Key words: mouse CCR1, monoclonal antibody, epitope mapping, alanine scanning, flow cytometry
1. Introduction
Chemokine receptors belong to class A seven transmembrane (7TM) receptors and play an essential role in guiding leukocyte trafficking in immune surveillance and inflammatory response [
1]. The cognate chemokines are named according to the sequence of the first two cysteines (CC, CXC, XC, or CX3C motif). The CC ligands (CCL1 to CCL28) are recognized by C-C chemokine receptor type 1 (CCR1) to CCR10 [
2]. Upon ligand binding, chemokine receptors typically activate G protein pathways and recruit β-arrestins [
3,
4].
CCR1 mediates inflammatory responses and plays an essential role in the development of autoimmune diseases [
1,
5]. It has been considered an attractive drug target for treating allergic and autoimmune diseases [
6]. Among the chemokine receptors, CCR1 possesses ligand promiscuity, which allows it to recognize at least nine human CC chemokines, including CCL3, CCL5, CCL7, CCL8, CCL13–16, and CCL23 [
2,
7,
8].
For the development of therapeutic agents in the chemokine system, the structural understanding of the chemokine receptor activation is essential. Among the CCR family members, CCR2 and CCR5 have been characterized in both inactive and active states [
9,
10,
11,
12], while active-state of CCR8 and CCR6 and inactive-state of CCR7 and CCR9 structures were also determined [
13,
14,
15,
16]. Furthermore, the CCL15-CCR1 complex showed a crucial sequence for ligand binding distinct from many other chemokine–receptor complexes, providing new insights into the mode of chemokine recognition [
13]. Moreover, the structures of CCR8 in complex with either the endogenous ligand CCL1 or the antagonistic monoclonal antibody (mAb) were solved, which provides the specific activation mechanism by CCL1 and inhibition by mAb [
17]. Therefore, anti-chemokine receptor mAbs with defined epitopes are useful for the analysis of specific structures.
An anti-mouse CCR1 (mCCR1) mAb (clone S15040E) has been used in various in vivo studies to identify mCCR1-positive cells using flow cytometry [
18,
19,
20]. However, the binding epitope has not been determined. To determine the epitopes of 7TM proteins, we have faced difficulty using conventional methods such as enzyme-linked immunosorbent assay. This study investigated the binding epitope of S15040E using flow cytometry-based approaches.
2. Materials and Methods
2.1 Plasmid Construction
pCAG-Ble-mCCR1 and pCAG-Ble-mouse CCR5 (mCCR5) were generated as previously described [
21,
22]. Chimeric mutants including mCCR5 (mCCR1p2–34), mCCR5 (mCCR1p92–107), mCCR5 (mCCR1p172–197), and mCCR5 (mCCR1p265–281) were produced with a PA16 tag at their N-terminus using the HotStar HiFidelity polymerase kit (Qiagen Inc., Hilden, Germany). Alanine (or glycine) substitutions in mCCR1 were conducted using QuikChange Lightning Site-Directed Mutagenesis Kits (Agilent Technologies Inc., Santa Clara, CA, USA).
2.2. Cell Lines
Chinese hamster ovary (CHO)-K1 cell was obtained from the America Type Culture Collection (ATCC, Manassas, VA, USA). The chimeric and the point mutant plasmids were transfected into CHO-K1 cells using the Neon Transfection System (Thermo Fisher Scientific Inc., Waltham, MA, USA).
2.3. Antibodies
An anti-mCCR1 mAb (clone S15040E) was purchased from BioLegend (San Diego, CA, USA). NZ-1 (an anti-PA16 tag mAb) was described previously [
23].
2.4. Flow Cytometry
Cells were harvested after brief exposure to 0.25% trypsin/1 mM ethylenediaminetetraacetic acid (Nacalai Tesque, Inc.). After washing with 0.1% bovine serum albumin in phosphate-buffered saline, cells were treated with S15040E (1 μg/mL) or NZ-1 (1 μg/mL) for 30 min at 4℃ and subsequently with Alexa Fluor 488-conjugated anti-rat IgG (1:2000; Cell Signaling Technology, Inc., Danvers, MA, USA). Fluorescence data were obtained using the SA3800 Cell Analyzer (Sony Corp., Tokyo, Japan).
