Submitted:
21 April 2025
Posted:
22 April 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Ethical Compliance
2.2. Virus Strains and Preparation
2.3. Infection in Mice
2.4. Infection and Transmission in Ferrets
2.5. Hemagglutination Inhibition (HI) Test
2.6. Histopathological and Immunohistochemical (IHC) Studies
2.7. Cytokine/Chemokine Quantification
2.8. Whole Genomic Sequencing of the Virus
2.9. Statistical Analysis
3. Results
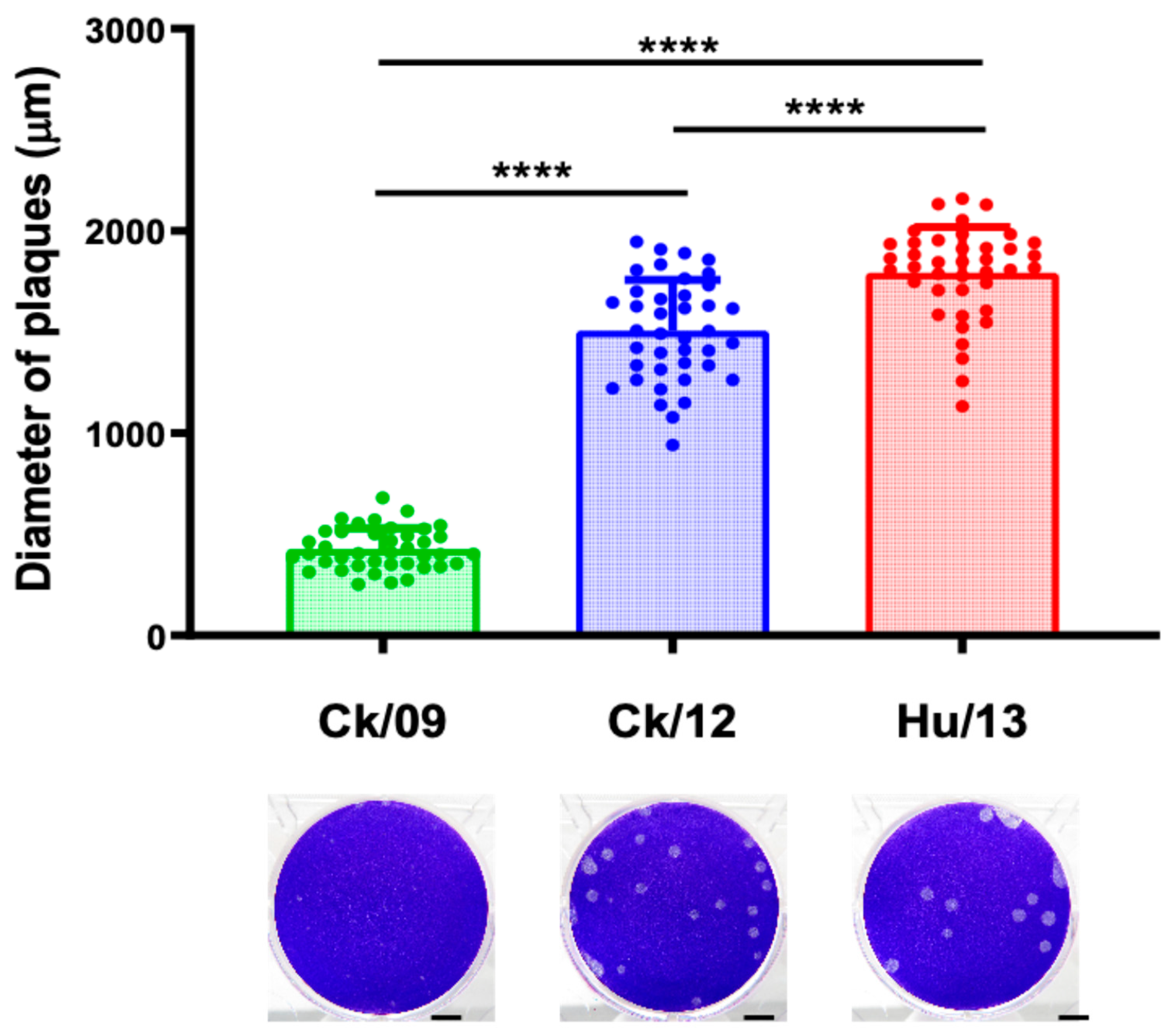
3.1. Viral Replication in MDCK Cells
3.2. Viral Replication and Pathogenicity in BALB/c Mice
3.2.1. Clinical Outcomes and Survival
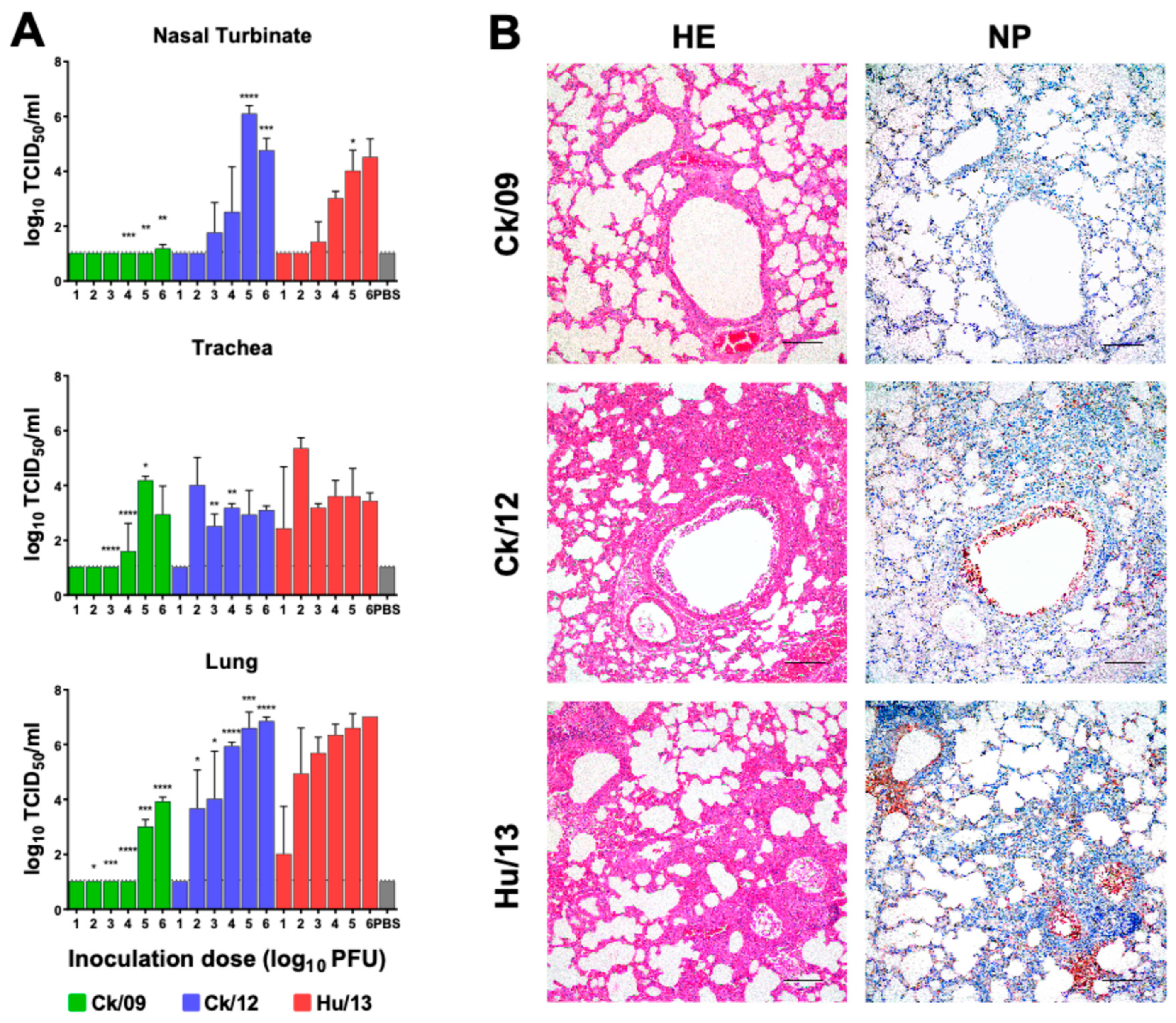
