Introduction
Approximately 29% of the total population is moderately or severely food insecure (FAO, 2023). Even though there has been some progress towards reducing chronic hunger in the last four years in regions such as Asia and Latin America and the Caribbean, projected numbers of undernourished indicate that the world is far off from achieving zero hunger by 2030 (FAO et al., 2024). In addition, global food supply and demand are expected to increase notably, especially in developing countries, because the world population is expected to grow by 1.6 billion people by 2050 (FAO, 2023). To meet these challenges, crop yield needs to increase by 70% by 2050 to satisfy the demands of a growing population (Hussain et al., 2021). Efforts to increase crop yield have included agronomical practices such as intercropping, breeding high-yield crops, better fertilizers and pesticides, and the development of stress-tolerant plants (Hussain et al., 2021). However, the implementation of these techniques is not sufficient to satisfy future food demands (Murchie et al., 2009).
While the majority of studies looking at boosting crop production have focused mainly on C3 species, the genetic factors controlling yield in C4 species are not well understood (Sales et al., 2021). C4 species such as maize, sorghum, and sugarcane are among the top ten crops with the highest annual global production, with sorghum being the fifth most important among cereal crops in terms of both annual metric production and yield (FAO, 2023). Sorghum (Sorghum bicolor L.) is known for its dual-purpose crop (staple food for humans and feed crop for livestock) and its greatest drought tolerance among C4 plants (Hossain et al., 2022), vital to secure future food security. The small relative genome makes sorghum an ideal model crop for crop genomic studies. Moreover, the availability of the sorghum genome sequences in public databases has made it possible to conduct different analyses, including genome-wide association (GWAS) analysis for the identification of molecular markers associated with key physiological traits.
To improve prediction accuracy in genomic studies, statistical models such as Bayesian Generalized Linear Regression (BGLR) have been widely used. BGLR offers flexibility through multiple Bayesian methods (e.g., BayesA, BayesB, BayesCπ, LASSO, and gBLUP) and allows users to model complex traits and marker effects with different prior assumptions (Pérez & de los Campos, 2014). The main advantage of BGLR is its ability to fit a range of models suited for various genetic architectures. However, it requires careful parameter tuning and expertise in Bayesian statistics, and it can be computationally demanding.
Alternatively, the Mining the Maximum Accuracy of Prediction (MMAP) platform was developed to automatically select the best genomic prediction method for a given trait by leveraging cloud computing (Huang et al., 2020). MMAP simplifies the prediction process, making it accessible to users with limited statistical backgrounds by selecting the optimal model based on prior data similarity. Nonetheless, MMAP’s effectiveness is contingent upon the breadth and quality of publicly available datasets in its database, and it has been infrequently cited compared to more established packages like BGLR and GAPIT, potentially limiting its acceptance. Currently, there is a significant gap in knowledge regarding the use of MMAP for genome prediction, particularly in plants. While traditional methods have been extensively benchmarked, the practical applicability and limitations of MMAP in plant breeding programs remain underexplored. Addressing this gap is critical for understanding whether cloud-based, automated platforms like MMAP can serve as viable alternatives for accelerating crop improvement. We hypothesize that MMAP can offer comparable or even superior prediction performance to traditional models like those in BGLR when predicting plant height in Sorghum bicolor, despite its current underutilization in plant genomic prediction studies.
Material and Methods
Plant Material
The number of sorghum accessions was obtained as outlined by Enyew et al. (2025), with the difference that only 309 accessions were used in this study.
Phenotyping Data
Plant height (cm; PH) was measured from each sorghum accession before the flowering stage.
SNP Selection and Genotyping
The 309 accessions were genotyped using SeqSNP. This genotyping method used 5000 SNP makers that were previously identified in the genetic diversity analysis of sorghum accessions by Enyew et al. (2022). The SNP markers are targeted on chromosomes 1 to 10. The majority of the SNP markers (93.7%) came from the sorghum DNP database SorGSD (
http://sorgsd.big.ac.cn), and the remaining (6.3%) were identified as outlined by Enyew et al. (2022). Among the 5000 SNP markers, the polymorphic SNP loci and bi-allelic loci were identified as described by Enyew et al. (2025). In addition, from the bi-allelic SNP loci group, further SNP filtering analysis through minimum allele frequency (MAF) was performed to identify high-quality SNP loci (3142 SNPs).
