Submitted:
03 April 2025
Posted:
03 April 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
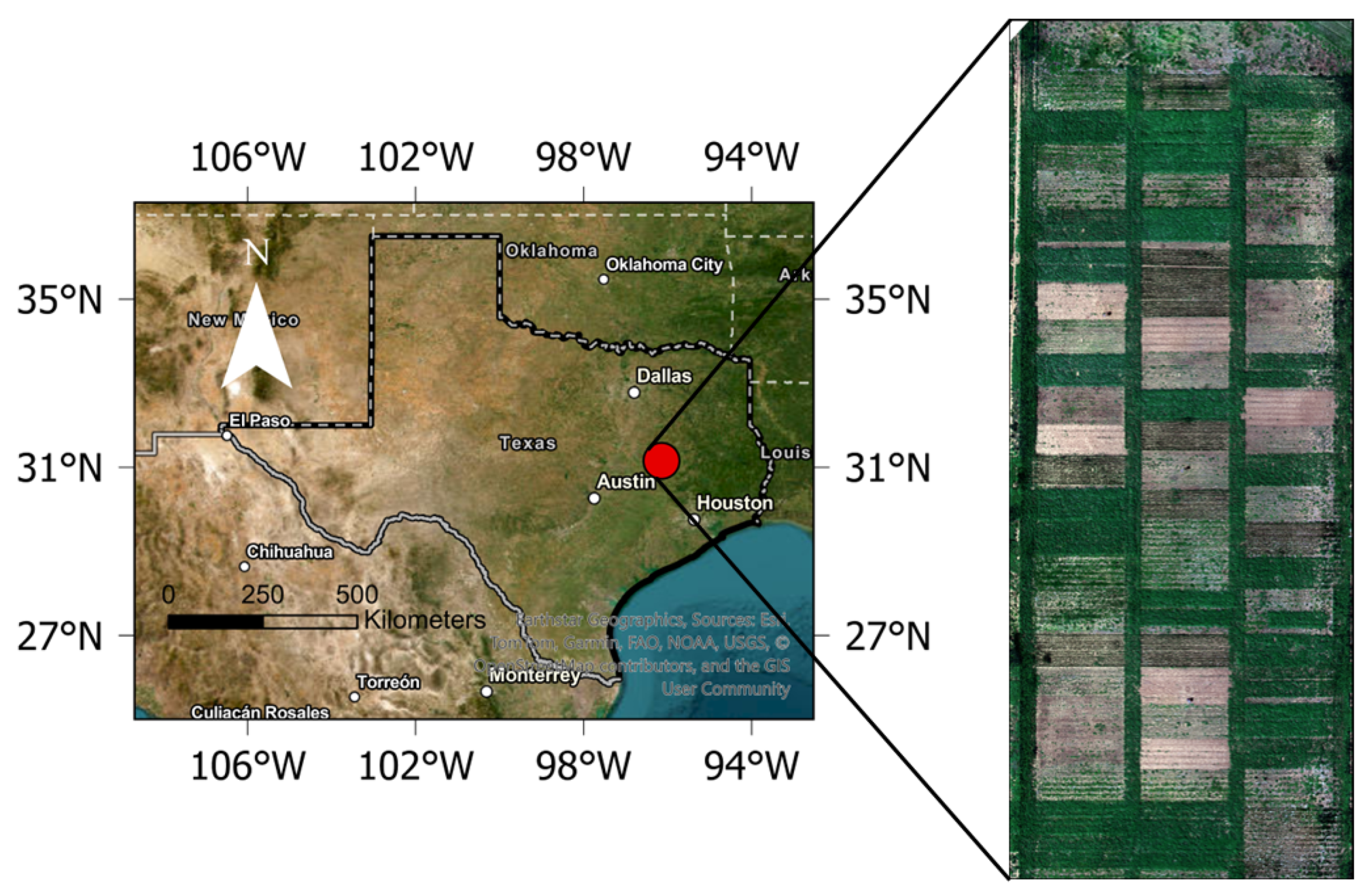
2.1. Study Site
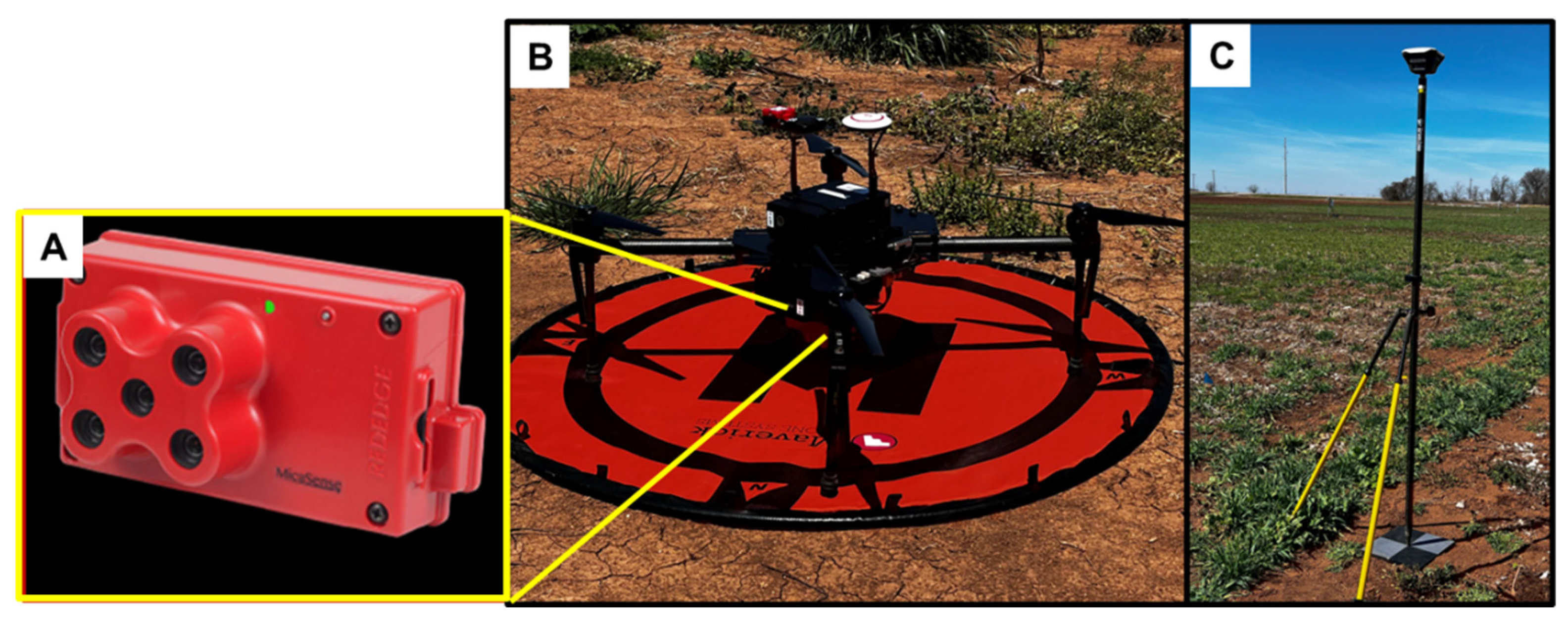
2.2. Image Acquisition
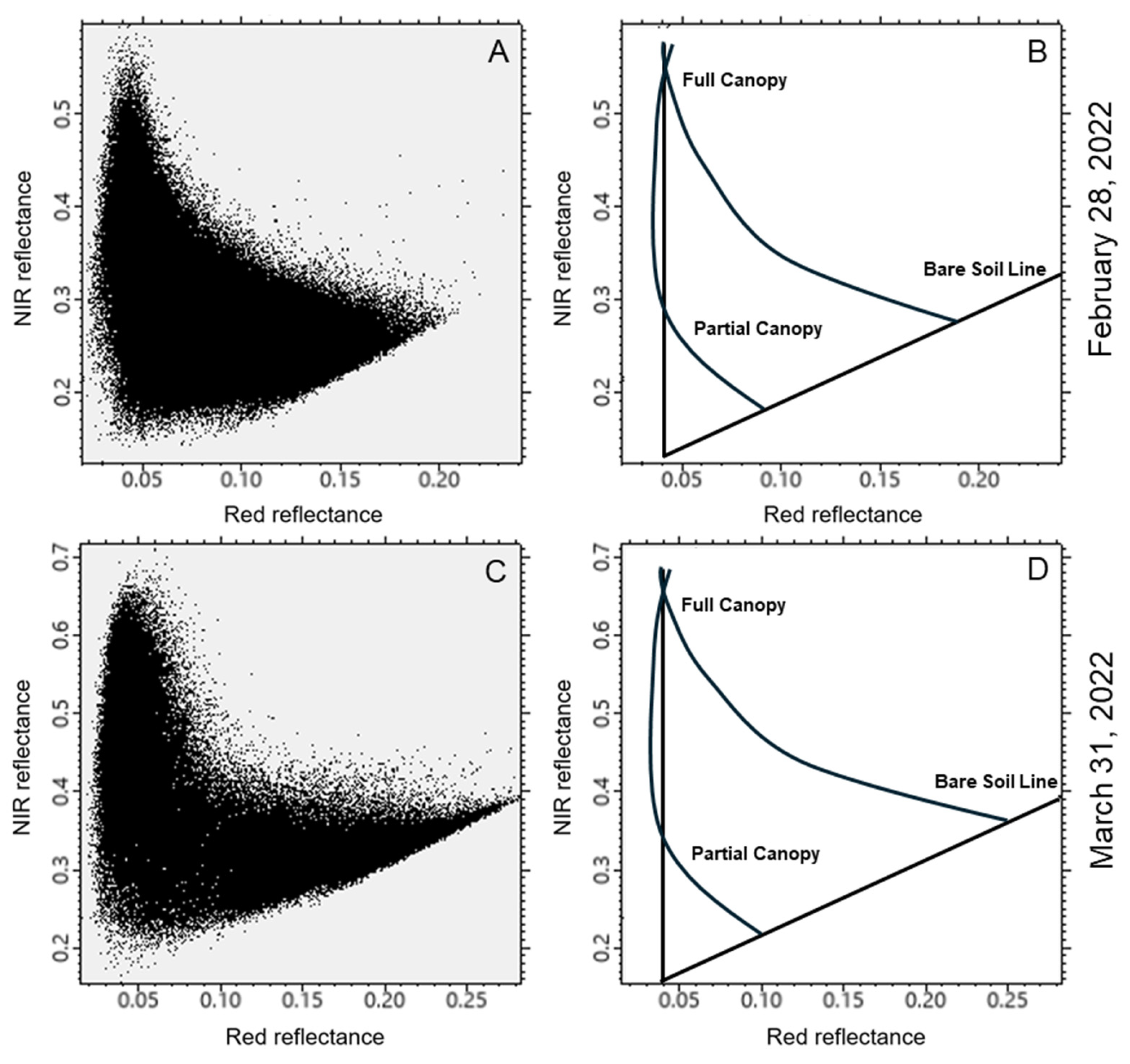
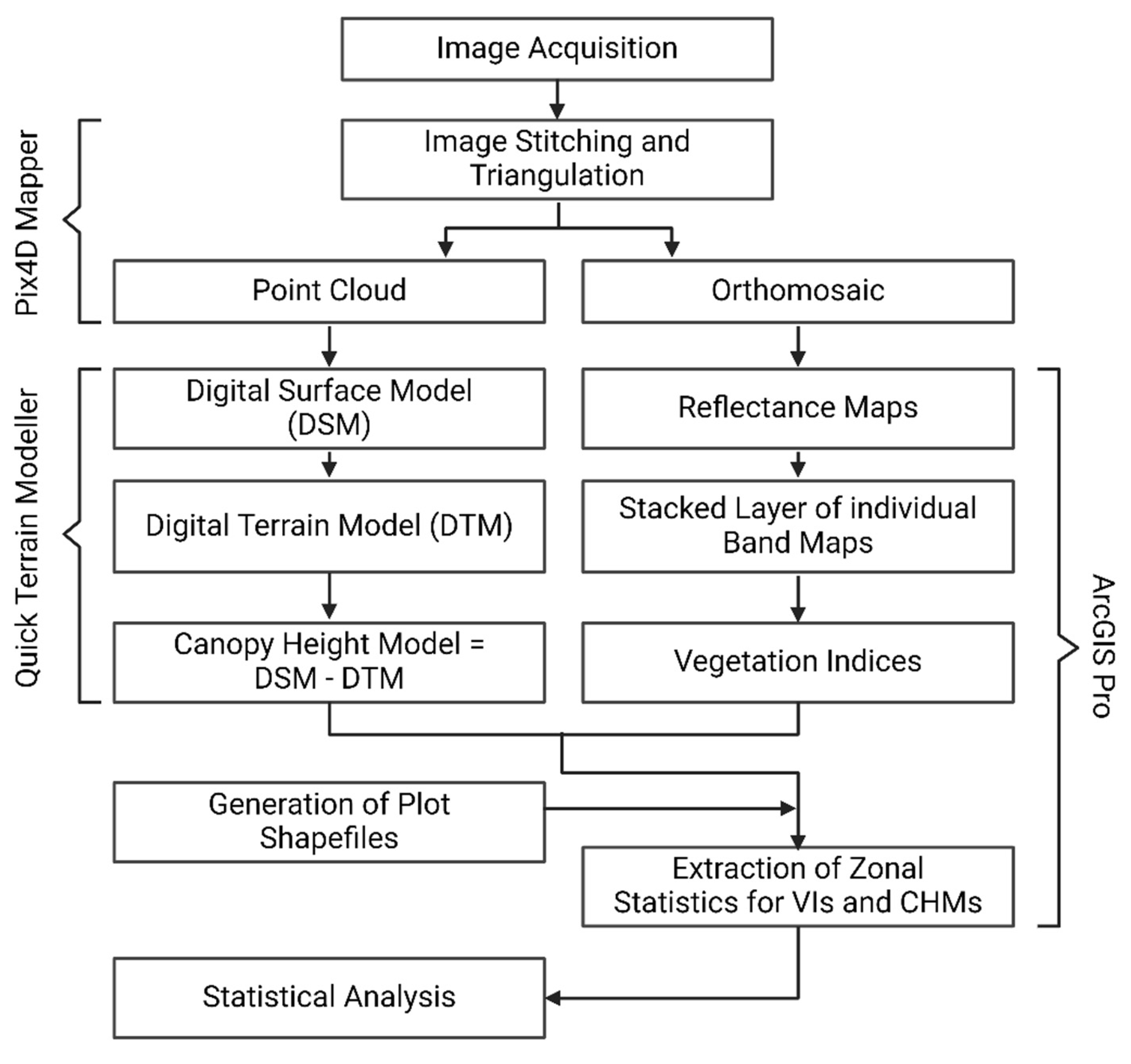
2.3. Image Processing Approach
2.4. Statistical Analyses
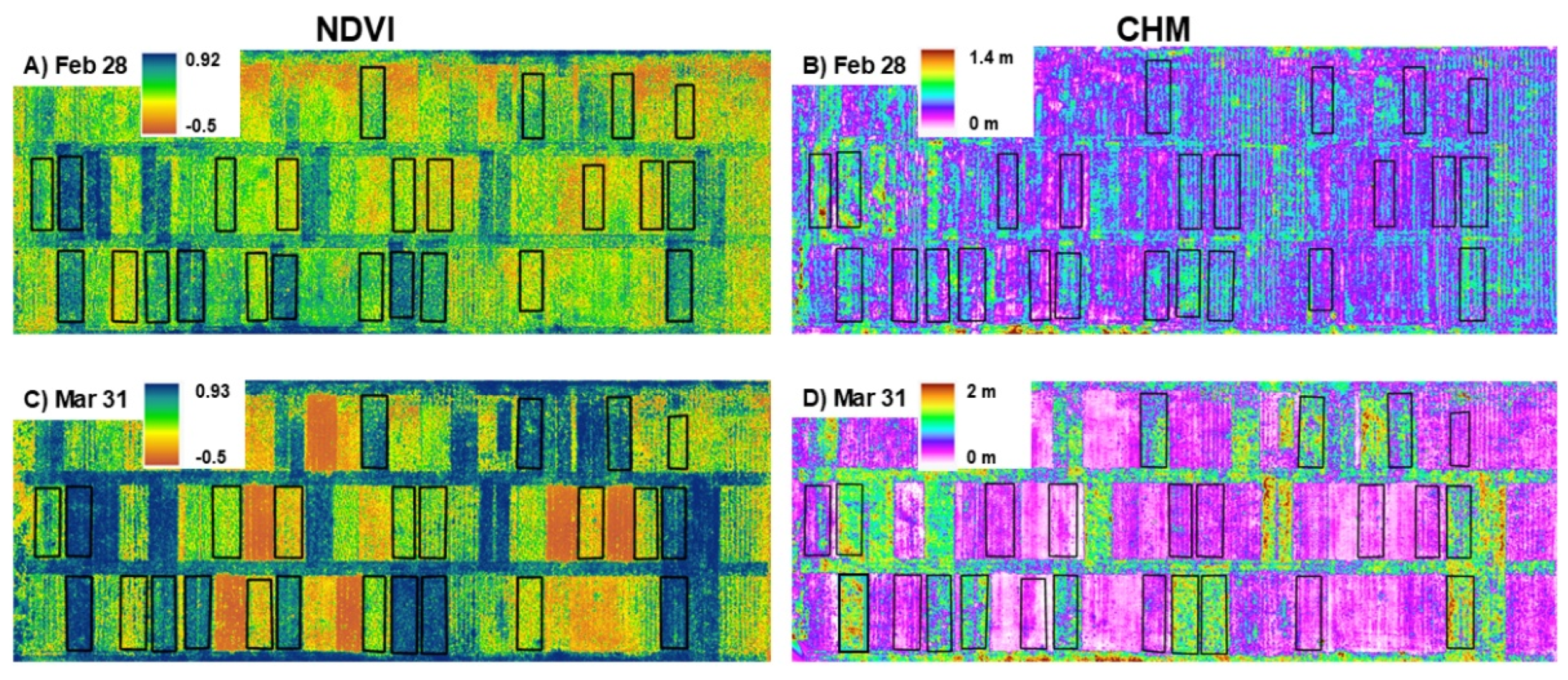
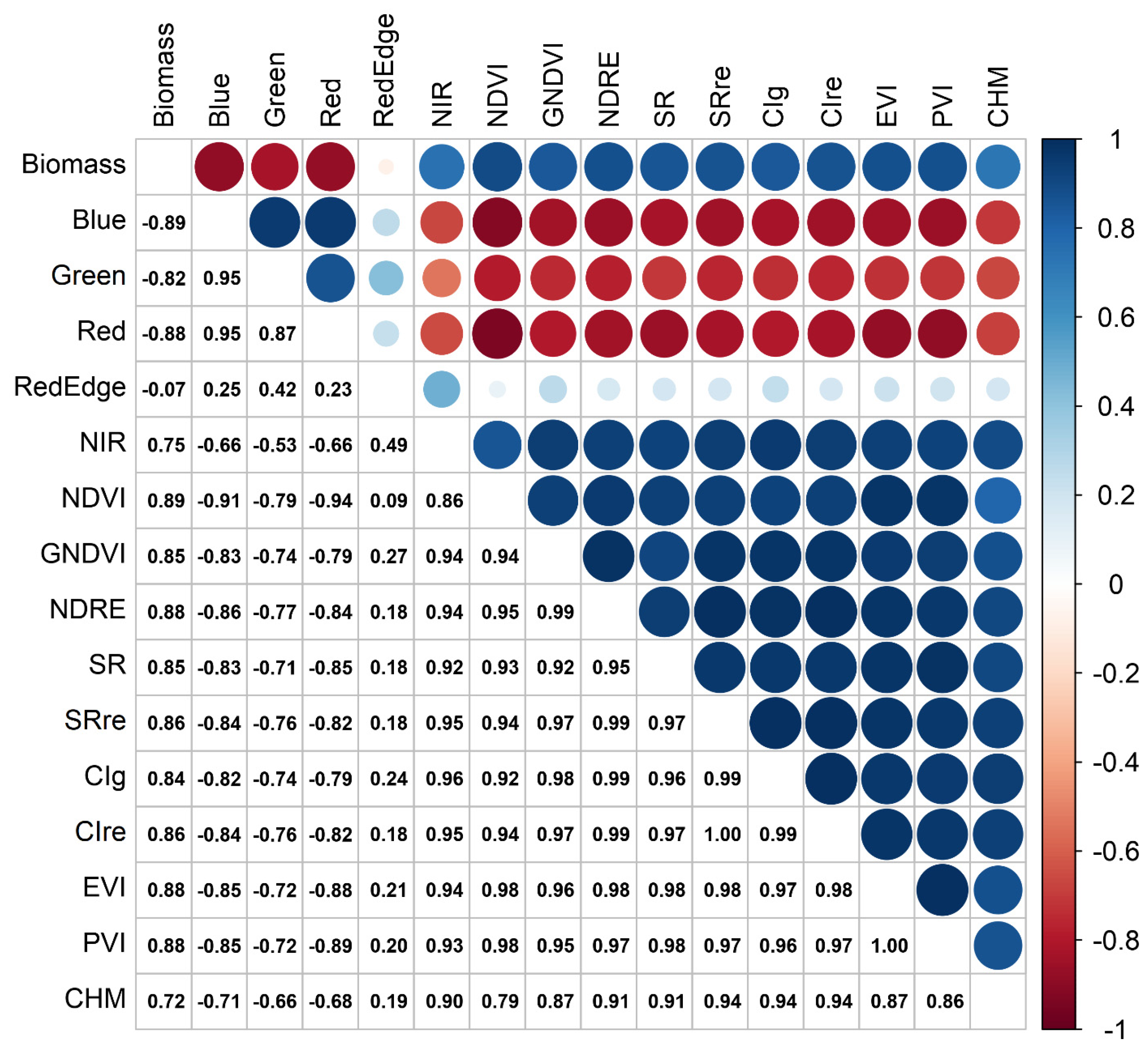
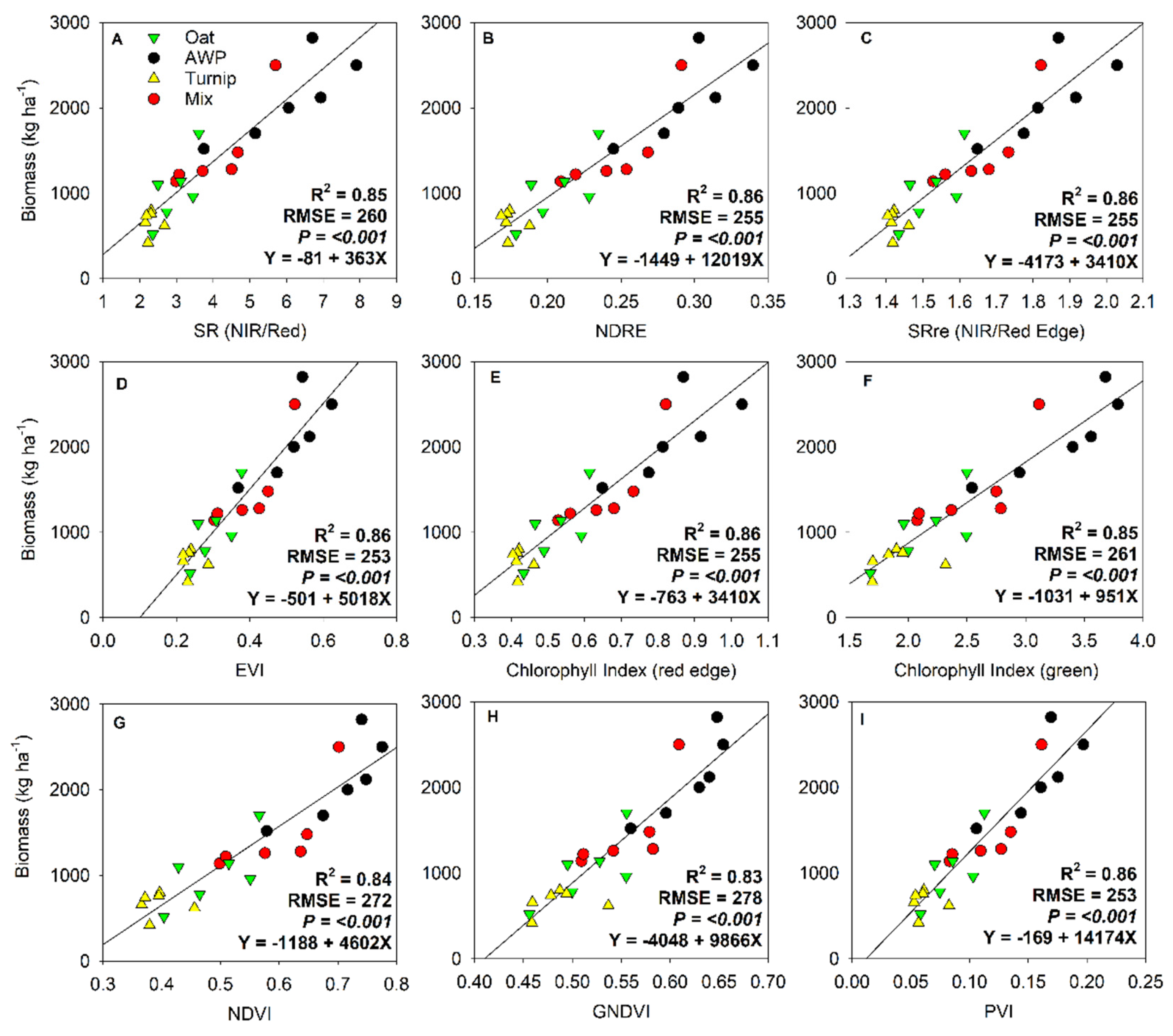
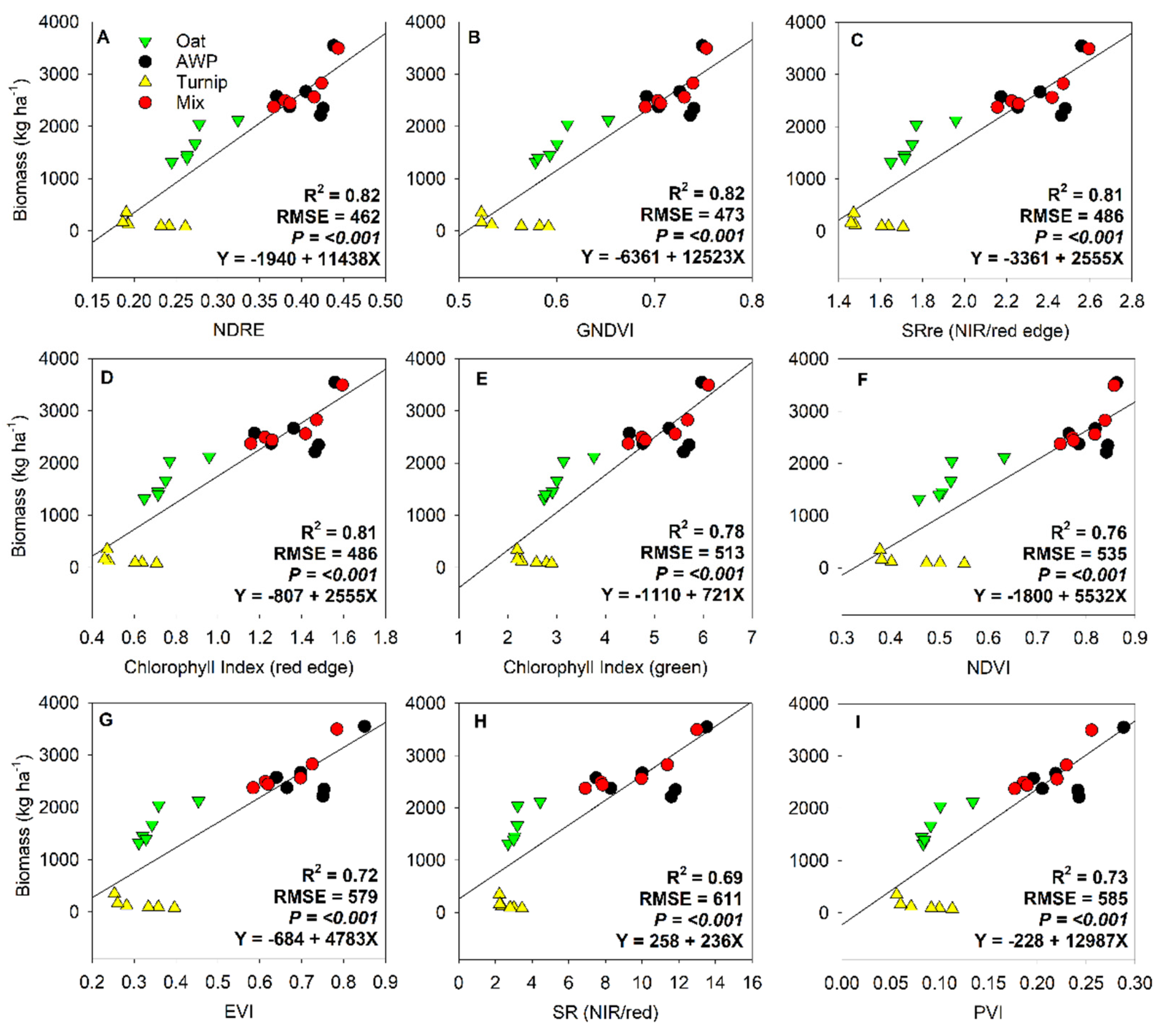
3. Results
4. Discussion
4.1. Temporal Differences in Biomass Estimation
4.2. Influence of Cover Crop Types on Biomass Estimation
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| UAV | Unmanned Aerial Vehicle |
| AWP | Austrian winter peas |
| NIR | Near Infrared |
| NDVI | Normalized Difference Vegetation Index |
| NDRE | Normalized Difference Red Edge Index |
| CIg | Chlorophyll Index Green |
| CIre | Chlorophyll Index Red Edge |
