarano, A., Maiano, L., Papa, L., & Amerini, I. (2024). Estimating optical flow: A comprehensive review of the state of the art. Computer Vision and Image Understanding, 104160. [CrossRef]
- Almagrabi, A. O., & Khan, R. A. (2024). Optimizing secure AI lifecycle model management with innovative generative AI strategies. IEEE Access. [CrossRef]
- Alowais, S. A., Alghamdi, S. S., Alsuhebany, N., Alqahtani, T., Alshaya, A. I., Almohareb, S. N.,... & Albekairy, A. M. (2023). Revolutionizing healthcare: the role of artificial intelligence in clinical practice. BMC medical education, 23(1), 689. [CrossRef]
- Argueta, N., Notari, E., & Szigeti, K. (2022). Role of pharmacogenomics in individualizing treatment for Alzheimer’s disease. CNS drugs, 36(4), 365-376. [CrossRef]
- Arrambide, G., Comabella, M., & Tur, C. (2024). Big data and artificial intelligence applied to blood and CSF fluid biomarkers in multiple sclerosis. Frontiers in Immunology, 15, 1459502. [CrossRef]
- Askr, H., Elgeldawi, E., Aboul Ella, H., Elshaier, Y. A., Gomaa, M. M., & Hassanien, A. E. (2023). Deep learning in drug discovery: an integrative review and future challenges. Artificial Intelligence Review, 56(7), 5975-6037. [CrossRef]
- Bakare, S. S., Adeniyi, A. O., Akpuokwe, C. U., & Eneh, N. E. (2024). Data privacy laws and compliance: a comparative review of the EU GDPR and USA regulations. Computer Science & IT Research Journal, 5(3), 528-543. [CrossRef]
- Bellomo, G., Indaco, A., Chiasserini, D., Maderna, E., Paolini Paoletti, F., Gaetani, L.,... & Di Fede, G. (2021). Machine learning driven profiling of cerebrospinal fluid core biomarkers in Alzheimer’s disease and other neurological disorders. Frontiers in neuroscience, 15, 647783. [CrossRef]
- Bettanti, A., Beccari, A. R., & Biccarino, M. (2024). Exploring the future of biopharmaceutical drug discovery: can advanced AI platforms overcome current challenges?. Discover Artificial Intelligence, 4(1), 1-16. [CrossRef]
- Bhunia, S. S., Saxena, M., & Saxena, A. K. (2021). Ligand- and structure-based virtual screening in drug discovery. In Biophysical and Computational Tools in Drug Discovery (pp. 281-339). Cham: Springer International Publishing.
- Catacutan, D. B., Alexander, J., Arnold, A., & Stokes, J. M. (2024). Machine learning in preclinical drug discovery. Nature Chemical Biology, 20(8), 960-973. [CrossRef]
- Chen, H., King, F. J., Zhou, B., Wang, Y., Canedy, C. J., Hayashi, J.,... & Zhou, Y. (2024). Drug target prediction through deep learning functional representation of gene signatures. Nature communications, 15(1), 1853. [CrossRef]
- Chiranjeevi, R., Keerthana, H., & Keerthana, G. (2024, August). Identification of Epileptic Seizures using Recurrent Neural Networks and Time Series Transformer. In 2024 7th International Conference on Circuit Power and Computing Technologies (ICCPCT) (Vol. 1, pp. 1546-1553). IEEE.
- Choudhury, C., Murugan, N. A., & Priyakumar, U. D. (2022). Structure-based drug repurposing: Traditional and advanced AI/ML-aided methods. Drug discovery today, 27(7), 1847-1861. [CrossRef]
- Chowdhary, K., & Chowdhary, K. R. (2020). Natural language processing. Fundamentals of artificial intelligence, 603-649.
- Colin, I. M., Szczepanski, L. W., Gérard, A. C., & Elosegi, J. A. (2023). Emerging evidence for the use of Antidiabetic Drugs, glucagon-like peptide 1 receptor agonists, for the treatment of Alzheimer’s Disease. touchREVIEWS in Endocrinology, 19(1), 16. [CrossRef]
- Courtman, M., Thurston, M., Wang, H., Banerjee, S., Streeter, A., McGavin, L.,... & Mullin, S. (2024). Deep learning classification of MRI differentiates brain changes in genetic and idiopathic Parkinson's disease. medRxiv, 2024-11.