BGLR (Bayesian Generalized Linear Regression)
The data was analyzed following the guidelines in the BGLR package version 1.1.4. (Pérez & de los Campos, 2014) from the R software environment. From the BGLR package, we used four different random sampling methods, such as Bayesian B, Cπ, least absolute shrinkage and selection operator (LASSO), and genomic best linear unbiased prediction (gBLUP).
MMAP (Minimum the Maximum Accuracy of Prediction)
The genotype and phenotype data from the 309 sorghum accessions were analyzed using a cloud computing platform to solve the problem by mining the maximum accuracy of predicting phenotypes (MMAP) from genotypes (Huang et al., 2020).
Results and Discussions
BGLR
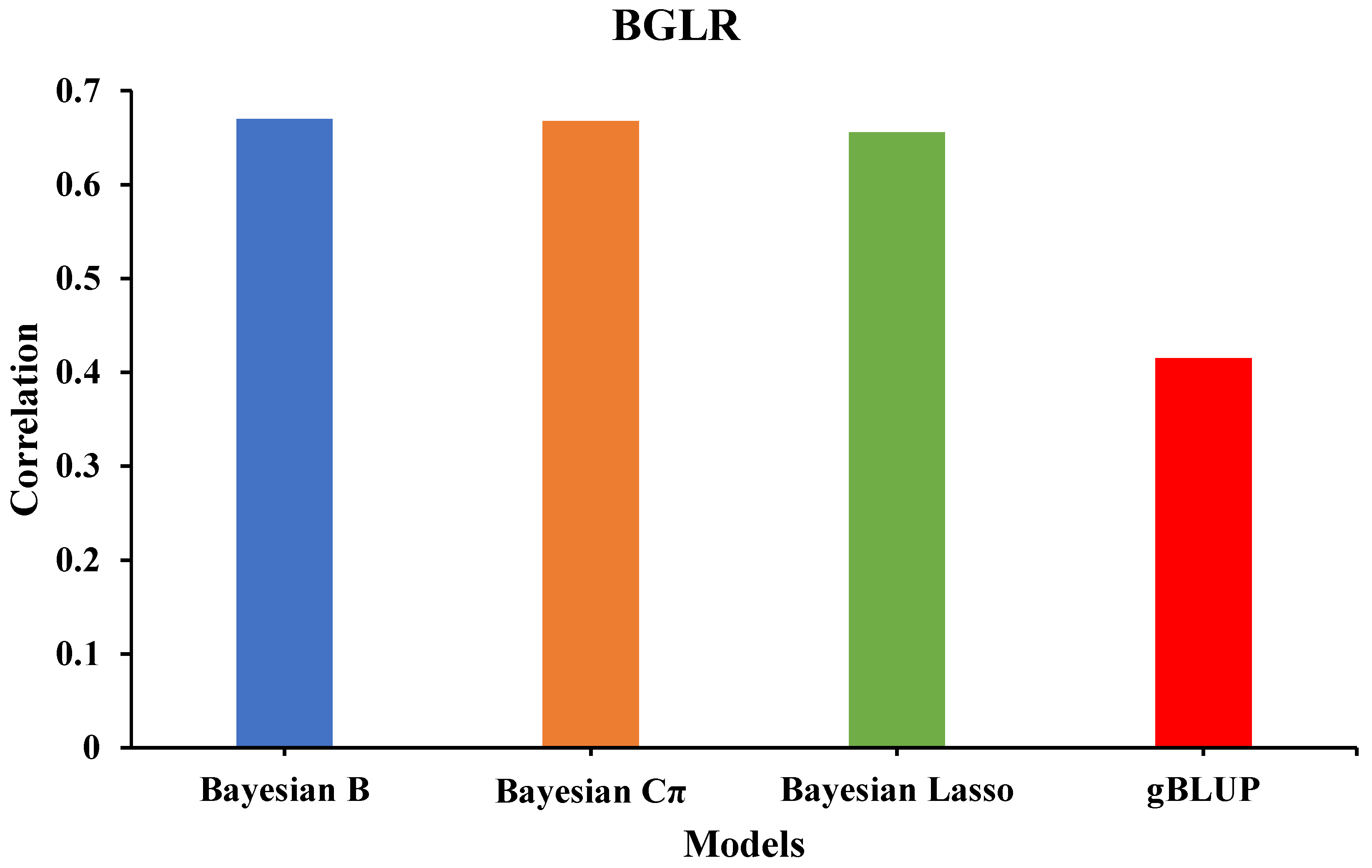
The predicted phenotype values for training data from 309 sorghum accessions were determined by using four different methods of the package BGLR. To determine which method has the highest correlation between the predicted and actual phenotype values, we calculated the instant accuracy values for each one of them. The analysis showed that Bayesian B, Cπ, and Lasso have a greater instant accuracy value compared to gBLUP (0.67, 0.67, 0.66 vs. 0.42; respectively) (
Figure 1). Lee et al. (2019) reported that when using sorghum seed traits to find associations between seed shape and genome-wide single-nucleotide polymorphisms, the prediction phenotype accuracy value obtained from gBLUP was ~0.50. This suggests that the magnitude of the instant accuracy varies by method and plant trait. Additionally, the type of trait used to predict the phenotype has an influence on the accuracy of the method. Even though our study has a lower instant accuracy value