| EVI | Enhanced Vegetation Index |
| GNDVI | Green Normalized Difference Vegetation Index |
| SR | Simple Ratio of Near-Infrared over Red |
| SRre | Simple Ratio of Near-Infrared over Red Edge |
| PVI | Perpendicular Vegetation Index |
| CHM | Canopy Height Model |
| GCP | Ground Control Point |
References
- Blanco-Canqui, H. and S. Ruis, Cover crop impacts on soil physical properties: A review. Soil Science Society of America Journal, 2020. 84(5): p. 1527-1576. [CrossRef]
- Daryanto, S., B.J. Fu, L.X. Wang, P.A. Jacinthe, and W.W. Zhao, Quantitative synthesis on the ecosystem services of cover crops. Earth-Science Reviews, 2018. 185: p. 357-373. [CrossRef]
- Jian, J., X. Du, M.S. Reiter, and R.D. Stewart, A meta-analysis of global cropland soil carbon changes due to cover cropping. Soil Biology and Biochemistry, 2020. 143: p. 107735. [CrossRef]
- Decker, H.L., A.V. Gamble, K.S. Balkcom, A.M. Johnson, and N.R. Hull, Cover crop monocultures and mixtures affect soil health indicators and crop yield in the southeast United States. Soil Science Society of America Journal, 2022. 86(5): p. 1312-1326. [CrossRef]
- MacLaren, C., P. Swanepoel, J. Bennett, J. Wright, and K. Dehnen-Schmutz, Cover crop biomass production is more important than diversity for weed suppression. Crop Science, 2019. 59(2): p. 733-748.
- Osipitan, O.A., A. Dille, Y. Assefa, E. Radicetti, A. Ayeni, and S.Z. Knezevic, Impact of Cover Crop Management on Level of Weed Suppression: A Meta-Analysis. Crop Science, 2019. 59(3): p. 833-842. [CrossRef]
- Darapuneni, M.K., et al., Growth characteristics of summer cover crop grasses and their relation to soil aggregate stability and wind erosion control in arid southwest. Applied Engineering in Agriculture, 2021. 37(1): p. 11-23. [CrossRef]
- Zuazo, V.H.D. and C.R.R. Pleguezuelo, Soil-Erosion and Runoff Prevention by Plant Covers: A Review. Sustainable Agriculture, ed. E. Lichtfouse, et al. 2009, Berlin: Springer-Verlag Berlin. 785-811.