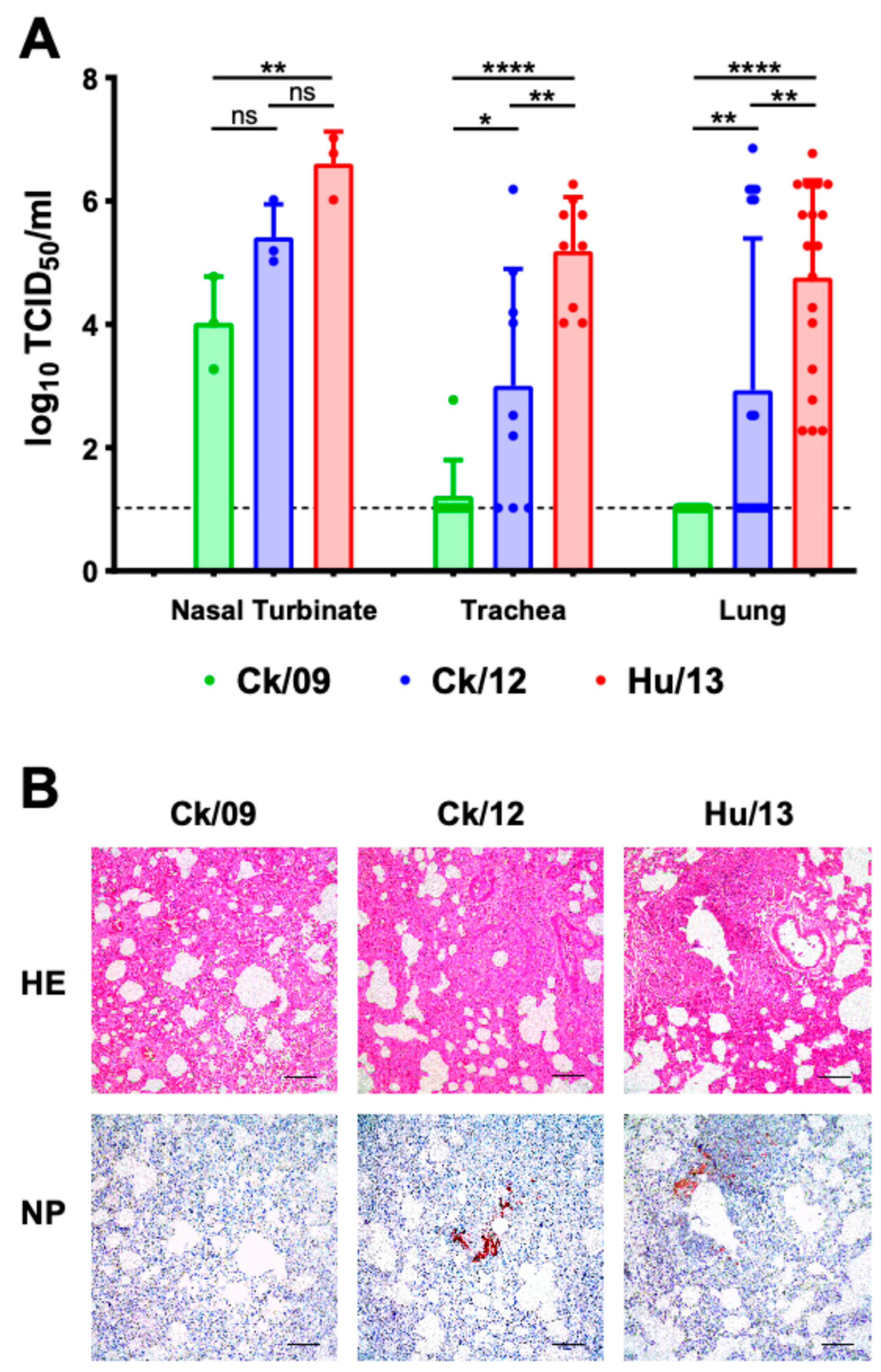
3.2.2. Viral Loads in Respiratory Tissues
3.2.3. Histopathology and Immune Responses
3.2.4. Seroconversion and Infectivity Parameters
3.3. Viral Infection and Transmission Dynamics in Ferrets
3.3.1. Clinical Observations
3.3.2. Viral Shedding and Transmission
3.3.3. Pathological Changes in Ferrets
3.3.4. Seroconversion
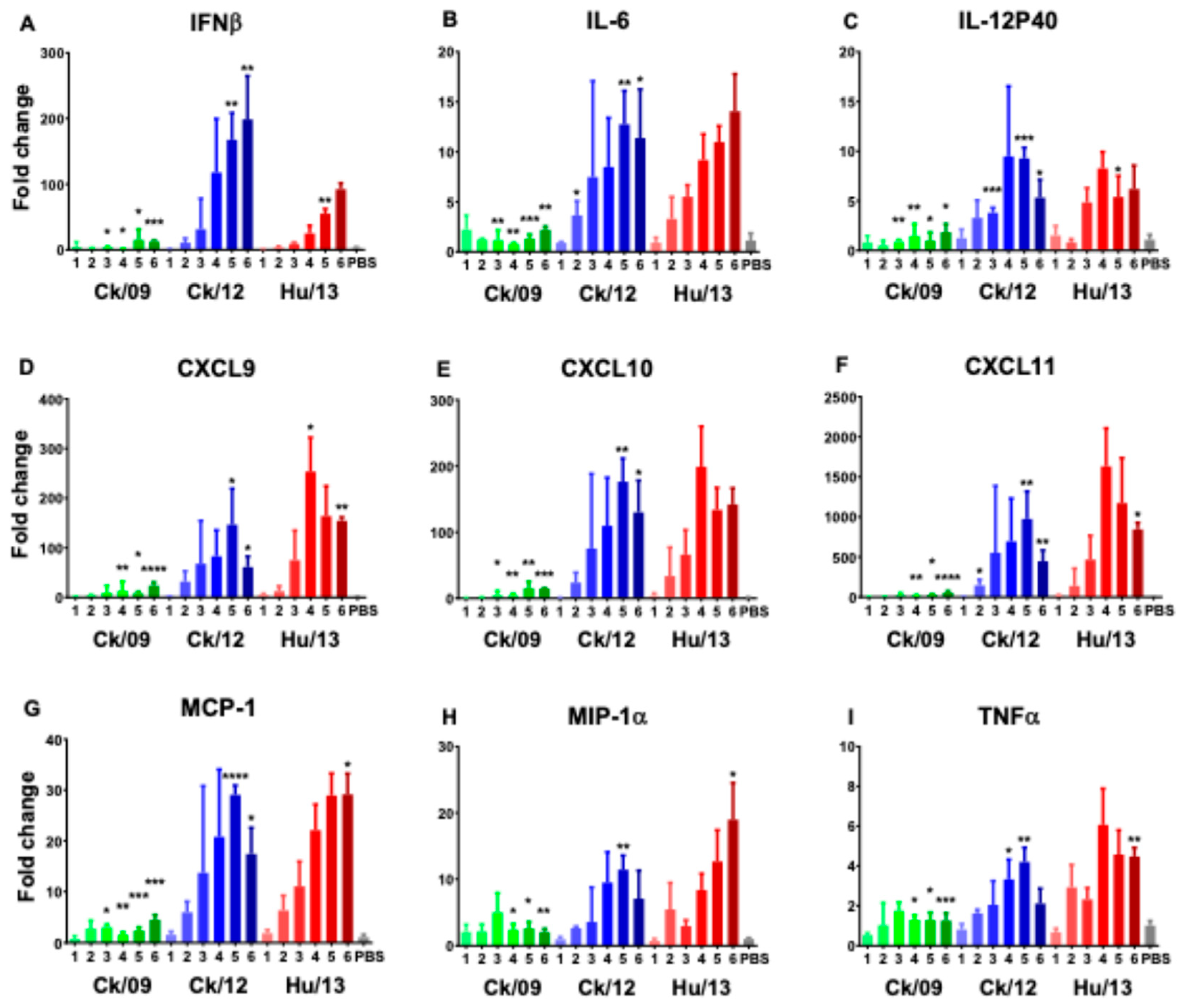
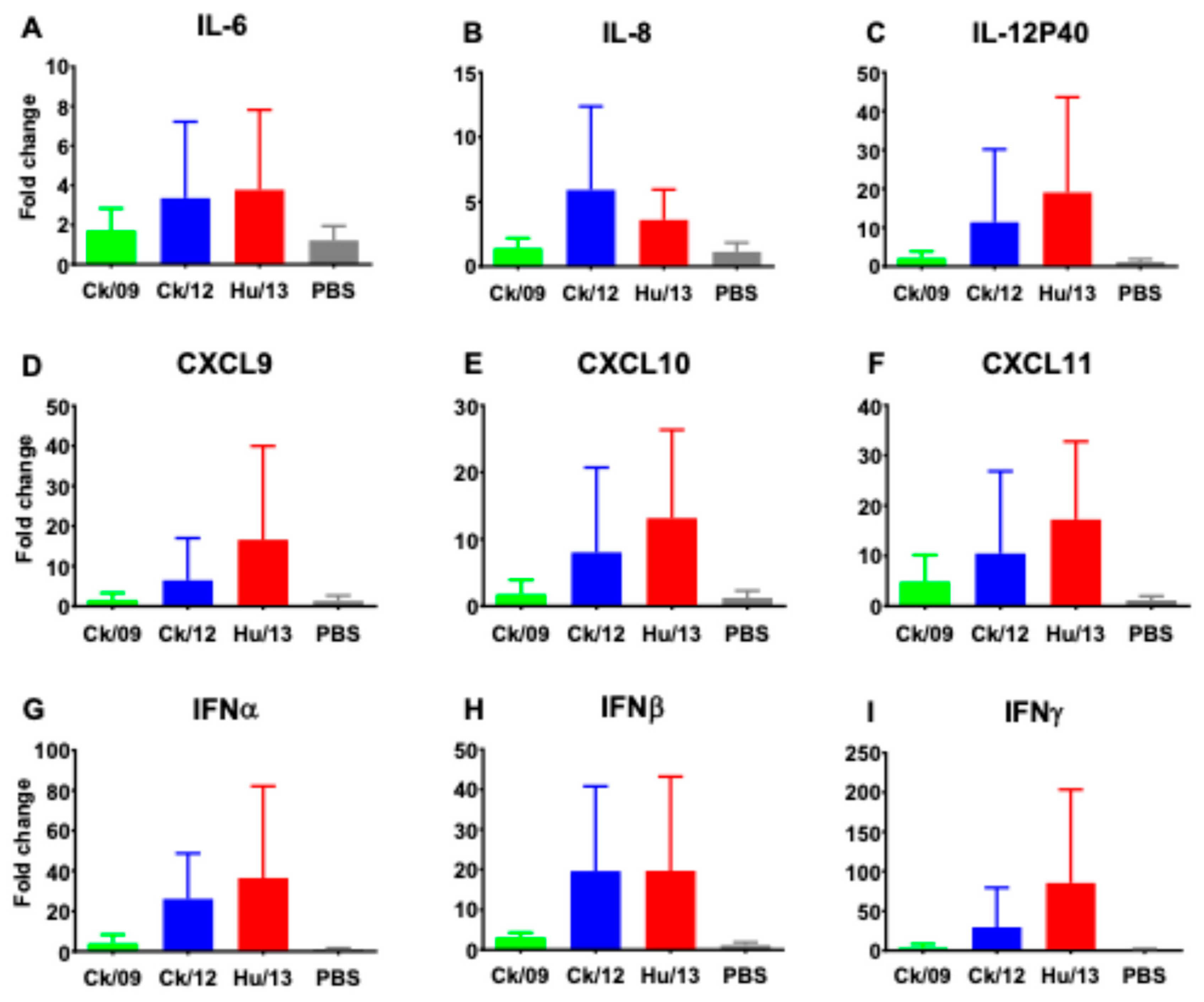
3.3.5. Proinflammatory Responses
3.3.6. Viral Mutations and Adaptation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AIV | avian influenza virus |
| Hu/13 | A/Taiwan/02/2013 |
