Submitted:
30 April 2025
Posted:
30 April 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Isolation and Phenotypic Identification of T. pyogenes
2.2. Antimicrobial Susceptibility Testing
2.3.16. S rRNA Gene Sequence Analysis
2.4. Detection of Virulence Genes
3. Results
3.1. Isolation and Phenotypic Identification of T. pyogenes
3.2. Antimicrobial Susceptibility Testing
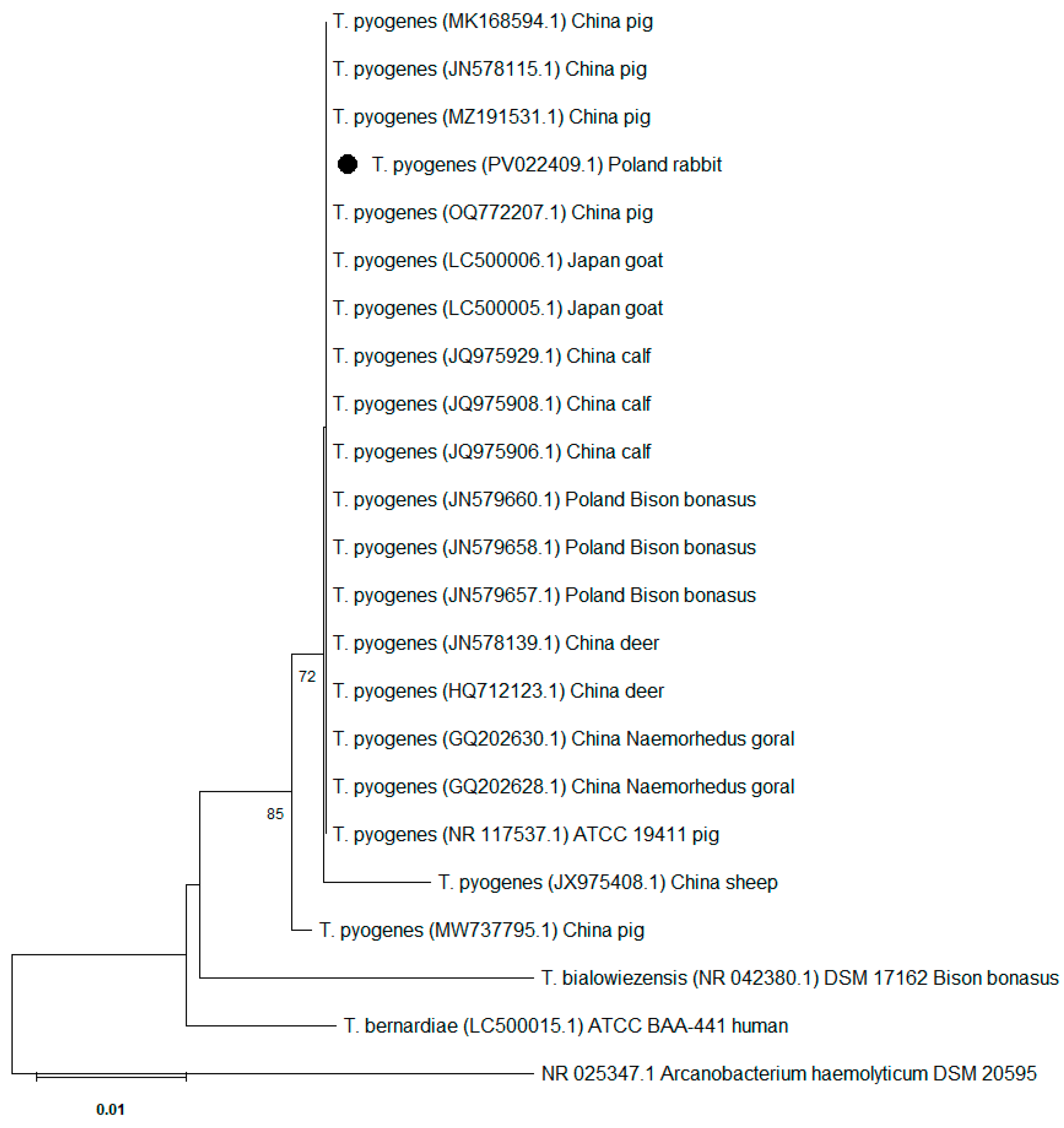
3.3. 16S rRNA Gene Sequence Analysis
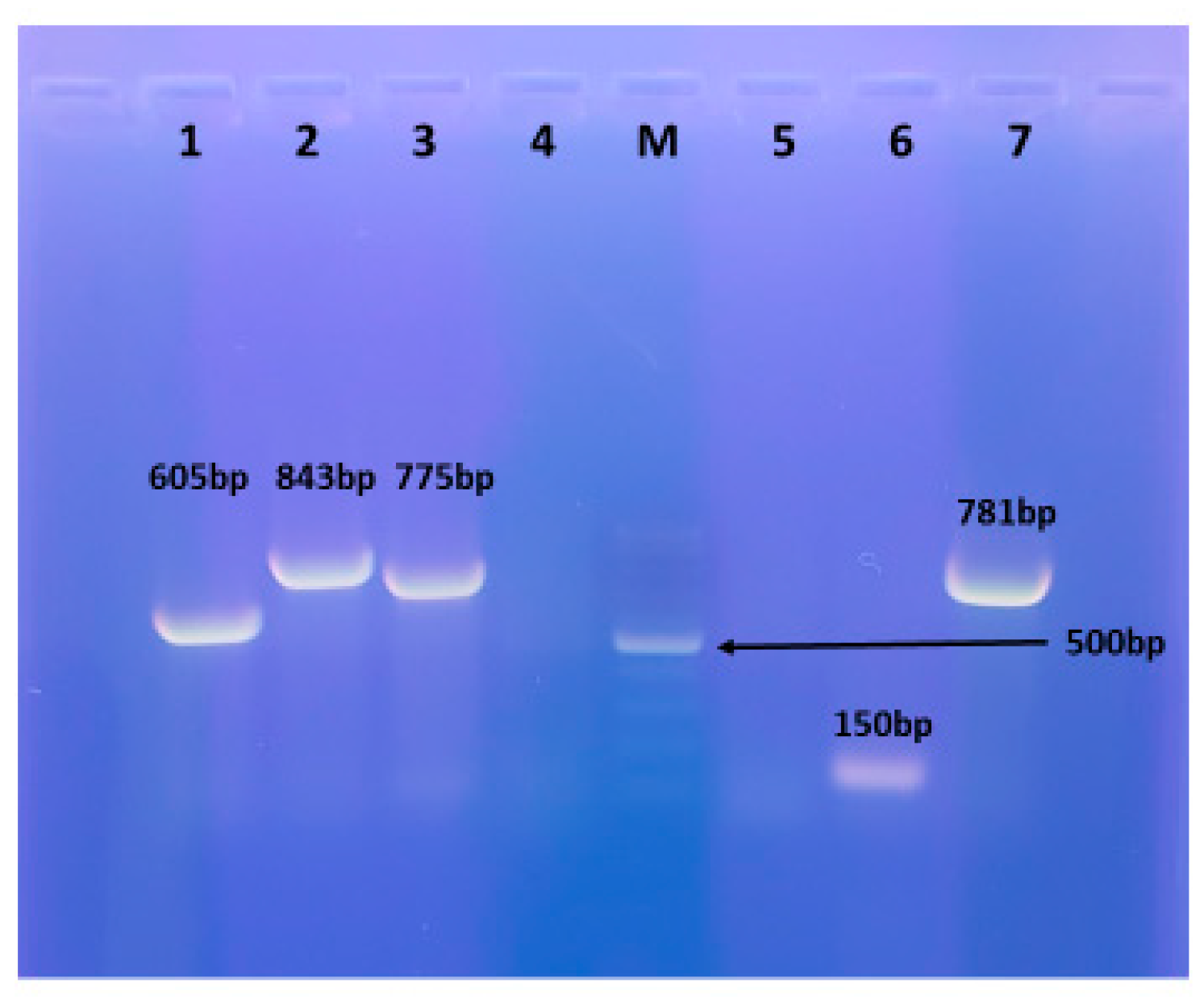
3.4. Detection of Virulence Genes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Taylor, W.M.; Beaufrère, H.; Mans, C.; Smith, D.A. Long-term outcome of treatment of dental abscesses with a wound-packing technique in pet rabbits: 13 cases (1998-2007). J. Am. Vet. Med. Assoc. 2010, 237, 1444-1449. [CrossRef]
- Levy, I.; Mans, C. Diagnosis and outcome of odontogenic abscesses in client-owned rabbits (Oryctolagus cuniculus): 72 cases (2011-2022). J. Am. Vet. Med. Assoc. 2024, 262, 658-664. [CrossRef]
- Rätsep, E.; Ludwig, L.; Dobromylskyj, M. Orofacial masses in domestic rabbits: a retrospective review of 120 cases from 2 institutions, 2000-2023. J. Vet. Diagn. Invest. 2024, 36, 724-729. [CrossRef]