for PH, which is a continuous variable, the greater accuracy for seed shape may have been due to no scaling being applied to the data, and the seed shape average was used instead. This indicates that seed size or variables that are continuous can be easier to predict (Lee et al., 2019).
The greater accuracy value for Bayesian B, Cπ, and Lasso compared to gBLUP can be related to the type of trait used for genomic selection. For instance, traits that have a high heritability, such as plant height, Bayesian Lasso perform better than compressed BLUP, and the opposite is observed when a trait has a low heritability (Wang et al., 2018). In addition, it has been reported that gBLUP is suited for polygenic traits (Huang et al., 2020). Since plant height is a polygenic trait and has a high heritability (Begna, 2025), using both methods can be the best approach for genomic selection, but the significant difference in their instant accuracy values limits the use of gBLUP for plant height in our sorghum accessions.
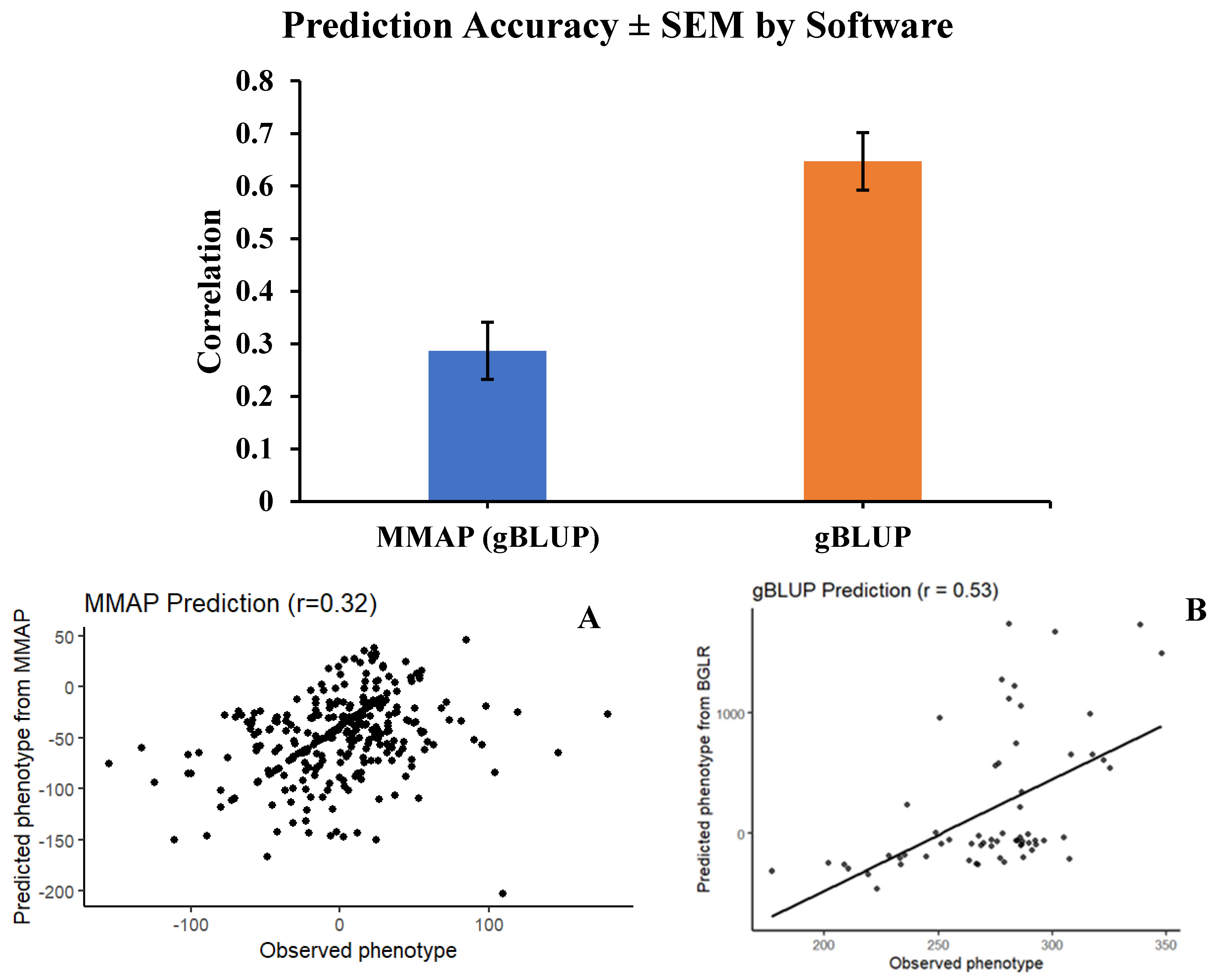
Prediction Accuracy of MAPP and BGLR
Using accuracy values can be a strong tool for choosing a suitable method for a given trait. However, the applicability of specific methods for genomic selection is dependent on the number of genes underlying the trait and its heritability (Huang et al., 2020). To overcome this limitation, the software Minimum the Maximum Accuracy of Prediction (MMAP) can be used to do the computational work and select the best prediction method for a particular trait (Huang et al., 2020).