- Díaz-Rodríguez, N., Del Ser, J., Coeckelbergh, M., de Prado, M. L., Herrera-Viedma, E., & Herrera, F. (2023). Connecting the dots in trustworthy Artificial Intelligence: From AI principles, ethics, and key requirements to responsible AI systems and regulation. Information Fusion, 99, 101896. [CrossRef]
- Duo, L., Liu, Y., Ren, J., Tang, B., & Hirst, J. D. (2024). Artificial intelligence for small molecule anticancer drug discovery. Expert Opinion on Drug Discovery, 1-16. [CrossRef]
- Eap, C. B., Gründer, G., Baumann, P., Ansermot, N., Conca, A., Corruble, E.,... & Hiemke, C. (2021). Tools for optimising pharmacotherapy in psychiatry (therapeutic drug monitoring, molecular brain imaging and pharmacogenetic tests): focus on antidepressants. The World Journal of Biological Psychiatry, 22(8), 561-628. [CrossRef]
- Ebrahimzadeh, E., Saharkhiz, S., Rajabion, L., Oskouei, H. B., Seraji, M., Fayaz, F.,... & Soltanian-Zadeh, H. (2022). Simultaneous electroencephalography-functional magnetic resonance imaging for assessment of human brain function. Frontiers in Systems Neuroscience, 16, 934266. [CrossRef]
- El-Assy, A. M., Amer, H. M., Ibrahim, H. M., & Mohamed, M. A. (2024). A novel CNN architecture for accurate early detection and classification of Alzheimer’s disease using MRI data. Scientific Reports, 14(1), 3463. [CrossRef]
- ElShawi, R., Sherif, Y., Al-Mallah, M., & Sakr, S. (2021). Interpretability in healthcare: A comparative study of local machine learning interpretability techniques. Computational Intelligence, 37(4), 1633-1650. [CrossRef]
- Fountzilas, E., Tsimberidou, A. M., Vo, H. H., & Kurzrock, R. (2022). Clinical trial design in the era of precision medicine. Genome medicine, 14(1), 101. [CrossRef]
- Gangwal, A., & Lavecchia, A. (2024). AI-Driven Drug Discovery for Rare Diseases. Journal of Chemical Information and Modeling. [CrossRef]
- Ghosh, P., Kundu, A., & Ganguly, D. (2025). From experimental studies to computational approaches: recent trends in designing novel therapeutics for amyloidogenesis. Journal of Materials Chemistry B.
- Glielmo, A., Husic, B. E., Rodriguez, A., Clementi, C., Noé, F., & Laio, A. (2021). Unsupervised learning methods for molecular simulation data. Chemical Reviews, 121(16), 9722-9758. [CrossRef]
- González, J., & Cirillo, E. (2020). Synthetic Observational Health Data with GANs: from slow adoption to a boom in medical research and ultimately digital twins?. arXiv preprint arXiv:2005.13510.
- González-González, M. A., Conde, S. V., Latorre, R., Thébault, S. C., Pratelli, M., Spitzer, N. C.,... & Ma, B. (2024). Bioelectronic medicine: a multidisciplinary roadmap from biophysics to precision therapies. Frontiers in Integrative Neuroscience, 18, 1321872. [CrossRef]
- Griñán-Ferré, C., Bellver-Sanchis, A., Guerrero, A., & Pallàs, M. (2024). Advancing personalized medicine in neurodegenerative diseases: The role of epigenetics and pharmacoepigenomics in pharmacotherapy. Pharmacological Research, 205, 107247. [CrossRef]
- Gupta, R., Srivastava, D., Sahu, M., Tiwari, S., Ambasta, R. K., & Kumar, P. (2021). Artificial intelligence to deep learning: machine intelligence approach for drug discovery. Molecular diversity, 25, 1315-1360. [CrossRef]
- Gutman, B., Shmilovitch, A. H., Aran, D., & Shelly, S. (2024). Twenty-Five Years of AI in Neurology: The Journey of Predictive Medicine and Biological Breakthroughs. JMIR Neurotechnology, 3(1), e59556. [CrossRef]
- Hassija, V., Chamola, V., Mahapatra, A., Singal, A., Goel, D., Huang, K.,... & Hussain, A. (2024). Interpreting black-box models: a review on explainable artificial intelligence. Cognitive Computation, 16(1), 45-74. [CrossRef]
- Hosseinian, S. M., & Mirzahossein, H. (2024). Efficiency and safety of traffic networks under the effect of autonomous vehicles. Iranian Journal of Science and Technology, Transactions of Civil Engineering, 48(4), 1861-1885. [CrossRef]
- Ibrahim, M., Khalil, Y. A., Amirrajab, S., Sun, C., Breeuwer, M., Pluim, J.,... & Dumontier, M. (2024). Generative AI for Synthetic Data Across Multiple Medical Modalities: A Systematic Review of Recent Developments and Challenges. arXiv preprint arXiv:2407.00116. [CrossRef]
- Inampudi, R. K., Surampudi, Y., & Kondaveeti, D. (2023). AI-driven real-time risk assessment for financial transactions: leveraging deep learning models to minimize fraud and improve payment compliance. Journal of Artificial Intelligence Research and Applications, 3(1), 716-758.