- Tyrrell, K.L.; Citron, D.M.; Jenkins, J.R.; Goldstein, E.J. Periodontal bacteria in rabbit mandibular and maxillary abscesses. J. Clin. Microbiol. 2002, 40, 1044-1047. [CrossRef]
- Gardhouse, S.; Sanchez-Migallon, Guzman, D.; Paul-Murphy, J.; Byrne, B.A.; Hawkins, M.G. Bacterial isolates and antimicrobial susceptibilities from odontogenic abscesses in rabbits: 48 cases. Vet. Rec. 2017, 181, 538. [CrossRef]
- Crăciun, S.; Novac, C.Ş.; Fiţ, N.I.; Bouari, C.M.; Bel, L.V.; Nadăş, G.C. Bacterial Diversity in Pet Rabbits: Implications for Public Health, Zoonotic Risks, and Antimicrobial Resistance. Microorganisms 2025, 13, 653. [CrossRef]
- Silva, E.; Gaivão, M.; Leitão, S.; Jost, B.H.; Carneiro, C.; Vilela, C.L.; Lopes, da Costa L.; Mateus, L. Genomic characterization of Arcanobacterium pyogenes isolates recovered from the uterus of dairy cows with normal puerperium or clinical metritis. Vet. Microbiol. 2008, 132, 111-118. [CrossRef]
- Rzewuska, M.; Kwiecień, E.; Chrobak-Chmiel, D.; Kizerwetter-Świda, M.; Stefańska, I.; Gieryńska, M. Pathogenicity and Virulence of Trueperella pyogenes: A Review. Int. J. Mol. Sci. 2019, 20, 2737. [CrossRef]