| Ck/09 | A/Chicken/Taiwan/CF19/2009 |
| Ck/12 | A/Chicken/Taiwan/2267/2012 |
| CULATR | Committee on the Use of Live Animals in Teaching and Learning |
| BSL-3 | Biosafety Level 3 |
| MDCK | Madin-Darby canine kidney |
| TCID50 | 50% tissue culture infectious dose |
| EID50 | 50% egg infectious dose |
| PBS | phosphate buffered saline |
| PFU | plaque-forming unit |
| MWCO | molecular weight cut-off |
| dpi | day(s) post-inoculation |
| MLD50 | Mouse median lethal dose |
| MID50 | 50% mouse infectious dose |
| IN | intranasally inoculated |
| DC | direct physical-contact |
| AC | airborne-contact |
| dpc | day(s) post-contact |
| HI | hemagglutination inhibition |
| IHC | immunohistochemical |
| HE | hematoxylin and eosin |
| NP | nucleoprotein |
| PCR | polymerase chain reaction |
| SD | standard deviation |
| ANOVA | analysis of variance |
| AUC | area under the curve |
| RIG-I | retinoic acid-inducible gene-I |
| ISGs | antiviral interferon-stimulated genes |
| CDC | Centers for Disease Control and Prevention |
| WHO | World Health Organization |
References
- CDC. Reported Human Infections with Avian Influenza A Viruses. Available online: https://www.cdc.gov/bird-flu/php/avian-flu-summary/reported-human-infections.html (accessed on 16 April, 2025).