- Jamali, A. A., Kusalik, A., & Wu, F. X. (2021). NMTF-DTI: a nonnegative matrix tri-factorization approach with multiple kernel fusion for drug-target interaction prediction. IEEE/ACM Transactions on Computational Biology and Bioinformatics, 20(1), 586-594. [CrossRef]
- Janiesch, C., Zschech, P., & Heinrich, K. (2021). Machine learning and deep learning. Electronic Markets, 31(3), 685-695.
- Javaid, M., Haleem, A., Singh, R. P., & Suman, R. (2023). Towards insighting cybersecurity for healthcare domains: A comprehensive review of recent practices and trends. Cyber Security and Applications, 1, 100016. [CrossRef]
- Jiang, J., Chen, L., Ke, L., Dou, B., Zhang, C., Feng, H.,... & Wei, G. (2024). A review of transformers in drug discovery and beyond. Journal of Pharmaceutical Analysis, 101081. [CrossRef]
- Jiménez-Luna, J., Grisoni, F., Weskamp, N., & Schneider, G. (2021). Artificial intelligence in drug discovery: recent advances and future perspectives. Expert opinion on drug discovery, 16(9), 949-959. [CrossRef]
- Jones, C., & Nemeroff, C. B. (2021). Precision psychiatry: Biomarker-guided tailored therapy for effective treatment and prevention in major depression. Major Depressive Disorder: Rethinking and Understanding Recent Discoveries, 535-563.
- Jones, K. M., Asher, A., Goben, A., Perry, M. R., Salo, D., Briney, K. A., & Robertshaw, M. B. (2020). “We're being tracked at all times”: Student perspectives of their privacy in relation to learning analytics in higher education. Journal of the Association for Information Science and Technology, 71(9), 1044-1059. [CrossRef]
- Kale, M. B., Wankhede, N. L., Pawar, R. S., Ballal, S., Kumawat, R., Goswami, M.,... & Koppula, S. (2024). AI-Driven Innovations in Alzheimer's Disease: Integrating Early Diagnosis, Personalized Treatment, and Prognostic Modelling. Ageing Research Reviews, 102497. [CrossRef]
- Kalusivalingam, A. K., Sharma, A., Patel, N., & Singh, V. (2021). Enhancing Drug Discovery and Repurposing through Transformer Models and Reinforcement Learning Algorithms. International Journal of AI and ML, 2(3).
- Karikari, E., & Koshechkin, K. A. (2023). Review on brain-computer interface technologies in healthcare. Biophysical Reviews, 15(5), 1351-1358. [CrossRef]
- Karimian, G., Petelos, E., & Evers, S. M. (2022). The ethical issues of the application of artificial intelligence in healthcare: a systematic scoping review. AI and Ethics, 2(4), 539-551. [CrossRef]
- Kasaraneni, R. K. (2021). AI-Enhanced Virtual Screening for Drug Repurposing: Accelerating the Identification of New Uses for Existing Drugs. Hong Kong Journal of AI and Medicine, 1(2), 129-161.
- Kaswan, K. S., Dhatterwal, J. S., & Ojha, R. P. (2024). AI in personalized learning. In Advances in Technological Innovations in Higher Education (pp. 103-117). CRC Press.