Figure 2 shows the accuracy values between two different software programs, BGLR and MMAP, where the gBLUP method was selected by MMAP as the best option to predict plant height in sorghum accessions. Although gBLUP was picked by MMAP, the predicted accuracy was still lower compared to gBLUP from the BGLR package (0.29 vs. 0.65, respectively). It is important to mention that the efficacy of MMAP in selecting the appropriate prediction method depends on the publicly available data on the platform, suggesting that its performance can improve as more data are uploaded to the platform. However, this could be a limitation because the majority of studies have used other packages such as BGLR and BLR for genomic selection, while MMAP has been cited only two times (Chen et al., 2022; Gupta & Sharma, 2023).
Conclusion
This study compared the predictive performance of BGLR and MMAP for estimating plant height in Sorghum bicolor. BGLR models, particularly Bayesian B, Cπ, and LASSO, outperformed gBLUP and showed greater prediction accuracy, consistent with expectations for traits with high heritability and polygenic architecture. Although MMAP selected gBLUP as the best method, its prediction accuracy was lower than that achieved using BGLR. The limited performance of MMAP may reflect the current size and diversity of its reference dataset, suggesting that its utility will likely improve as more genomic data becomes available. Overall, while traditional statistical frameworks like BGLR currently provide higher accuracy, MMAP represents a promising cloud-based alternative for genomic prediction, especially for users seeking simplified and automated solutions. Future studies should continue to evaluate MMAP across a broader range of traits and species to assess its potential for plant breeding applications fully.