- Kwiecień, E.; Stefańska, I.; Kizerwetter-Świda, M.; Chrobak-Chmiel, D.; Didkowska, A.; Bielecki, W.; Olech, W.; Krzysztof Anusz, K.; Rzewuska, M. Prevalence and Genetic Diversity of Trueperella pyogenes Isolated from Infections in European Bison (Bison bonasus). Animals (Basel) 2022, 12, 1825. [CrossRef]
- Kwiecień, E.; Stefańska, I.; Kizerwetter-Świda, M.; Chrobak-Chmiel, D.; Czopowicz, M.; Moroz-Fik, A.; Mickiewicz, M.; Biernacka, K.; Bagnicka, E.; Kaba, J.; Rzewuska, M. Genetic diversity and virulence properties of caprine Trueperella pyogenes isolates. BMC Vet. Res. 2024, 20, 395. [CrossRef]
- Nagib, S.; Glaeser, S.P.; Eisenberg; T.; Sammra, O.; Lämmler, C.; Kämpfer, P.; Schauerte, N.; Geiger, C.; Kaim, U.; Prenger-Berninghoff, E.; Becker, A.; Abdulmawjood, A. Fatal infection in three Grey Slender Lorises (Loris lydekkerianus nordicus) caused by clonally related Trueperella pyogenes. BMC Vet. Res. 2017, 13, 273. [CrossRef]
- Ahmed, M.F.E.; Alssahen, M.; Lämmler, C.; Eisenberg, T.; Plötz, M.; Abdulmawjood, A. Studies on Trueperella pyogenes isolated from an okapi (Okapia johnstoni) and a royal python (Python regius). BMC Vet. Res. 2020, 16, 292. https://doi:10.1186/s12917-020-02508-y.