- Knutson, C., Bontha, M., Bilbrey, J. A., & Kumar, N. (2022). Decoding the protein–ligand interactions using parallel graph neural networks. Scientific reports, 12(1), 7624. [CrossRef]
- Krishnamurthy, K., & Pradhan, R. K. (2024). Emerging perspectives of synaptic biomarkers in ALS and FTD. Frontiers in Molecular Neuroscience, 16, 1279999. [CrossRef]
- Ladin, K., Cuddeback, J., Duru, O. K., Goel, S., Harvey, W., Park, J. G.,... & Kent, D. M. (2024). Guidance for unbiased predictive information for healthcare decision-making and equity (GUIDE): considerations when race may be a prognostic factor. NPJ Digital Medicine, 7(1), 290. [CrossRef]
- Lappala, A. (2024). The next revolution in computational simulations: Harnessing AI and quantum computing in molecular dynamics. Current Opinion in Structural Biology, 89, 102919. [CrossRef]
- Lian, X., Zhu, J., Lv, T., Hong, X., Ding, L., Chu, W.,... & Pan, X. (2024). Advancing the Boundary of Pre-trained Models for Drug Discovery: Interpretable Fine-Tuning Empowered by Molecular Physicochemical Properties. IEEE Journal of Biomedical and Health Informatics. [CrossRef]
- Liu, G., Singha, M., Pu, L., Neupane, P., Feinstein, J., Wu, H. C.,... & Brylinski, M. (2021). GraphDTI: a robust deep learning predictor of drug-target interactions from multiple heterogeneous data. Journal of cheminformatics, 13, 1-17. [CrossRef]
- Lobo, P., Morais, P., Murray, P., & Vilaça, J. L. (2024). Trends and Innovations in Wearable Technology for Motor Rehabilitation, Prediction, and Monitoring: A Comprehensive Review. Sensors, 24(24), 7973. [CrossRef]
- Mahapatra C. (2025). Artificial intelligence for diagnosing bladder pathophysiology: An updated review and future prospects. Bladder. 2025: e21200042 . [CrossRef]
- Mahapatra, C. (2017, April). Computational study of inward rectifying ion channel in urinary bladder over activity: student research abstract. In Proceedings of the Symposium on Applied Computing (pp. 31-32). [CrossRef]
- Mahapatra, C. (2017, December). A computational model of action potential in the mouse detrusor smooth muscle cell. In 2017 Winter Simulation Conference (WSC) (pp. 4634-4635). IEEE. [CrossRef]
- Mahapatra, C. (2018, April). Simulation study of transient receptor potential current in urinary bladder over activity: student research abstract. In Proceedings of the 33rd Annual ACM Symposium on Applied Computing (pp. 74-75). [CrossRef]
- Mahapatra, C. (2021). Computational Study of Action Potential Generation in Urethral Smooth Muscle Cell. In Computational Advances in Bio and Medical Sciences: 10th International Conference, ICCABS 2020, Virtual Event, December 10-12, 2020, Revised Selected Papers 10 (pp. 26-32). Springer International Publishing. [CrossRef]
- Mahapatra, C. (2025). Exploring the nexus between medical gases and ion channel function: implications for therapeutic innovation. Medical Gas Research, 10-4103. [CrossRef]
- Mahapatra, C. (2025). Recent advances in medical gas sensing with artificial intelligence-enabled technology. Medical Gas Research, 15(2), 318-326. [CrossRef]
- Mahapatra, C., & Kaur, A. (2024). Abstract 2221 In silico electrophysiological study reveals Ibrutinib, an important therapeutic agent for B-Cell lymphoma causes cardiac toxicity by inhibiting sodium current. Journal of Biological Chemistry, 300(3). [CrossRef]
- Mahapatra, C., & Kumar, R. (2024). Biophysical Mechanisms of Vaginal Smooth Muscle Contraction: The Role of the Membrane Potential and Ion Channels. Pathophysiology, 31(2), 225-243. [CrossRef]
- Mahapatra, C., & Manchanda, R. (2015). Computational studies on bladder smooth muscle: modeling ion channels and their role in generating electrical activity. Biophysical Journal, 108(2), 588a. [CrossRef]
- Mahapatra, C., & Manchanda, R. (2016, August). Computational simulation of spike patterns in STN neuron using Izhikevich model. In 2016 International Conference on Inventive Computation Technologies (ICICT) (Vol. 2, pp. 1-4). IEEE. [CrossRef]
- Mahapatra, C., & Manchanda, R. (2017). Simulation of In Vitro-Like Electrical Activities in Urinary Bladder Smooth Muscle Cells. Journal of Biomimetics, Biomaterials and Biomedical Engineering, 33, 45-51. [CrossRef]
- Mahapatra, C., & Manchanda, R. (2019). Modeling Vas Deferens Smooth Muscle Electrophysiology: Role of Ion Channels in Generating Electrical Activity. In Soft Computing for Problem Solving: SocProS 2017, Volume 2 (pp. 655-663). Springer Singapore. [CrossRef]
- Mahapatra, C., & Manchanda, R. (2019). Modeling Vas Deferens Smooth Muscle Electrophysiology: Role of Ion Channels in Generating Electrical Activity. In Soft Computing for Problem Solving: SocProS 2017, Volume 2 (pp. 655-663). Springer Singapore. [CrossRef]
- Mahapatra, C., & Manchanda, R. (2019). Modulating Properties of Hyperpolarization-Activated Cation Current in Urinary Bladder Smooth Muscle Excitability: A Simulation Study. In Recent Findings in Intelligent Computing Techniques: Proceedings of the 5th ICACNI 2017, Volume 1 (pp. 261-266). Springer Singapore. [CrossRef]
- Mahapatra, C., & Manchanda, R. (2019). Modulating Properties of Hyperpolarization-Activated Cation Current in Urinary Bladder Smooth Muscle Excitability: A Simulation Study. In Recent Findings in Intelligent Computing Techniques: Proceedings of the 5th ICACNI 2017, Volume 1 (pp. 261-266). Springer Singapore. [CrossRef]
- Mahapatra, C., & Manchanda, R. (2019, July). Computational studies on ureter smooth muscle: modeling ion channels and their role in generating electrical activity. In Proceedings of the 2019 Summer Simulation Conference (pp. 1-6).