- Bai, X.X., et al., Responses of soil carbon sequestration to climate-smart agriculture practices: A meta-analysis. Global Change Biology, 2019. 25(8): p. 2591-2606. [CrossRef]
- Blanco-Canqui, H., Cover crops and carbon sequestration: Lessons from US studies. Soil Science Society of America Journal, 2022. 86(3): p. 501-519. [CrossRef]
- Kuyah, S., et al., Allometric equations for estimating biomass in agricultural landscapes: II. Belowground biomass. Agriculture Ecosystems & Environment, 2012. 158: p. 225-234. [CrossRef]
- Qi, Y.L., W. Wei, C.G. Chen, and L.D. Chen, Plant root-shoot biomass allocation over diverse biomes: A global synthesis. Global Ecology and Conservation, 2019. 18: p. 14. [CrossRef]
- Bai, G., K. Koehler-Cole, D. Scoby, V.R. Thapa, A. Basche, and Y.F. Ge, Enhancing estimation of cover crop biomass using field-based high-throughput phenotyping and machine learning models. Frontiers in Plant Science, 2024. 14: p. 12. [CrossRef]
- Xia, Y.S., K.Y. Guan, K. Copenhaver, and M. Wander, Estimating cover crop biomass nitrogen credits with Sentinel-2 imagery and sites covariates. Agronomy Journal, 2021. 113(2): p. 1084-1101. [CrossRef]
- Xu, M., C.G. Lacey, and S.D. Armstrong, The feasibility of satellite remote sensing and spatial interpolation to estimate cover crop biomass and nitrogen uptake in a small watershed. Journal of Soil and Water Conservation, 2018. 73(6): p. 682-692. [CrossRef]
- Bendini, H.D., et al., Estimating Winter Cover Crop Biomass in France Using Optical Sentinel-2 Dense Image Time Series and Machine Learning. Remote Sensing, 2024. 16(5): p. 24. [CrossRef]
- Goffart, D., Y. Curnel, V. Planchon, J.P. Goffart, and P. Defourny, Field-scale assessment of Belgian winter cover crops biomass based on Sentinel-2 data. European Journal of Agronomy, 2021. 126: p. 15. [CrossRef]
- Shi, Y., et al., Unmanned aerial vehicles for high-throughput phenotyping and agronomic research. PloS one, 2016. 11(7): p. e0159781.
- Acorsi, M.G., F.D.A. Miranda, M. Martello, D.A. Smaniotto, and L.R. Sartor, Estimating Biomass of Black Oat Using UAV-Based RGB Imaging. Agronomy-Basel, 2019. 9(7): p. 14. [CrossRef]
- Kümmerer, R., P.O. Noack, and B. Bauer, Using High-Resolution UAV Imaging to Measure Canopy Height of Diverse Cover Crops and Predict Biomass. Remote Sensing, 2023. 15(6): p. 18. [CrossRef]
- Roth, L. and B. Streit, Predicting cover crop biomass by lightweight UAS-based RGB and NIR photography: an applied photogrammetric approach. Precision Agriculture, 2018. 19(1): p. 93-114. [CrossRef]
- Yuan, M., J. Burjel, J. Isermann, N. Goeser, and C. Pittelkow, Unmanned aerial vehicle–based assessment of cover crop biomass and nitrogen uptake variability. Journal of Soil and Water Conservation, 2019. 74(4): p. 350-359.