3. Results
3.1. Determination of the Epitope of an Anti-mCCR1 mAb, S15040E, by Flow Cytometry Using Chimeric Proteins
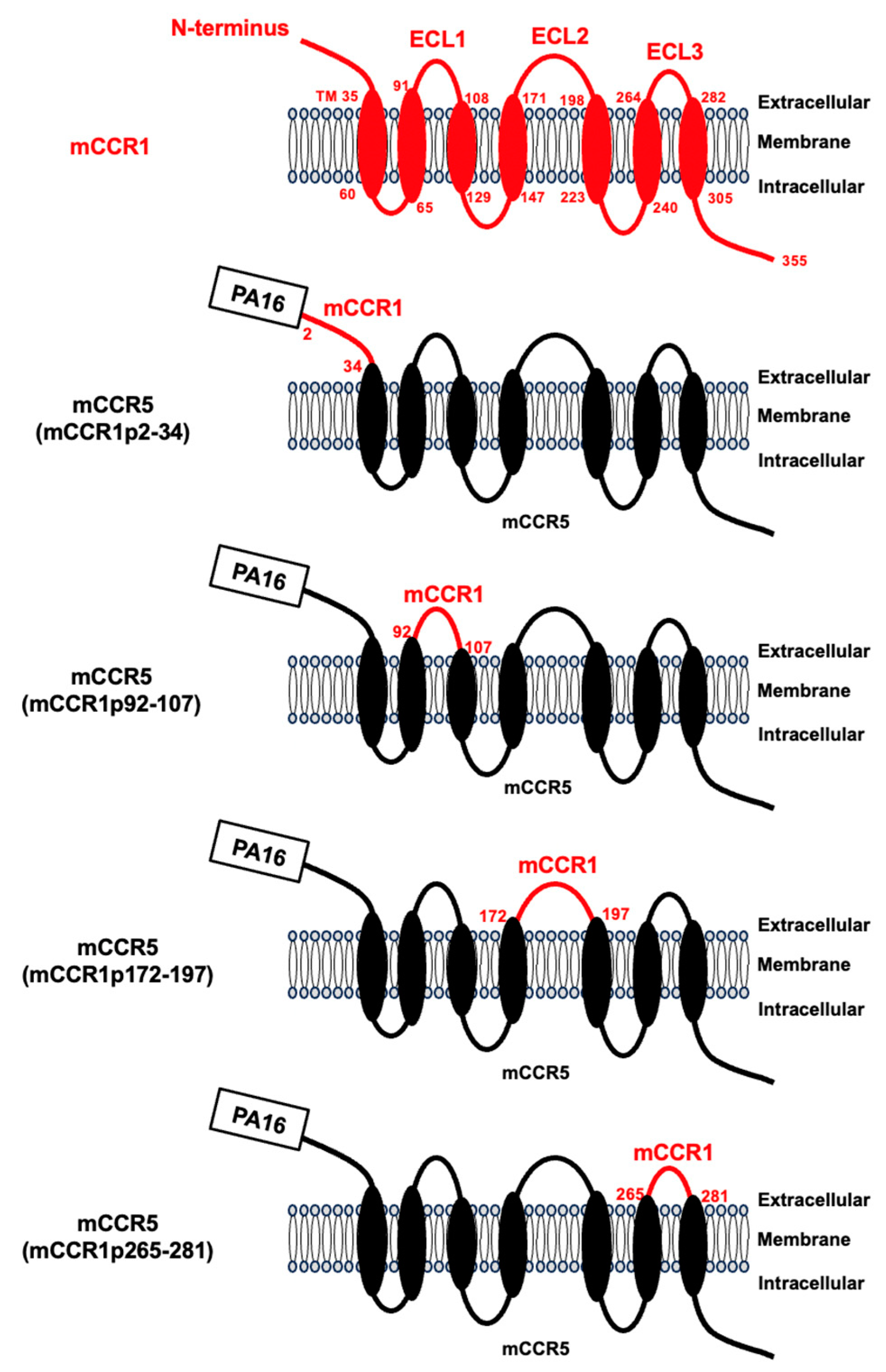
An anti-mCCR1 mAb (clone S15040E) is applicable for flow cytometry. To investigate the binding epitope of S15040E, we focused on four extracellular regions of mCCR1, including the N-terminal region (aa 2–34), extracellular loop 1 (ECL1; a 92–107), ECL2 (aa 172–197), and ECL3 (aa 265–281). The four extracellular regions of mCCR1 were substituted into the corresponding regions of mCCR5, which possesses a similar structure to mCCR1. As shown in