- Mahapatra, C., & Samuilik, I. (2024). A Mathematical Model of Spontaneous Action Potential Based on Stochastics Synaptic Noise Dynamics in Non-Neural Cells. Mathematics, 12(8), 1149. [CrossRef]
- Mahapatra, C., & Thakkar, R. (2024). In Silico Electrophysiological Investigation of Transient Receptor Potential Melastatin-4 Ion Channel Biophysics to Study Detrusor Overactivity. International Journal of Molecular Sciences, 25(13), 6875. [CrossRef]
- Mahapatra, C., Adam, S., & Gaidhane, P. (2024, December). Influence of Glucose on Cardiac Pacemaking Electrophysiology through Potassium Ion Channel Biophysics: A Computational Study. In 2024 IEEE 21st India Council International Conference (INDICON) (pp. 1-5). IEEE. [CrossRef]
- Mahapatra, C., Brain, K. L., & Manchanda, R. (2016, August). Computational study of ATP gated Potassium ion channel in urinary bladder over activity. In 2016 International Conference on Inventive Computation Technologies (ICICT) (Vol. 2, pp. 1-4). IEEE. [CrossRef]
- Mahapatra, C., Brain, K. L., & Manchanda, R. (2018, October). Computational study of Hodgkin-Huxley type calcium-dependent potassium current in urinary bladder over activity. In 2018 IEEE 8th international conference on computational advances in bio and medical sciences (ICCABS) (pp. 1-4). IEEE. [CrossRef]
- Mahapatra, C., Dave, V., & Manchanda, R. (2017). A Mathematical Modeling of Voltage-gated Calcium ion channel-based Calcium Transient Response in Urinary Bladder Smooth Muscle Cell. International Journal of Pure and Applied Mathematics, 117(9), 71-75.
- Mahapatra, C., Gawad, J., Bonde, C., & Palkar, M. B. (2025). Bioelectric Membrane Potential and Breast Cancer: Advances in Neuroreceptor Pharmacology for Targeted Therapeutic Strategies. Receptors, 4(2), 9. [CrossRef]
- Mahapatra, C., Kishore, A., Gawad, J., Al-Emam, A., Kouzeiha, R. A., & Rusho, M. A. (2025). Review of electrophysiological models to study membrane potential changes in breast cancer cell transformation and tumor progression. Frontiers in Physiology, 16, 1536165. [CrossRef]
- Mahapatra, C., Thakkar, R., & Kumar, R. (2024). Modulatory Impact of Oxidative Stress on Action Potentials in Pathophysiological States: A Comprehensive Review. Antioxidants, 13(10), 1172. [CrossRef]
- Mak, K. K., Wong, Y. H., & Pichika, M. R. (2024). Artificial intelligence in drug discovery and development. Drug discovery and evaluation: safety and pharmacokinetic assays, 1461-1498.