- Kharel, T.P., A.B. Bhandari, P. Mubvumba, H.L. Tyler, R.S. Fletcher, and K.N. Reddy, Mixed-Species Cover Crop Biomass Estimation Using Planet Imagery. Sensors, 2023. 23(3): p. 16. [CrossRef]
- Prabhakara, K., W.D. Hively, and G.W. McCarty, Evaluating the relationship between biomass, percent groundcover and remote sensing indices across six winter cover crop fields in Maryland, United States. International Journal of Applied Earth Observation and Geoinformation, 2015. 39: p. 88-102. [CrossRef]
- Shafian, S., et al., Unmanned aerial systems-based remote sensing for monitoring sorghum growth and development. PloS one, 2018. 13(5): p. e0196605.
- Roth, R.T., et al., Prediction of Cereal Rye Cover Crop Biomass and Nutrient Accumulation Using Multi-Temporal Unmanned Aerial Vehicle Based Visible-Spectrum Vegetation Indices. Remote Sensing, 2023. 15(3): p. 18. [CrossRef]
- Holzhauser, K., T. Räbiger, T. Rose, H. Kage, and I. Kühling, Estimation of Biomass and N Uptake in Different Winter Cover Crops from UAV-Based Multispectral Canopy Reflectance Data. Remote Sensing, 2022. 14(18): p. 17. [CrossRef]
- Sangjan, W., R.J. McGee, and S. Sankaran, Optimization of UAV-Based Imaging and Image Processing Orthomosaic and Point Cloud Approaches for Estimating Biomass in a Forage Crop. Remote Sensing, 2022. 14(10): p. 19. [CrossRef]
- Elhakeem, A., L. Bastiaans, S. Houben, T. Couwenberg, D. Makowski, and W. van der Werf, Do cover crop mixtures give higher and more stable yields than pure stands? Field Crops Research, 2021. 270: p. 10. [CrossRef]
- Salehin, S.M.U., et al., Cover crops in organic cotton influence greenhouse gas emissions and soil microclimate. Agronomy Journal, 2025. 117(1): p. e21735. DOI: doi.org/10.1002/agj2.21735.
- SRCC. Southern Regional Climate Center: Now Data. 2023 May 25, 2021]; Available from: https://www.srcc.tamu.edu/services/nowdata/.
- Soil Survey Staff, N.R.C.S., United State Department of Agriculture, Web Soil Survey. 2022.
- Hinzmann, T., J.L. Schonberger, M. Pollefeys, and R. Siegwart, Mapping on the fly: Real-time 3D dense reconstruction, digital surface map and incremental orthomosaic generation for unmanned aerial vehicles, in Field and Service Robotics: Results of the 11th International Conference. 2018.
- ESRI. ArcGIS Desktop: Release 10.8.1. 2020.
- Hunt, E.R., C.S.T. Daughtry, J.U.H. Eitel, and D.S. Long, Remote Sensing Leaf Chlorophyll Content Using a Visible Band Index. Agronomy Journal, 2011. 103(4): p. 1090-1099. [CrossRef]
- Nagler, P.L., R.L. Scott, C. Westenburg, J.R. Cleverly, E.P. Glenn, and A.R. Huete, Evapotranspiration on western US rivers estimated using the Enhanced Vegetation Index from MODIS and data from eddy covariance and Bowen ratio flux towers. Remote Sensing of Environment, 2005. 97(3): p. 337-351. [CrossRef]
- Gitelson, A.A., Y.J. Kaufman, and M.N. Merzlyak, Use of a green channel in remote sensing of global vegetation from EOS-MODIS. Remote Sensing of Environment, 1996. 58(3): p. 289-298. [CrossRef]
- Rouse, J.W., R.H. Haas, J.A. Schell, and D.W. Deering, Monitoring vegetation systems in the Great Plains with ERTS. NASA Spec. Publ, 1974. 351(1): p. 309.
- Chen, J.M., Evaluation of vegetation indices and a modified simple ratio for boreal applications. Canadian Journal of Remote Sensing, 1996. 22(3): p. 229-242.
- Xie, Q., et al., Vegetation indices combining the red and red-edge spectral information for leaf area index retrieval. IEEE Journal of selected topics in applied earth observations and remote sensing, 2018. 11(5): p. 1482-1493.
- Richardson, A.J. and C.L. Wiegand, Distinguishing vegetation from soil background information. Photogrammetric Engineering and Remote Sensing, 1977. 43(12): p. 1541-1552.
- Maas, S.J., Linear mixture modeling approach for estimating cotton canopy ground cover using satellite multispectral imagery. Remote Sensing of Environment, 2000. 72(3): p. 304-308. [CrossRef]
- GeoCue Group Inc. Quick Terrain Modeller: Version 8.4. 2024.
- R core Team. R: A language and environment for statistical computing. 2024; Available from: https://www.R-project.org.
- Wei, T. and V. Simko, R package “corrplot’: Visualization of a correlation Matrix (version 0.92). 2021.
- Hebbali A., olsrr: Tools for Building OLS Regression Models. 2024.
- Fox, J. and S. Weisberg, An R companion to applied regression. Third ed. 2019, Thousand Oaks, CA: Sage.