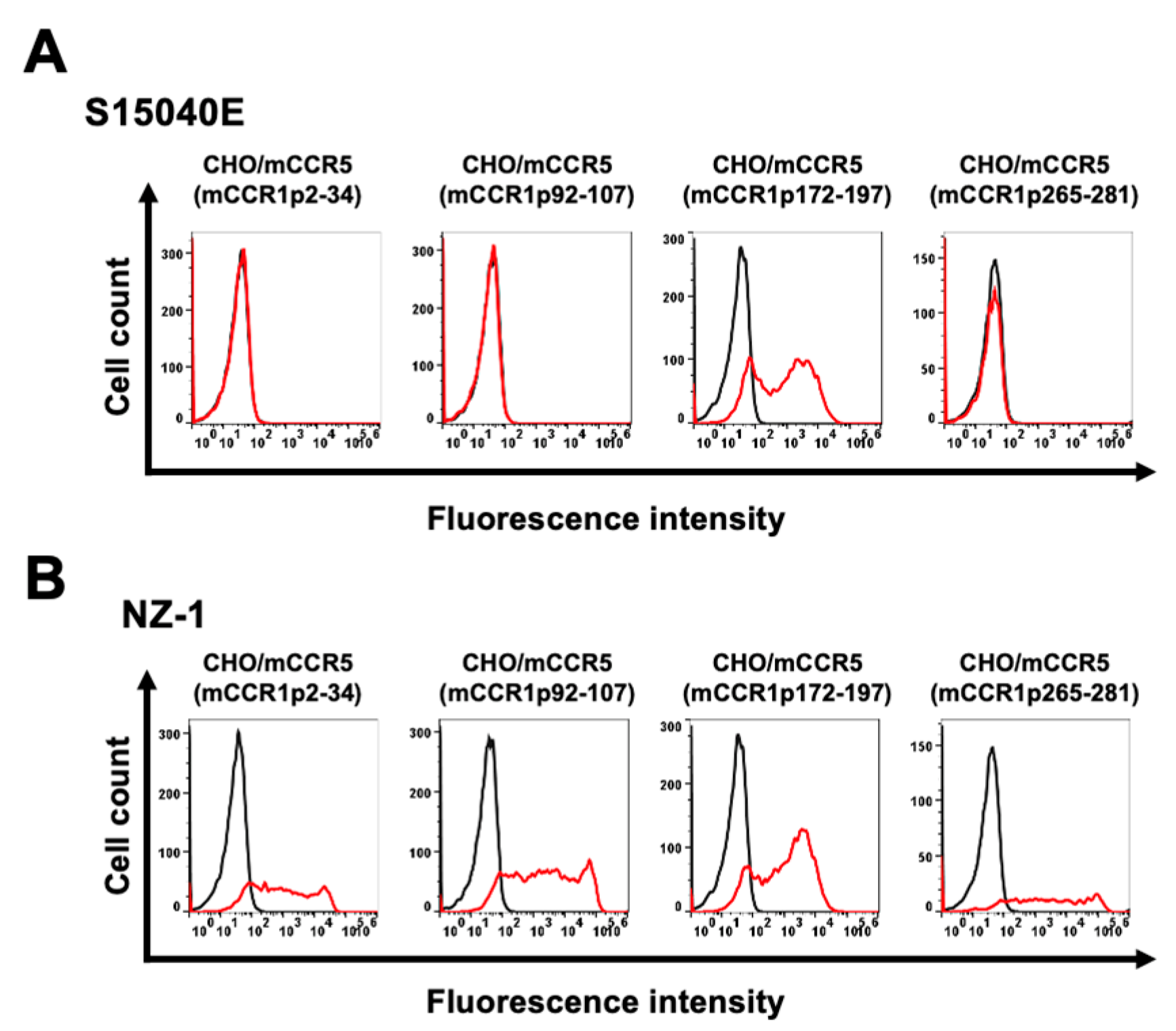
Figure 1, plasmids encoding mCCR5 (mCCR1p2–34), mCCR5 (mCCR1p92–107), mCCR5 (mCCR1p172–197), and mCCR5 (mCCR1p265–281) were generated. The chimeric proteins were transiently expressed on CHO-K1 cells, and the reactivities to S15040E were analyzed using flow cytometry (
Figure 2A). S15040E reacted with mCCR5 (mCCR1p172–197), but not with other chimeric proteins (
Figure 2A). The cell surface expression of each mutant was confirmed by an anti-PA16 tag mAb, NZ-1 (
Figure 2B). These results indicated that S15040E recognizes the ECL2 of mCCR1.