- Maristany, A. J., Sa, B. C., Murray, C., Subramaniam, A. B., & Oldak, S. E. (2024). Psychiatric manifestations of neurological diseases: a narrative review. Cureus, 16(7), e64152. [CrossRef]
- Martinelli, D. D. (2022). Generative machine learning for de novo drug discovery: A systematic review. Computers in Biology and Medicine, 145, 105403. [CrossRef]
- Maveyraud, L., & Mourey, L. (2020). Protein X-ray crystallography and drug discovery. Molecules, 25(5), 1030. [CrossRef]
- Meng, Z., Zhang, Y., Yang, L., Yuan, F., Wang, J., Chen, J.,... & Zang, G. (2024). Application of advanced biosensors in nervous system diseases. Interdisciplinary Medicine, e20240024. [CrossRef]
- Mir, R. H., Shah, A. J., Mohi-Ud-Din, R., Pottoo, F. H., Dar, M. A., Jachak, S. M., & Masoodi, M. H. (2021). Natural Anti-inflammatory compounds as Drug candidates in Alzheimer’s disease. Current medicinal chemistry, 28(23), 4799-4825. [CrossRef]
- Nayarisseri, A., Khandelwal, R., Tanwar, P., Madhavi, M., Sharma, D., Thakur, G.,... & Singh, S. K. (2021). Artificial intelligence, big data and machine learning approaches in precision medicine & drug discovery. Current drug targets, 22(6), 631-655. [CrossRef]
- Nimgampalle, M., Chakravarthy, H., Sharma, S., Shree, S., Bhat, A. R., Pradeepkiran, J. A., & Devanathan, V. (2023). Neurotransmitter systems in the etiology of major neurological disorders: Emerging insights and therapeutic implications. Ageing Research Reviews, 101994. [CrossRef]
- Obaido, G., Mienye, I. D., Egbelowo, O. F., Emmanuel, I. D., Ogunleye, A., Ogbuokiri, B.,... & Aruleba, K. (2024). Supervised machine learning in drug discovery and development: Algorithms, applications, challenges, and prospects. Machine Learning with Applications, 17, 100576. [CrossRef]
- Oliver, M., Hamilton, A., & Ingram, K. (2022). Best Practices for Working Creatively with Personal Data.
- Ononamadu, C. J., & Ibrahim, A. (2021). Molecular docking and prediction of ADME/drug-likeness properties of potentially active antidiabetic compounds isolated from aqueous-methanol extracts of Gymnema sylvestre and Combretum micranthum. BioTechnologia, 102(1), 85. [CrossRef]
- Oyanna, V. O., & Clarke, J. D. (2024). Mechanisms of intestinal pharmacokinetic natural product-drug interactions. Drug Metabolism Reviews, 56(3), 285-301. [CrossRef]
- Oyebanjo, O. T., Adetuyi, B. O., Adeoye, A. D., Adetuyi, O. A., Oni, P. G., & Ogunlana, O. O. (2024). Neuropharmacology and neurotherapeutics: Advancing the understanding and treatment of neurological disorders. In Biochemical and Molecular Pharmacology in Drug Discovery (pp. 403-425). Elsevier.
- Paliwal, A., Jain, S., Kumar, S., Wal, P., Khandai, M., Khandige, P. S.,... & Srivastava, S. (2024). Predictive Modelling in pharmacokinetics: from in-silico simulations to personalized medicine. Expert Opinion on Drug Metabolism & Toxicology, 20(4), 181-195. [CrossRef]
- Parvathy GA, Kamaraj BA, Sah BI, Maheshwari AA, Alexander AI, Dixit VI, Mumtaz HA, Saqib MU. Emerging artificial intelligence-aided diagnosis and management methods for ischemic strokes and vascular occlusions: A comprehensive review. World Neurosurgery: X. 2024 Feb 25:100303. [CrossRef]
- Patidar, N., Mishra, S., Jain, R., Prajapati, D., Solanki, A., Suthar, R.,... & Patel, H. (2024). Transparency in AI Decision Making: A Survey of Explainable AI Methods and Applications. Advances of Robotic Technology, 2(1).
- Perera, N., Dehmer, M., & Emmert-Streib, F. (2020). Named entity recognition and relation detection for biomedical information extraction. Frontiers in cell and developmental biology, 8, 673. [CrossRef]
- Poweleit, E. A., Vinks, A. A., & Mizuno, T. (2023). Artificial intelligence and machine learning approaches to facilitate therapeutic drug management and model-informed precision dosing. Therapeutic drug monitoring, 45(2), 143-150. [CrossRef]
- Prasanna, D. L., & Tripathi, S. L. (2022). Machine and deep-learning techniques for text and speech processing. Machine learning algorithms for signal and image processing, 115-128.
- Pu, Z., Shi, C. L., Jeon, C. O., Fu, J., Liu, S. J., Lan, C.,... & Jia, B. (2024). ChatGPT and generative AI are revolutionizing the scientific community: A Janus-faced conundrum. iMeta, 3(2), e178. [CrossRef]
- Raikar, A. S., Andrew, J., Dessai, P. P., Prabhu, S. M., Jathar, S., Prabhu, A.,... & Raikar, G. V. S. (2024). Neuromorphic computing for modeling neurological and psychiatric disorders: implications for drug development. Artificial Intelligence Review, 57(12), 318. [CrossRef]
- Rashid, A. B., & Kausik, A. K. (2024). AI revolutionizing industries worldwide: A comprehensive overview of its diverse applications. Hybrid Advances, 100277.