- Alexander, D.J. An overview of the epidemiology of avian influenza. Vaccine 2007, 25, 5637–5644. [Google Scholar] [CrossRef] [PubMed]
- Lam, T.T.; Wang, J.; Shen, Y.; Zhou, B.; Duan, L.; Cheung, C.L.; Ma, C.; Lycett, S.J.; Leung, C.Y.; Chen, X.; et al. The genesis and source of the H7N9 influenza viruses causing human infections in China. Nature 2013, 502, 241–244. [Google Scholar] [CrossRef]
- Ma, C.; Lam, T.T.; Chai, Y.; Wang, J.; Fan, X.; Hong, W.; Zhang, Y.; Li, L.; Liu, Y.; Smith, D.K.; et al. Emergence and evolution of H10 subtype influenza viruses in poultry in China. J. Virol. 2015, 89, 3534–3541. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.; Li, Y.; Huang, S.; Wen, F. Global distribution, receptor binding, and cross-species transmission of H6 influenza viruses: risks and implications for humans. J Virol. 2023, 97, e01370–23. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.; Zhu, H.; Fan, X.; Wang, J.; Cheung, C.L.; Duan, L.; Hong, W.; Liu, Y.; Li, L.; Smith, D.K.; et al. Establishment and lineage replacement of H6 influenza viruses in domestic ducks in southern China. J. Virol. 2012, 86, 6075–6083. [Google Scholar] [CrossRef]
- Wang, G.; Deng, G.; Shi, J.; Luo, W.; Zhang, G.; Zhang, Q.; Liu, L.; Jiang, Y.; Li, C.; Sriwilaijaroen, N; et al. H6 influenza viruses pose a potential threat to human health. J. Virol. 2014, 88, 3953–3964. [Google Scholar] [CrossRef]
- Lee, M.S.; Chang, P.C.; Shien, J.H.; Cheng, M.C.; Chen, C.L.; Shieh, H.K. Genetic and pathogenic characterization of H6N1 avian influenza viruses isolated in Taiwan between 1972 and 2005. Avian Dis. 2006, 50, 561–571. [Google Scholar] [CrossRef]
- Wei, S.H.; Yang, J.R.; Wu, H.S.; Chang, M.C.; Lin, J.S.; Lin, C.Y.; Liu, Y.L.; Lo, Y.C.; Yang, C.H.; Chuang, J.H.; et al. Human infection with avian influenza A H6N1 virus: an epidemiological analysis. Lancet Respir Med. 2013, 1, 771–778. [Google Scholar] [CrossRef]
- Lin, H.T.; Wang, C.H.; Chueh, L.L.; Su, B.L.; Wang, L.C. Influenza A(H6N1) Virus in Dogs, Taiwan. Emerg Infect Dis. 2015, 21, 2154–2157. [Google Scholar] [CrossRef]
- Tsai, S.K.; Shih, C.H.; Chang, H.W.; Teng, K.H.; Hsu, W.E.; Lin, H.J.; Lin, H.Y.; Huang, C.H.; Chen, H.W.; Wang, L.C. Replication of a Dog-Origin H6N1 Influenza Virus in Cell Culture and Mice. Viruses 2020, 12, 704. [Google Scholar] [CrossRef]
- Lee, C.C.; Zhu, H.; Huang, P.Y.; Peng, L.; Chang, Y.C.; Yip, C.H.; Li, Y.T.; Cheung, C.L.; Compans, R.; Yang, C.; et al. Emergence and evolution of avian H5N2 influenza viruses in chickens in Taiwan. J Virol. 2014, 88, 5677–5686. [Google Scholar] [CrossRef] [PubMed]
- Ni, F.; Kondrashkina, E.; Wang, Q. Structural and Functional Studies of Influenza Virus A/H6 Hemagglutinin. PLoS One 2015, 10, e0134576. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Qi, J.; Bi, Y.; Zhang, W.; Wang, M.; Zhang, B.; Wang, M.; Liu, J.; Yan, J.; Shi, Y.; et al. Adaptation of avian influenza A (H6N1) virus from avian to human receptor-binding preference. EMBO J. 2015, 34, 1661–1673. [Google Scholar] [CrossRef]
- Yang, H.; Carney, P.J.; Chang, J.C.; Villanueva, J.M.; Stevens, J. Structure and receptor binding preferences of recombinant hemagglutinins from avian and human H6 and H10 influenza A virus subtypes. J Virol. 2015, 89, 4612–4623. [Google Scholar] [CrossRef]
- Tzarum, N.; de Vries, R.P.; Zhu, X.; Yu, W.; McBride, R.; Paulson, J.C.; Wilson, I.A. Structure and receptor binding of the hemagglutinin from a human H6N1 influenza virus. Cell Host Microbe 2015, 17, 369–376. [Google Scholar] [CrossRef]
- Yuan, J.; Zhang, L.; Kan, X.; Jiang, L.; Yang, J.; Guo, Z.; Ren, Q. Origin and molecular characteristics of a novel 2013 avian influenza A(H6N1) virus causing human infection in Taiwan. Clin Infect Dis. 2013, 57, 1367–1368. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Shi, Y.; Wu, Y.; Liu, D.; Gao, G.F. Origin and molecular characterization of the human-infecting H6N1 influenza virus in Taiwan. Protein Cell 2013, 4, 846–853. [Google Scholar] [CrossRef]
- WHO Global Influenza Surveillance Network. Manual for the laboratory diagnosis and virological surveillance of influenza. Available online: https://www.who.int/publications/i/item/manual-for-the-laboratory-diagnosis-and-virological-surveillance-of-influenza (accessed on 27 February, 2025).