3.2. Determination of the S15040E Epitope by Flow Cytometry Using Alanine Scanning
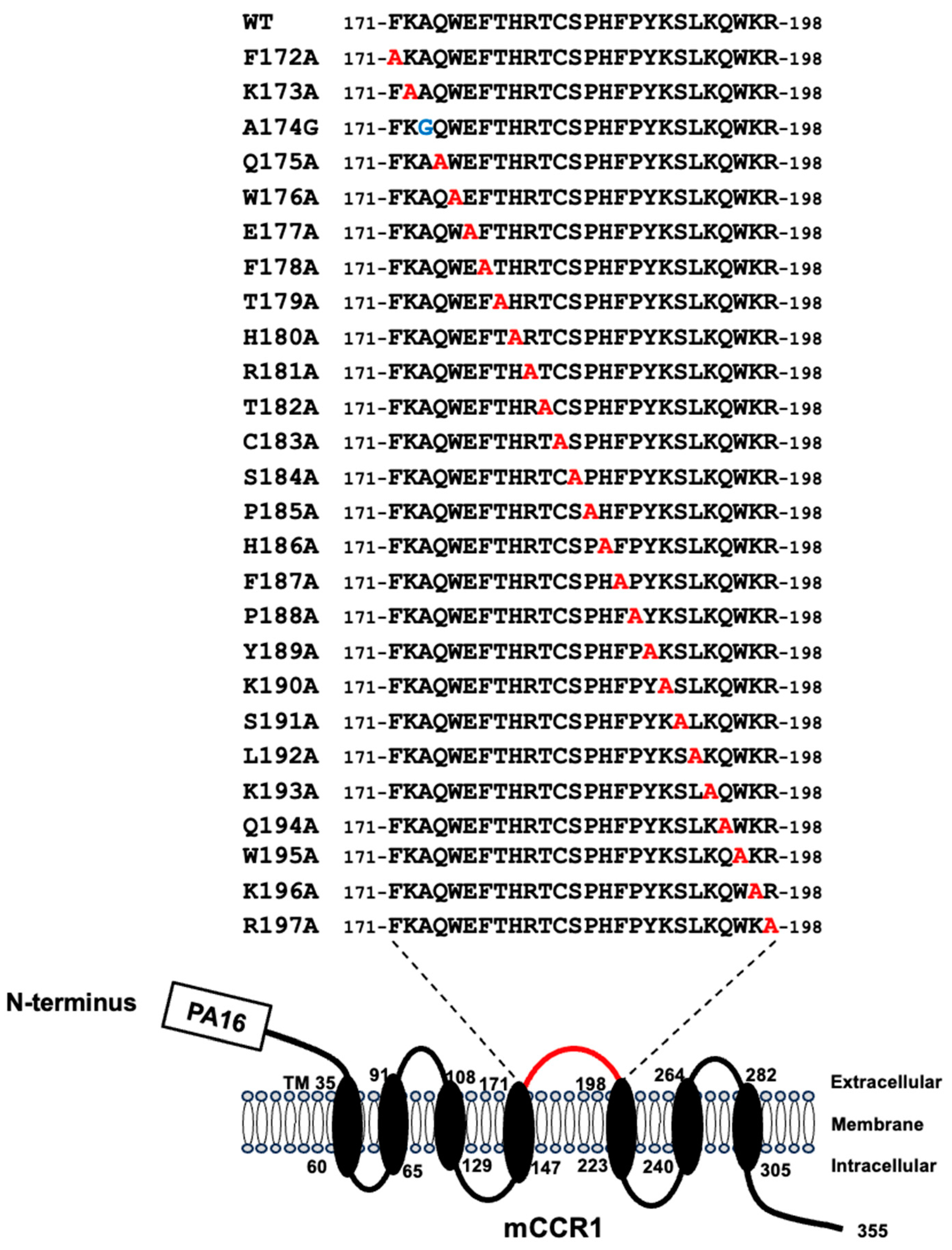
Next, alanine scanning was conducted in the ECL2 of mCCR1. Twenty-six alanine (or glycine) substituted mutants of mCCR1 were constructed (
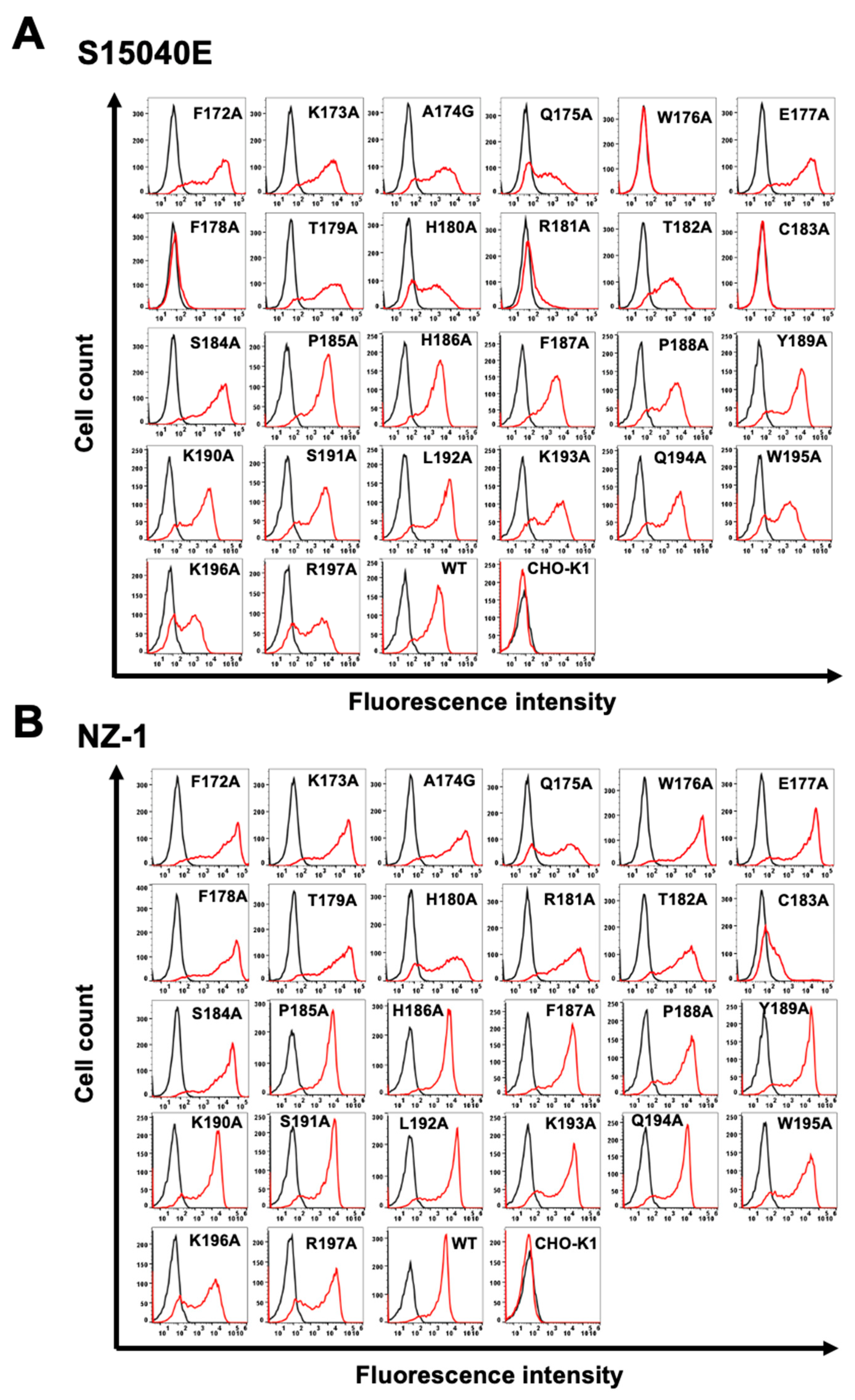
Figure 3), and the mutant proteins were transiently expressed on CHO-K1 cells. The reactivity against S15040E was assessed using flow cytometry. As shown in
Figure 4A, S15040E did not react with four mutants (W176A, F178A, R181A, and C183A). In contrast, S15040E reacted with the other twenty-two mutants. The cell surface expression of each mutant was confirmed by NZ-1 (
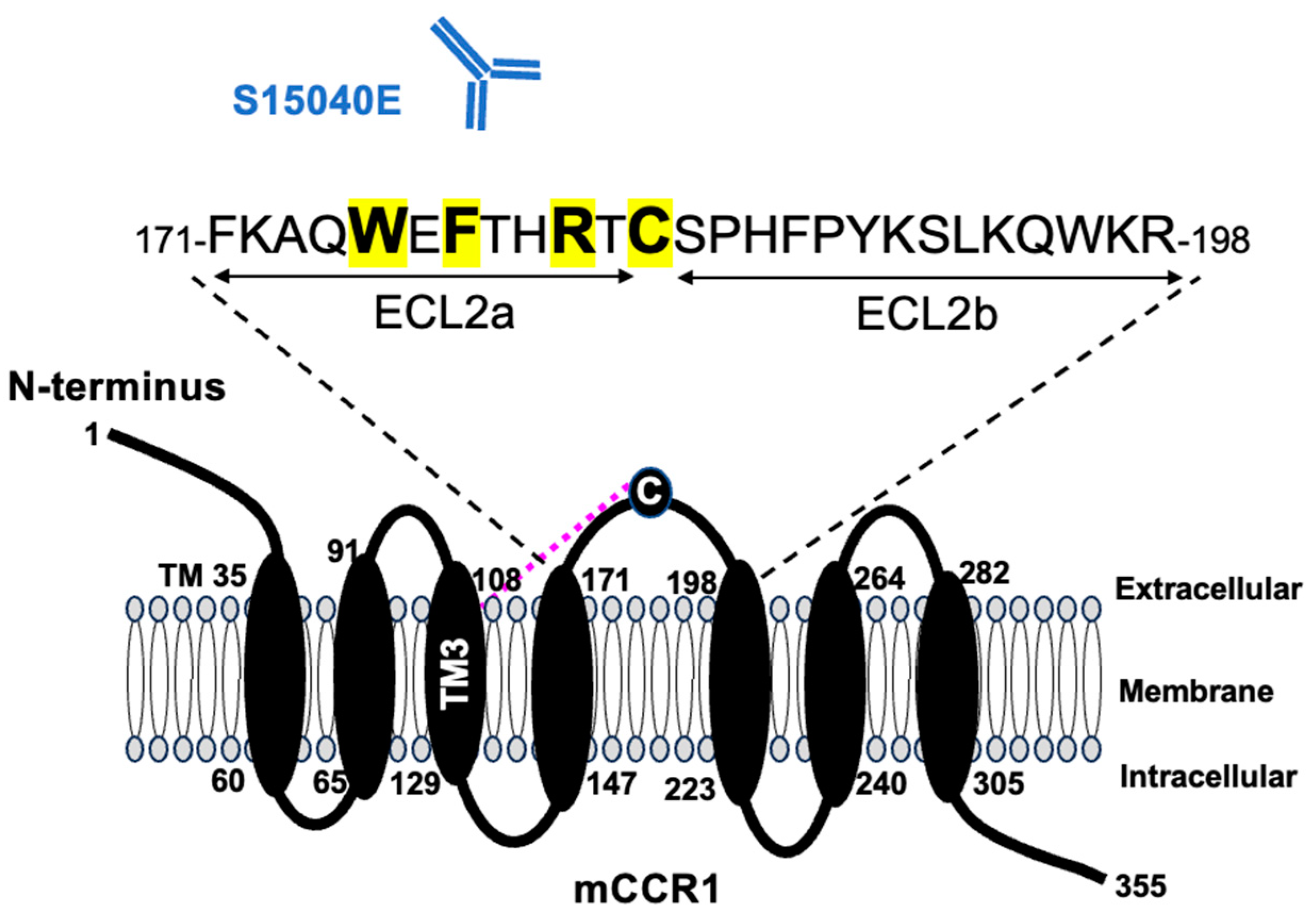
Figure 4B). These results showed that Trp176, Phe178, Arg181, and Cys183 of mCCR1 are important for S15040E binding.
4. Discussion
This study demonstrated the flow cytometry-based epitope mapping of an anti-mouse CCR1 mAb (S15040E) using the chimeric proteins (Figs. 1 and 2). Furthermore, we determined that the Trp176, Phe178, Arg181, and Cys183 in ECL2 are essential for the recognition by S15040E in alanine scanning (