- RothRichard, H., & DingJun, B. (2020). From neurons to cognition: technologies for precise recording of neural activity underlying behavior. BME frontiers.
- Sadybekov, A. V., & Katritch, V. (2023). Computational approaches streamlining drug discovery. Nature, 616(7958), 673-685. [CrossRef]
- Salas-Nuñez, L. F., Barrera-Ocampo, A., Caicedo, P. A., Cortes, N., Osorio, E. H., Villegas-Torres, M. F., & González Barrios, A. F. (2024). Machine Learning to Predict Enzyme–Substrate Interactions in Elucidation of Synthesis Pathways: A Review. Metabolites, 14(3), 154. [CrossRef]
- Schauperl, M., & Denny, R. A. (2022). AI-based protein structure prediction in drug discovery: impacts and challenges. Journal of Chemical Information and Modeling, 62(13), 3142-3156. [CrossRef]
- Schmidgall, S., Ziaei, R., Achterberg, J., Kirsch, L., Hajiseyedrazi, S., & Eshraghian, J. (2024). Brain-inspired learning in artificial neural networks: a review. APL Machine Learning, 2(2). [CrossRef]
- Serrano, D. R., Luciano, F. C., Anaya, B. J., Ongoren, B., Kara, A., Molina, G.,... & Lalatsa, A. (2024). Artificial intelligence (AI) applications in drug discovery and drug delivery: Revolutionizing personalized medicine. Pharmaceutics, 16(10), 1328. [CrossRef]
- Singh, N. K., Maiti, N. J., Mishra, M., Raj, S., Rakshit, G., Ghosh, R., & Roy, S. (2025). Virtual Screening and Lead Discovery. Computational Methods for Rational Drug Design, 97-121.
- Singh, S., Gupta, H., Sharma, P., & Sahi, S. (2024). Advances in Artificial Intelligence (AI)-assisted approaches in drug screening. Artificial Intelligence Chemistry, 2(1), 100039. [CrossRef]
- Soleimani, G., Nitsche, M. A., Bergmann, T. O., Towhidkhah, F., Violante, I. R., Lorenz, R.,... & Ekhtiari, H. (2023). Closing the loop between brain and electrical stimulation: towards precision neuromodulation treatments. Translational psychiatry, 13(1), 279. [CrossRef]
- Srinivasan, E., Chandrasekhar, G., Chandrasekar, P., Anbarasu, K., Vickram, A. S., Karunakaran, R.,... & Srikumar, P. S. (2021). Alpha-synuclein aggregation in Parkinson's disease. Frontiers in medicine, 8, 736978.
- Sucharitha, P., Reddy, K. R., Satyanarayana, S. V., & Garg, T. (2022). Absorption, distribution, metabolism, excretion, and toxicity assessment of drugs using computational tools. In Computational approaches for novel therapeutic and diagnostic designing to mitigate SARS-CoV-2 infection (pp. 335-355). Academic Press.
- Sultana, O. F., Bandaru, M., Islam, M. A., & Reddy, P. H. (2024). Unraveling the complexity of human brain: Structure, function in healthy and disease states. Ageing Research Reviews, 100, 102414. [CrossRef]
- Syed, F. M., & ES, F. K. (2024). AI in Securing Pharma Manufacturing Systems Under GxP Compliance. International Journal of Machine Learning Research in Cybersecurity and Artificial Intelligence, 15(1), 448-472.
- Taherdoost, H., & Ghofrani, A. (2024). AI and the Evolution of Personalized Medicine in Pharmacogenomics. Intelligent Pharmacy.
- Thatoi, P., Choudhary, R., Shiwlani, A., Qureshi, H. A., & Kumar, S. (2023). Natural Language Processing (NLP) in the Extraction of Clinical Information from Electronic Health Records (EHRs) for Cancer Prognosis. International Journal, 10(4), 2676-2694.