- CLSI, 2017. Methods for Antimicrobial Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria Isolated From Animals. 1st ed. CLSI supplement VET06. Clinical and Laboratory Standards Institute; 2017, Wayne, PA.
- CLSI, 2024. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals, 6th ed. CLSI standard VET01. Clinical and Laboratory Standards Institute, 2024, Wayne PA.
- Alexeeva, I.; Elliott, E.J.; Rollins, S.; Gasparich, G.E.; Lazar, J.; Rohwer, R.G. Absence of Spiroplasma or other bacterial 16s rRNA genes in brain tissue of hamsters with scrapie. J. Clin. Microbiol. 2006, 44, 91-97. [CrossRef]
- Kumar, S.; Stecher, G.; Suleski, M.; Sanderford, M.; Sharm,. S.; Tamura, K. MEGA12: Molecular Evolutionary Genetic Analysis version 12 for adaptive and green computing. Mol. Biol. Evol. 2024. [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406-425. [CrossRef]
- Felsenstein, J. Confidence Limits in Phylogenies: An Approach Using the Bootstrap. Evolution 1985, 39, 783-791. [CrossRef]
- Tamura, K.; Nei, M.; Kumar, S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. USA 2004, 101, 30, 11030-11035. [CrossRef]
- Shahbazfar, A.A.; Kolahian, S.; Mohammadpour, H.; Helan, J. Multi abscessation with multinodular abscesses in a New Zealand white rabbit (Oryctolagus cuniculus) following Arcanobacterium pyogenes infection. Revue. Méd. Vét. 2013;164, 23-26.
- Fernández, M.; Garcias, B.; Duran, I.; Molina-López, R.A.; Darwich ,L. Current Situation of Bacterial Infections and Antimicrobial Resistance Profiles in Pet Rabbits in Spain. Vet. Sci. 2023, 10, 352. [CrossRef]
- Hijazin, M.; Ulbegi-Mohyla, H.; Alber, J.; Lämmler, C.; Hassan, A.A.; Abdulmawjood, A.; Prenger-Berninghoff, E.; Weiss, R.; Zschöck, M. Molecular identification and further characterization of Arcanobacterium pyogenes isolated from bovine mastitis and from various other origins. J. Dairy Sci. 2011, 94, 1813-9. [CrossRef]
- Planas, J.; Ester, Pintado, E.; Verdés, J.; Lourdes, Abarca, M.; Martorell, J. Rabbit with polyuria and polydipsia. 2020. J. Exot. Pet. Med. 2020, 35, 4-8. [CrossRef]
- Minich, D.J.; Marrow, J.C.; Sadar, M.J.; Borsdorf, M.C. High incidence of complications following intraoral extractions and treatment of periapical infections in the management of domestic rabbit (Oryctolagus cuniculus) dental disease (51 cases). J. Am. Vet. Med. Assoc. 2024, 263, 193-198. [CrossRef]
- Rogovskyy, A.S.; Lawhon, S.; Kuczmanski, K.; Gillis, D.C.; Wu, J.; Hurley, H.; Rogovska, Y.V.; Konganti, K.; Yang, C.Y.; Duncan, K. Phenotypic and genotypic characteristics of Trueperella pyogenes isolated from ruminants. J. Vet. Diagn. Invest. 2018, 30, 348-353. https://doi:10.1177/1040638718762479.
- Magossi, G.; Gzyl, K.E.; Holman, D.B.; Nagaraja, T.G.; Amachawadi, R.; Amat, S. Genomic and metabolic characterization of Trueperella pyogenes isolated from domestic and wild animals. Appl. Environ. Microbiol. 2025, 91, e0172524. [CrossRef]
- Flenghi, L.; Mazouffre, M.; Le, Loc'h, A.; Le, Loc'h, G.; Bulliot, C. Normal bacterial flora of the oral cavity in healthy pet rabbits (Oryctolagus cuniculus). Vet. Med. Sci. 2023, 9, 1621-1626. [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