Figure 3 and
Figure 4). We previously determined the epitope of an anti-mouse CCR8 mAb, C
8Mab-2 [
24]. Our strategy for epitope identification would contribute to the understanding of mAb-epitope interaction.
The 7TM receptors have conserved disulfide bridge between transmembrane helix 3 (TM3) and ECL2 [
25]. The Cys183 is well conserved and sole cysteine in ECL2, which forms a disulfide bridge with TM3. We previously confirmed that S15040E could not detect mCCR1 in western blotting [
21]. Therefore, S15040E recognizes the conformational epitope, which depends on the disulfide bridge between TM3 and ECL2.
Figure 5 summarizes the epitope of S15040E.
ECL2 is essential for interaction with chemokines and is the largest region covering the activation-associated receptor binding pocket. ECL2 is divided into two parts before and after the disulfide bridge (ECL2a and ECL2b, respectively). Both parts involve the chemokine signaling selectivity and pharmacological activity [
26,
27,
28,
29]. The Trp176, Phe178, and Arg181 are in the ECL2a of mCCR1. In human CCR1, the ECL2a is essential for the recognition of CCL15 [
13]. Therefore, S15040E may possess the neutralization activity to the ECL2a-bound ligands.
G protein-coupled receptors can transduce intracellular signaling through G proteins and β-arrestins. "Balanced" agonists or antagonists can activate or inhibit these signaling pathways. In contrast, specific pathways can be selectively triggered in a "biased" response. The biased responses can arise from biased ligands or biased receptors, all of which can drive preferential activation of either G protein- or β-arrestin-mediated pathways [
30]. CCR1 is known to be a biased receptor that can selectively activate G proteins or β-arrestin pathways by diverse CCL15 isoforms [
13,
31]. Further structural analysis of the S15040E-mCCR1 complex may provide new insights into the mechanism of biased response and the development of therapeutic drugs.