References
- Begna, T. (2025). Phenotypic variability analysis of key sorghum (Sorghum bicolor (L) Moench) genotypes under dry lowland areas. Reproduction and Breeding, 5(2), 79-87. [CrossRef]
- Chen, C. J., Rutkoski, J., Schnable, J. C., Murray, S. C., Wang, L., Jin, X., Stich, B., Crossa, J., Hayes, B. J., & Zhang, Z. (2022). Role of the Genomics–Phenomics–Agronomy Paradigm in Plant Breeding. In Plant Breeding Reviews (pp. 627-673). [CrossRef]
- Enyew, M., Feyissa, T., Carlsson, A. S., Tesfaye, K., Hammenhag, C., Seyoum, A., & Geleta, M. (2022). Genome-wide analyses using multi-locus models revealed marker-trait associations for major agronomic traits in Sorghum bicolor. Frontiers in Plant Science, 13, 999692-999692. [CrossRef]
- Enyew, M., Geleta, M., Tesfaye, K., Seyoum, A., Feyissa, T., Alemu, A., Hammenhag, C., & Carlsson, A. S. (2025). Genome-wide association study and genomic prediction of root system architecture traits in Sorghum (Sorghum bicolor (L.) Moench) at the seedling stage. BMC plant biology, 25(1), 69-69. [CrossRef]
- FAO. (2023). FAOSTAT statistical database. Rome, Italy. Accessed on August 5. https://www.fao.org/faostat/en/#data/FS.
- FAO, IFAD, UNICEF, WFP, & WHO. (2024). The State of Food Security and Nutrition in the World 2024—Financing to end hunger, food insecurity and malnutrition in all its forms . [CrossRef]
- Gupta, U., & Sharma, R. (2023, 3-4 March 2023). A Study of Cloud-Based Solution for Data Analytics in Healthcare. 6th International Conference on Information Systems and Computer Networks (ISCON).
- Hossain, M. S., Islam, M. N., Rahman, M. M., Mostofa, M. G., & Khan, M. A. R. (2022). Sorghum: A prospective crop for climatic vulnerability, food and nutritional security. Journal of Agriculture and Food Research, 8, 100300. [CrossRef]
- Huang, W., Zheng, P., Cui, Z., Li, Z., Gao, Y., Yu, H., Tang, Y., Yuan, X., & Zhang, Z. (2020). MMAP: a cloud computing platform for mining the maximum accuracy of predicting phenotypes from genotypes. Bioinformatics, 37(9), 1324-1326. [CrossRef]
- Hussain, S., Ulhassan, Z., Brestic, M., Zivcak, M., Weijun, Z., Allakhverdiev, S. I., Yang, X., Safdar, M. E., Yang, W., & Liu, W. (2021). Photosynthesis research under climate change. Photosynthesis Research, 150(1-3), 5-19. [CrossRef]
- Lee, S., Dang, C., Choy, Y., Do, C., Cho, K., Kim, J., Kim, Y., & Lee, J. (2019). Comparison of genome-wide association and genomic prediction methods for milk production traits in Korean Holstein cattle. Asian-Australasian Journal of Animal Sciences, 32(7), 913-921.
- Murchie, E. H., Pinto, M., & Horton, P. (2009). Agriculture and the new challenges for photosynthesis research. The New phytologist, 181(3), 532-552. [CrossRef]
- Pérez, P., & de los Campos, G. (2014). Genome-Wide Regression and Prediction with the BGLR Statistical Package. Genetics, 198(2), 483-495. [CrossRef]
- Sales, C. R. G., Wang, Y., Evers, J. B., & Kromdijk, J. (2021). Improving C4 photosynthesis to increase productivity under optimal and suboptimal conditions. Journal of Experimental Botany, 72(17), 5942-5960. [CrossRef]
- Wang: J., Zhou, Z., Zhang, Z., Li, H., Liu, D., Zhang, Q., Bradbury, P. J., Buckler, E. S., & Zhang, Z. (2018). Expanding the BLUP alphabet for genomic prediction is adaptable to the genetic architectures of complex traits. Heredity, 121(6), 648-662. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).