- Reed, L. J.; Muench, H. A simple method of estimating fifty percent endpoints. Am J Hyg. 1938, 27, 493–497. [Google Scholar]
- Li, L.; Chen, R.; Yan, Z.; Cai, Q.; Guan, Y.; Zhu, H. Experimental infection of rats with influenza A viruses: Implications for murine rodents in influenza A virus ecology. Viruses 2025, 17, 495. [Google Scholar] [CrossRef]
- Chen, P.; Jin, Z.; Peng, L.; Zheng, Z.; Cheung, Y.M.; Guan, J.; Chen, L.; Huang, Y.; Fan, X.; Zhang, Z.; et al. Characterization of an Emergent Chicken H3N8 Influenza Virus in Southern China: a Potential Threat to Public Health. J Virol. 2023, 97, e0043423. [Google Scholar] [CrossRef]
- Giulietti, A.; Overbergh, L.; Valckx, D.; Decallonne, B.; Bouillon, R.; Mathieu, C. An overview of real-time quantitative PCR: applications to quantify cytokine gene expression. Methods 2001, 25, 386–401. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Friggeri, A.; Yang, Y.; Park, Y.J.; Tsuruta, Y.; Abraham, E. miR-147, a microRNA that is induced upon Toll-like receptor stimulation, regulates murine macrophage inflammatory responses. Proc Natl Acad Sci U S A. 2009, 106, 15819–15824. [Google Scholar] [CrossRef]
- Martinez-Gil, L.; Goff, P.H.; Hai, R.; Garcia-Sastre, A.; Shaw, M.L.; Palese, P. A Sendai virus-derived RNA agonist of RIG-I as a virus vaccine adjuvant. J Virol. 2013, 87, 1290–1300. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Ma, X. Interferon regulatory factor 8 regulates RANTES gene transcription in cooperation with interferon regulatory factor-1, NF-kappaB, and PU.1. J Biol Chem. 2006, 281, 19188–19195. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.; Kemper, A.; McCarthy, K.D.; Pytel, P.; Wang, J.P.; Campbell, I.L.; Utset, M.F.; Popko, B. Interferon-gamma induced medulloblastoma in the developing cerebellum. J Neurosci. 2004, 24, 10074–10083. [Google Scholar] [CrossRef]
- Arima, Y.; Harada, M.; Kamimura, D.; Park, J.H.; Kawano, F.; Yull, F.E.; Kawamoto, T.; Iwakura, Y.; Betz, U.A.; Marquez, G.; et al. Regional neural activation defines a gateway for autoreactive T cells to cross the blood-brain barrier. Cell 2012, 148, 447–457. [Google Scholar] [CrossRef]
- Yoshida, S.; Yoshida, A.; Ishibashi, T.; Elner, S.G.; Elner, V.M. Role of MCP-1 and MIP-1alpha in retinal neovascularization during postischemic inflammation in a mouse model of retinal neovascularization. J Leukoc Biol. 2003, 73, 137–144. [Google Scholar] [CrossRef]
- Svitek, N.; von Messling, V. Early cytokine mRNA expression profiles predict Morbillivirus disease outcome in ferrets. Virology 2007, 362, 404–410. [Google Scholar] [CrossRef]
- Maines, T.R.; Belser, J.A.; Gustin, K.M.; van Hoeven, N.; Zeng, H.; Svitek, N.; von Messling, V.; Katz, J.M.; Tumpey, T.M. Local innate immune responses and influenza virus transmission and virulence in ferrets. J Infect Dis. 2012, 205, 474–485. [Google Scholar] [CrossRef]