Author Contributions
Ayaka Okada: Investigation; Hiroyuki Suzuki: Writing – original draft; Tomohiro Tanaka: Investigation, Funding acquisition; Mika K. Kaneko: Conceptualization; Yukinari Kato: Conceptualization, Funding acquisition, Project administration, Writing – review and editing; All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported in part by Japan Agency for Medical Research and Development (AMED) under Grant Numbers: JP25am0521010 (to Y.K.), JP25ama121008 (to Y.K.), JP25ama221339 (to Y.K.), and JP25bm1123027 (to Y.K.), and by the Japan Society for the Promotion of Science (JSPS) Grants-in-Aid for Scientific Research (KAKENHI) grant nos. 24K18268 (to T.T.) and 25K10553 (to Y.K.).
Informed Consent Statement
Not applicable.
Data Availability Statement
All related data and methods are presented in this paper. Additional inquiries should be addressed to the corresponding authors.
Conflicts of Interest
The authors declare no conflict of interest involving this article.
References
- Griffith, J.W.; Sokol, C.L.; Luster, A.D. Chemokines and chemokine receptors: positioning cells for host defense and immunity. Annu Rev Immunol 2014, 32, 659–702. [Google Scholar] [CrossRef] [PubMed]
- Stone, M.J.; Hayward, J.A.; Huang, C.; Z, E.H.; Sanchez, J. Mechanisms of Regulation of the Chemokine-Receptor Network. Int J Mol Sci 2017, 18. [Google Scholar] [CrossRef]
- Defea, K. Beta-arrestins and heterotrimeric G-proteins: collaborators and competitors in signal transduction. Br J Pharmacol, S: (Suppl 1).
- Weis, W.I.; Kobilka, B.K. The Molecular Basis of G Protein-Coupled Receptor Activation. Annu Rev Biochem 2018, 87, 897–919. [Google Scholar] [CrossRef]
- Tian, Q.; Yan, Z.; Guo, Y.; Chen, Z.; Li, M. Inflammatory Role of CCR1 in the Central Nervous System. Neuroimmunomodulation 2024, 31, 173–182. [Google Scholar] [CrossRef]
- Scholten, D.J.; Canals, M.; Maussang, D.; et al. Pharmacological modulation of chemokine receptor function. Br J Pharmacol 2012, 165, 1617–1643. [Google Scholar] [CrossRef] [PubMed]
- Murphy, P.M.; Baggiolini, M.; Charo, I.F.; et al. International union of pharmacology. XXII. Nomenclature for chemokine receptors. Pharmacol Rev 2000, 52, 145–176. [Google Scholar] [CrossRef]
- Schall, T.J.; Proudfoot, A.E. Overcoming hurdles in developing successful drugs targeting chemokine receptors. Nat Rev Immunol 2011, 11, 355–363. [Google Scholar] [CrossRef]
- Shao, Z.; Tan, Y.; Shen, Q.; et al. Molecular insights into ligand recognition and activation of chemokine receptors CCR2 and CCR3. Cell Discov 2022, 8, 44. [Google Scholar] [CrossRef] [PubMed]
- Isaikina, P.; Tsai, C.J.; Dietz, N.; et al. Structural basis of the activation of the CC chemokine receptor 5 by a chemokine agonist. Sci Adv 2021, 7. [Google Scholar] [CrossRef]
- Zheng, Y.; Qin, L.; Zacarías, N.V.; et al. Structure of CC chemokine receptor 2 with orthosteric and allosteric antagonists. Nature 2016, 540, 458–461. [Google Scholar] [CrossRef]
- Tan, Q.; Zhu, Y.; Li, J.; et al. Structure of the CCR5 chemokine receptor-HIV entry inhibitor maraviroc complex. Science 2013, 341, 1387–1390. [Google Scholar] [CrossRef] [PubMed]
- Shao, Z.; Shen, Q.; Yao, B.; et al. Identification and mechanism of G protein-biased ligands for chemokine receptor CCR1. Nat Chem Biol 2022, 18, 264–271. [Google Scholar] [CrossRef]
- Wasilko, D.J.; Johnson, Z.L.; Ammirati, M.; et al. Structural basis for chemokine receptor CCR6 activation by the endogenous protein ligand CCL20. Nat Commun 2020, 11, 3031. [Google Scholar] [CrossRef] [PubMed]
- Jaeger, K.; Bruenle, S.; Weinert, T.; et al. Structural Basis for Allosteric Ligand Recognition in the Human CC Chemokine Receptor 7. Cell 2019, 178, 1222–1230.e1210. [Google Scholar] [CrossRef]