- Tolosa, E., Vila, M., Klein, C., & Rascol, O. (2020). LRRK2 in Parkinson disease: challenges of clinical trials. Nature Reviews Neurology, 16(2), 97-107. [CrossRef]
- Trezza, A., Visibelli, A., Roncaglia, B., Spiga, O., & Santucci, A. (2024). Unsupervised Learning in Precision Medicine: Unlocking Personalized Healthcare through AI. Applied Sciences, 14(20), 9305. [CrossRef]
- Tsuji, S., Hase, T., Yachie-Kinoshita, A., Nishino, T., Ghosh, S., Kikuchi, M.,... & Tanaka, H. (2021). Artificial intelligence-based computational framework for drug-target prioritization and inference of novel repositionable drugs for Alzheimer’s disease. Alzheimer's Research & Therapy, 13(1), 92. [CrossRef]
- van Nuland, A. J., Helmich, R. C., Dirkx, M. F., Zach, H., Toni, I., Cools, R., & den Ouden, H. E. (2020). Effects of dopamine on reinforcement learning in Parkinson’s disease depend on motor phenotype. Brain, 143(11), 3422-3434. [CrossRef]
- Varrone, A., Bundgaard, C., & Bang-Andersen, B. (2022). PET as a translational tool in drug development for neuroscience compounds. Clinical Pharmacology & Therapeutics, 111(4), 774-785. [CrossRef]
- Vatansever, S., Schlessinger, A., Wacker, D., Kaniskan, H. Ü., Jin, J., Zhou, M. M., & Zhang, B. (2021). Artificial intelligence and machine learning-aided drug discovery in central nervous system diseases: State-of-the-arts and future directions. Medicinal research reviews, 41(3), 1427-1473.
- Vicidomini, C., Fontanella, F., D’Alessandro, T., & Roviello, G. N. (2024). A Survey on Computational Methods in Drug Discovery for Neurodegenerative Diseases. Biomolecules, 14(10), 1330. [CrossRef]
- Wang, B., Xie, Q., Pei, J., Chen, Z., Tiwari, P., Li, Z., & Fu, J. (2023). Pre-trained language models in biomedical domain: A systematic survey. ACM Computing Surveys, 56(3), 1-52. [CrossRef]
- Wang, Y., Liu, L., & Wang, C. (2023). Trends in using deep learning algorithms in biomedical prediction systems. Frontiers in Neuroscience, 17, 1256351. [CrossRef]
- Wysocki, O., Davies, J. K., Vigo, M., Armstrong, A. C., Landers, D., Lee, R., & Freitas, A. (2023). Assessing the communication gap between AI models and healthcare professionals: Explainability, utility and trust in AI-driven clinical decision-making. Artificial Intelligence, 316, 103839. [CrossRef]
- Xiong, J., Xiong, Z., Chen, K., Jiang, H., & Zheng, M. (2021). Graph neural networks for automated de novo drug design. Drug discovery today, 26(6), 1382-1393. [CrossRef]
- Yang, S., & Kar, S. (2023). Application of artificial intelligence and machine learning in early detection of adverse drug reactions (ADRs) and drug-induced toxicity. Artificial Intelligence Chemistry, 100011. [CrossRef]
- Yang, X., & Zhu, C. (2024). Industrial expert systems review: A comprehensive analysis of typical applications. IEEE Access. [CrossRef]
- Yang, X., Lou, J., Shan, W., Ding, J., Jin, Z., Hu, Y.,... & Xu, J. (2021). Pathophysiologic role of neurotransmitters in digestive diseases. Frontiers in physiology, 12, 567650. [CrossRef]
- Yingngam, B., Navabhatra, A., & Sillapapibool, P. (2024). AI-Driven Decision-Making Applications in Pharmaceutical Sciences. In Using Traditional Design Methods to Enhance AI-Driven Decision Making (pp. 1-63). IGI Global.
- Zhang, R., Guo, J., Chen, L., Fan, Y., & Cheng, X. (2021). A review on question generation from natural language text. ACM Transactions on Information Systems (TOIS), 40(1), 1-43. [CrossRef]
- Zhu, T., Ye, D., Wang, W., Zhou, W., & Philip, S. Y. (2020). More than privacy: Applying differential privacy in key areas of artificial intelligence. IEEE Transactions on Knowledge and Data Engineering, 34(6), 2824-2843. [CrossRef]
- Zou, M., Zhou, H., Gu, L., Zhang, J., & Fang, L. (2024). Therapeutic Target Identification and Drug Discovery Driven by Chemical Proteomics. Biology, 13(8), 555. [CrossRef]
- Zuo, C., Ge, J., Wang, J., Watanabe, Y., & Tian, M. (2024). Molecular imaging for neurological diseases. In Transpathology (pp. 247-258). Elsevier.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).