- Zhou, B.; Donnelly, M.E.; Scholes, D.T.; St George, K.; Hatta, M.; Kawaoka, Y.; Wentworth, D.E. Single-reaction genomic amplification accelerates sequencing and vaccine production for classical and Swine origin human influenza a viruses. J Virol. 2009, 83, 10309–10313. [Google Scholar] [CrossRef]
- Schmieder, R.; Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Belser, J.A.; Maines, T.R. Adaptation of H9N2 Influenza Viruses to Mammalian Hosts: A Review of Molecular Markers. Viruses. 2020, 12, 541. [Google Scholar] [CrossRef]
- Li, B.; Su, G.; Xiao, C.; Zhang, J.; Li, H.; Sun, N.; Lao, G.; Yu, Y.; Ren, X.; Qi, W.; et al. The PB2 co-adaptation of H10N8 avian influenza virus increases the pathogenicity to chickens and mice. Transbound Emerg Dis. 2022, 69, 1794–1803. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.; Cui, H.; Teng, Q.; Li, L.; Shi, Y.; Li, X.; Yang, J.; Liu, Q.; Deng, J.; Li, Z. Evolution and pathogenicity of H6 avian influenza viruses isolated from Southern China during 2011 to 2017 in mice and chickens. Sci Rep. 2020, 10, 20583. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, M.; Tian, J.; Bai, X.; Li, Y. Genetic characteristics and pathogenicity of novel reassortant H6 viruses isolated from wild birds in China. Vet Microbiol. 2021, 254, 108978. [Google Scholar] [CrossRef]
- Xu, X.; Chen, Q.; Tan, M.; Liu, J.; Li, X.; Yang, L.; Shu, Y.; Wang, D.; Zhu, W. Epidemiology, evolution, and biological characteristics of H6 avian influenza viruses in China. Emerg Microbes Infect. 2023, 12, 2151380. [Google Scholar] [CrossRef]
- Kuchipudi, S.V.; Nelli, R.; White, G.A.; Bain, M.; Chang, K.C.; Dunham, S. Differences in influenza virus receptors in chickens and ducks: Implications for interspecies transmission. J Mol Genet Med. 2009, 3, 143–151. [Google Scholar] [CrossRef]
- Webster, R.G.; Bean, W.J.; Gorman, O.T.; Chambers, T.M.; Kawaoka, Y. Evolution and ecology of influenza A viruses. Microbiol. Rev. 1992, 56, 152–179. [Google Scholar] [CrossRef]
- Barber, M.R.W.; Aldridge, J.R., Jr.; Webster, R.G.; Magor, K.E. Association of RIG-I with innate immunity of ducks to influenza. Proc Natl Acad Sci U S A. 2010, 107, 5913–5918. [Google Scholar] [CrossRef]
- Li, J.; Zu Dohna, H.; Cardona, C.J.; Miller, J.; Carpenter, T.E. Emergence and Genetic Variation of Neuraminidase Stalk Deletions in Avian Influenza Viruses. PLoS One. 2011, 6, e14722. [Google Scholar] [CrossRef]
- Yamaji, R.; Yamada, S.; Le, M.Q; Li, C.; Chen, H.; Qurnianingsi, E.; Nidom, C.A.; Ito, M.; Sakai-Tagawa, Y.; Kawaoka, Y. Identification of PB2 mutations responsible for the efficient replication of H5N1 influenza viruses in human lung epithelial cells. J Virol. 2015, 89, 3947–3956. [Google Scholar] [CrossRef] [PubMed]