- Oswald, C.; Rappas, M.; Kean, J.; et al. Intracellular allosteric antagonism of the CCR9 receptor. Nature 2016, 540, 462–465. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Sun, Y.; Janezic, E.; et al. Structural basis of antibody inhibition and chemokine activation of the human CC chemokine receptor 8. Nat Commun 2023, 14, 7940. [Google Scholar] [CrossRef]
- Di Pilato, M.; Kfuri-Rubens, R.; Pruessmann, J.N.; et al. CXCR6 positions cytotoxic T cells to receive critical survival signals in the tumor microenvironment. Cell 2021, 184, 4512–4530.e4522. [Google Scholar] [CrossRef]
- Du, X.; Li, F.; Zhang, C.; et al. Eosinophil-derived chemokine (hCCL15/23, mCCL6) interacts with CCR1 to promote eosinophilic airway inflammation. Signal Transduct Target Ther 2021, 6, 91. [Google Scholar] [CrossRef]
- Niu, B.; Tian, T.; Wang, L.; et al. CCL9/CCR1 axis-driven chemotactic nanovesicles for attenuating metastasis of SMAD4-deficient colorectal cancer by trapping TGF-β. Acta Pharm Sin B 2024, 14, 3711–3729. [Google Scholar] [CrossRef]
- Ouchida, T.; Isoda, Y.; Nakamura, T.; et al. Establishment of a Novel Anti-Mouse CCR1 Monoclonal Antibody C(1)Mab-6. Monoclon Antib Immunodiagn Immunother 2024, 43, 67–74. [Google Scholar] [CrossRef]
- Suzuki, H.; Tanaka, T.; Li, G.; et al. Development of a Sensitive Anti-Mouse CCR5 Monoclonal Antibody for Flow Cytometry. Monoclon Antib Immunodiagn Immunother 2024, 43, 96–100. [Google Scholar] [CrossRef]
- Kato, Y.; Kaneko, M.K.; Kuno, A.; et al. Inhibition of tumor cell-induced platelet aggregation using a novel anti-podoplanin antibody reacting with its platelet-aggregation-stimulating domain. Biochem Biophys Res Commun 2006, 349, 1301–1307. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, H.; Suzuki, H.; Tanaka, T.; Kaneko, M.K.; Kato, Y. Epitope Mapping of an Anti-Mouse CCR8 Monoclonal Antibody C(8)Mab-2 Using Flow Cytometry. Monoclon Antib Immunodiagn Immunother 2024, 43, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Larsen, O.; van der Velden, W.J.C.; Mavri, M.; et al. Identification of a conserved chemokine receptor motif that enables ligand discrimination. Sci Signal 2022, 15, eabg7042. [Google Scholar] [CrossRef]
- Jørgensen, A.S.; Larsen, O.; Uetz-von Allmen, E.; et al. Biased Signaling of CCL21 and CCL19 Does Not Rely on N-Terminal Differences, but Markedly on the Chemokine Core Domains and Extracellular Loop 2 of CCR7. Front Immunol 2019, 10, 2156. [Google Scholar] [CrossRef]
- Barington, L.; Rummel, P.C.; Lückmann, M.; et al. Role of Conserved Disulfide Bridges and Aromatic Residues in Extracellular Loop 2 of Chemokine Receptor CCR8 for Chemokine and Small Molecule Binding. J Biol Chem 2016, 291, 16208–16220. [Google Scholar] [CrossRef] [PubMed]
- Rummel, P.C.; Thiele, S.; Hansen, L.S.; et al. Extracellular disulfide bridges serve different purposes in two homologous chemokine receptors, CCR1 and CCR5. Mol Pharmacol 2013, 84, 335–345. [Google Scholar] [CrossRef]
- Blanpain, C.; Doranz, B.J.; Bondue, A.; et al. The core domain of chemokines binds CCR5 extracellular domains while their amino terminus interacts with the transmembrane helix bundle. J Biol Chem 2003, 278, 5179–5187. [Google Scholar] [CrossRef]
- Smith, J.S.; Lefkowitz, R.J.; Rajagopal, S. Biased signalling: from simple switches to allosteric microprocessors. Nat Rev Drug Discov 2018, 17, 243–260. [Google Scholar] [CrossRef]
- Barnes, P.J. Chemokine receptor CCR1: new target for asthma therapy. Trends Pharmacol Sci 2022, 43, 539–541. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).