- Cox, A.; Schmierer, J.; D’Angelo, J.; Smith, A.; Levenson, D.; Treanor, J.; Kim, B.; Dewhurst, S. A Mutated PB1 Residue 319 Synergizes with the PB2 N265S Mutation of the Live Attenuated Influenza Vaccine to Convey Temperature Sensitivity. Viruses 2020, 12, 1246. [Google Scholar] [CrossRef] [PubMed]
- Kong, H.; Ma, S.; Wang, J.; Gu, C.; Wang, Z.; Shi, J.; Deng, G.; Guan, Y.; Chen, H. Identification of Key Amino Acids in the PB2 and M1 Proteins of H7N9 Influenza Virus That Affect Its Transmission in Guinea Pigs. J Virol. 2019, 94, e01180–19. [Google Scholar] [CrossRef]
- Bussey, K.A.; Desmet, E.A.; Mattiacio, J.L.; Hamilton, A.; Bradel-Tretheway, B.; Bussey, H.E.; Kim, B.; Dewhurst, S.; Takimoto, T. PA residues in the 2009 H1N1 pandemic influenza virus enhance avian influenza virus polymerase activity in mammalian cells. J Virol. 2011, 85, 7020–8. [Google Scholar] [CrossRef] [PubMed]
- Song, M.S.; Pascua, P.N.Q.; Lee, J.H.; Baek, Y.H.; Lee, O.J.; Kim, C.J.; Kim, H.; Webby, R.J.; Webster, R.G.; Choi, Y.K. The polymerase acidic protein gene of influenza a virus contributes to pathogenicity in a mouse model. J Virol. 2009, 83, 12325–12335. [Google Scholar] [CrossRef]
- Taft, A.S.; Ozawa, M.; Fitch, A.; Depasse, J.V.; Halfmann, P.J.; Hill-Batorski, L.; Hatta, M.; Friedrich, T.C.; Lopes, T.J.S.; Maher, E.A.; et al. Identification of mammalian-adapting mutations in the polymerase complex of an avian H5N1 influenza virus. Nat Commun. 2015, 6, 7491. [Google Scholar] [CrossRef]
- Yu, Z.; Cheng, K.; Xin, Y.; Sun, W.; Li, X.; Huang, J.; Zhang, K.; Yang, S.; Wang, T.; Zheng, X.; et al. Multiple amino acid substitutions involved in the adaptation of H6N1 avian influenza virus in mice. Vet Microbiol. 2014, 174, 316–321. [Google Scholar] [CrossRef]
- Kamal, R.P. , Alymova, I.V., York, I.A. Evolution and Virulence of Influenza A Virus Protein PB1-F2. Int J Mol Sci. 2017, 19, 96. [Google Scholar] [CrossRef]
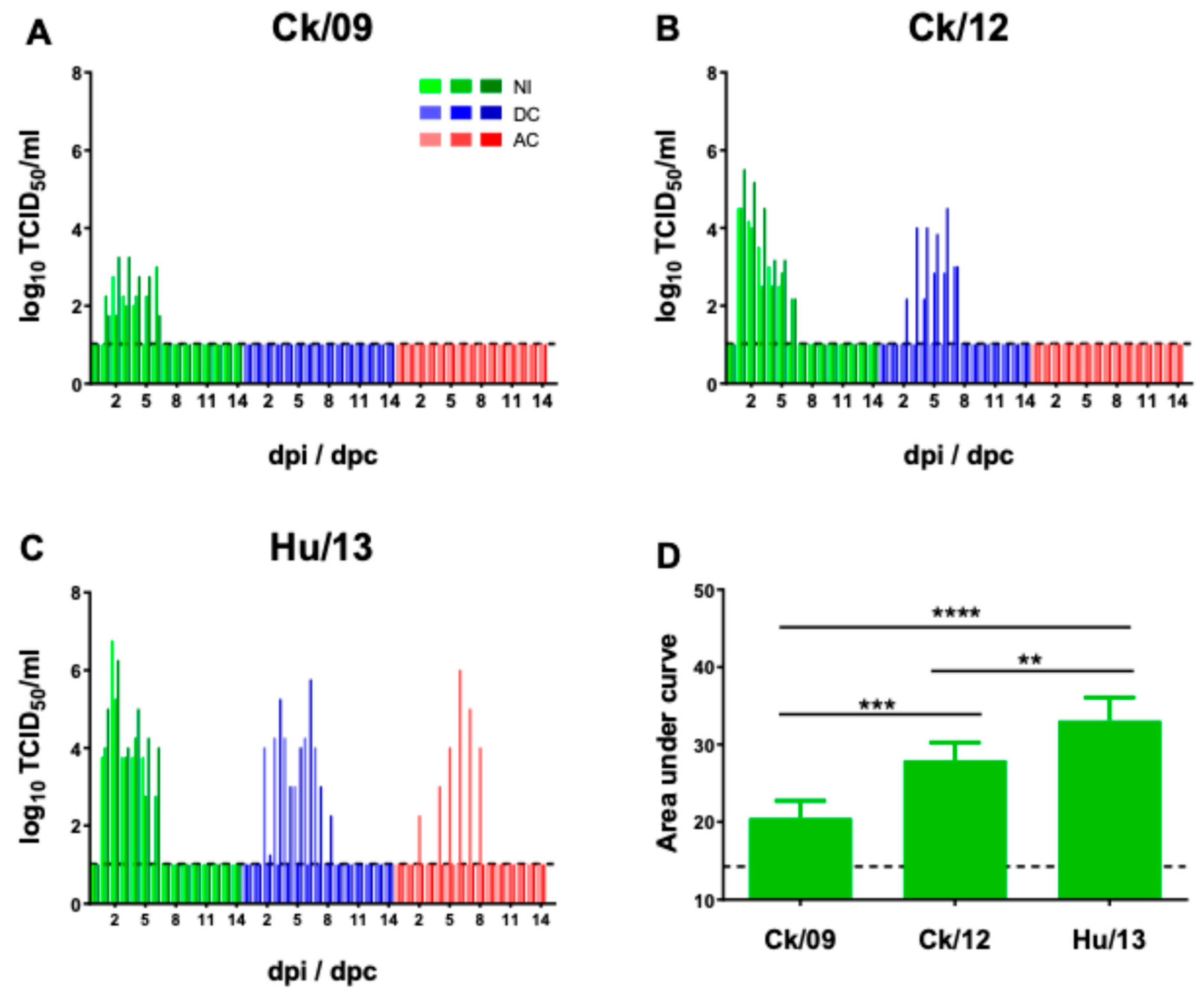
| Ferret Groups | Virus | Shedding of virus | Seroconversion (HI titers at 14 dpi/dpc) |
||
|---|---|---|---|---|---|
| Animal numbers | Peak titers (log10TCID50/mL) |
Duration (days) | |||
|
Intranasally inoculated (IN) |
Ck/09 | 3/3 | 2.8, 3.0, 3.3 | 3, 6, 6 | 320 ,640, 640 |
| Ck/12 | 3/3 | 4.5, 4.5, 5.5 | 5, 6, 6 | 640, 640, 1280 | |
| Hu/13 | 3/3 | 5.3, 6.3, 6.8 | 5, 6, 6 | 1280, 1280, 1280 | |
|
Direct physical contact (DC) |
Ck/09 | 0/3 | -, -, - | -, -, - | -, -, - |
| Ck/12 | 2/3 | 3.0, 4.5, - | 4, 6, - | 640, 640, - | |
| Hu/13 | 2/3 | 4.3, 5.8, - | 6, 7, - | 640, 1280, 1280 | |
|
Airborne contact (AC) |
Ck/09 | 0/3 | -, -, - | -, -, - | -, -, - |
| Ck/12 | 0/3 | -, -, - | -, -, - | -, -, - | |
| Hu/13 | 1/3 | 6.0, -, - | 7, -, - | 1280, 1280, - | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