- Buchhart, C. and U. Schmidhalter, Daytime and seasonal reflectance of maize grown in varying compass directions. Frontiers in Plant Science, 2022. 13: p. 13. [CrossRef]
- Zeng, L.J. and C.C. Chen, Using remote sensing to estimate forage biomass and nutrient contents at different growth stages. Biomass & Bioenergy, 2018. 115: p. 74-81. [CrossRef]
- Hansen, P.M. and J.K. Schjoerring, Reflectance measurement of canopy biomass and nitrogen status in wheat crops using normalized difference vegetation indices and partial least squares regression. Remote Sensing of Environment, 2003. 86(4): p. 542-553. [CrossRef]
- Kanke, Y., B. Tubaña, M. Dalen, and D. Harrell, Evaluation of red and red-edge reflectance-based vegetation indices for rice biomass and grain yield prediction models in paddy fields. Precision Agriculture, 2016. 17(5): p. 507-530. [CrossRef]
- Hatfield, J.L. and J.H. Prueger, Value of Using Different Vegetative Indices to Quantify Agricultural Crop Characteristics at Different Growth Stages under Varying Management Practices. Remote Sensing, 2010. 2(2): p. 562-578. [CrossRef]
- Martin, K.L., et al., Expression of variability in corn as influenced by growth stage using optical sensor measurements. Agronomy Journal, 2007. 99(2): p. 384-389. [CrossRef]
- Barboza, T.O.C., M. Ardigueri, G.F.C. Souza, M.A.J. Ferraz, J.R.F. Gaudencio, and A.F. dos Santos, Performance of Vegetation Indices to Estimate Green Biomass Accumulation in Common Bean. Agriengineering, 2023. 5(2): p. 840-854. [CrossRef]
- Gitelson, A.A., A. Viña, T.J. Arkebauer, D.C. Rundquist, G. Keydan, and B. Leavitt, Remote estimation of leaf area index and green leaf biomass in maize canopies -: art. no. 1248. Geophysical Research Letters, 2003. 30(5): p. 4. [CrossRef]
- Miller, J.O., A.L. Shober, and J. Taraila, Assessing relationships of cover crop biomass and nitrogen content to multispectral imagery. Agronomy Journal, 2024. 116(3): p. 1417-1427. [CrossRef]
- Kross, A., H. McNairn, D. Lapen, M. Sunohara, and C. Champagne, Assessment of RapidEye vegetation indices for estimation of leaf area index and biomass in corn and soybean crops. International Journal of Applied Earth Observation and Geoinformation, 2015. 34: p. 235-248. [CrossRef]
- Lu, N., et al., Improved estimation of aboveground biomass in wheat from RGB imagery and point cloud data acquired with a low-cost unmanned aerial vehicle system. Plant Methods, 2019. 15: p. 16. [CrossRef]
- Yue, J.B., et al., Estimation of Winter Wheat Above-Ground Biomass Using Unmanned Aerial Vehicle-Based Snapshot Hyperspectral Sensor and Crop Height Improved Models. Remote Sensing, 2017. 9(7): p. 19. [CrossRef]
- Feng, H.K., et al. Height and biomass inversion of winter wheat based on canopy height model. in 38th IEEE International Geoscience and Remote Sensing Symposium (IGARSS). 2018. Valencia, SPAIN: Ieee.
- Zhang, H.F., et al., Estimation of Grassland Canopy Height and Aboveground Biomass at the Quadrat Scale Using Unmanned Aerial Vehicle. Remote Sensing, 2018. 10(6): p. 19. [CrossRef]
- Jin, X.L., et al., Combined Multi-Temporal Optical and Radar Parameters for Estimating LAI and Biomass in Winter Wheat Using HJ and RADARSAR-2 Data. Remote Sensing, 2015. 7(10): p. 13251-13272. [CrossRef]
- Dube, T., C. Shoko, and T.W. Gara, Remote sensing of aboveground grass biomass between protected and non-protected areas in savannah rangelands. African Journal of Ecology, 2021. 59(3): p. 687-695. [CrossRef]
- Mutanga, O. and A.K. Skidmore, Narrow band vegetation indices overcome the saturation problem in biomass estimation. International Journal of Remote Sensing, 2004. 25(19): p. 3999-4014. [CrossRef]
- Shi, H., et al., Assessing the ability of MODIS EVI to estimate terrestrial ecosystem gross primary production of multiple land cover types. Ecological Indicators, 2017. 72: p. 153-164. [CrossRef]
- Jiang, J.L., et al., Phenotyping a diversity panel of quinoa using UAV-retrieved leaf area index, SPAD-based chlorophyll and a random forest approach. Precision Agriculture, 2022. 23(3): p. 961-983. [CrossRef]
- Corti, M., et al., Improved estimation of herbaceous crop aboveground biomass using UAV-derived crop height combined with vegetation indices. Precision Agriculture, 2023. 24(2): p. 587-606. [CrossRef]
- Biewer, S., T. Fricke, and M. Wachendorf, Determination of Dry Matter Yield from Legume-Grass Swards by Field Spectroscopy. Crop Science, 2009. 49(5): p. 1927-1936. [CrossRef]
- Pulina, A., V. Rolo, A. Hernández-Esteban, G. Seddaiu, P.P. Roggero, and G. Moreno, Long-term legacy of sowing legume-rich mixtures in Mediterranean wooded grasslands. Agriculture Ecosystems & Environment, 2023. 348: p. 10. [CrossRef]

| Band Name | Center Wavelength (nm) | Bandwidth (nm) |
| Blue | 475 | 32 |
| Green | 560 | 27 |
| Red | 668 | 14 |
| Red Edge | 717 | 12 |
| Near Infrared | 842 | 57 |
| Vegetation Index | Abbreviation | Band Formula | Reference |
| Chlorophyll Index Green | Clg | [35] | |
| Chlorophyll Index Red Edge | Clre | [35] | |
| Enhanced Vegetation Index | EVI | [36] | |
| Green Normalized Difference Vegetation Index | GNDVI | [37] | |
| Normalized Difference Vegetation Index | NDVI | [38] | |
| Normalized Difference Red Edge Index | NDRE | [35] | |
| Simple Ratio | SR | [39] | |
| Simple Ratio Red Edge | SRre | [40] | |
| Perpendicular vegetation index | PVI | [41] |
| Vegetation Indices | February 28 | March 31 |
| Correlation coefficients | ||
| Blue band | -0.83 | -0.91 |
| Green band | -0.65 | -0.87 |
| Red band | -0.89 | -0.90 |
| Red edge band | 0.62 | -0.55 |
| NIR band | 0.91 | 0.76 |
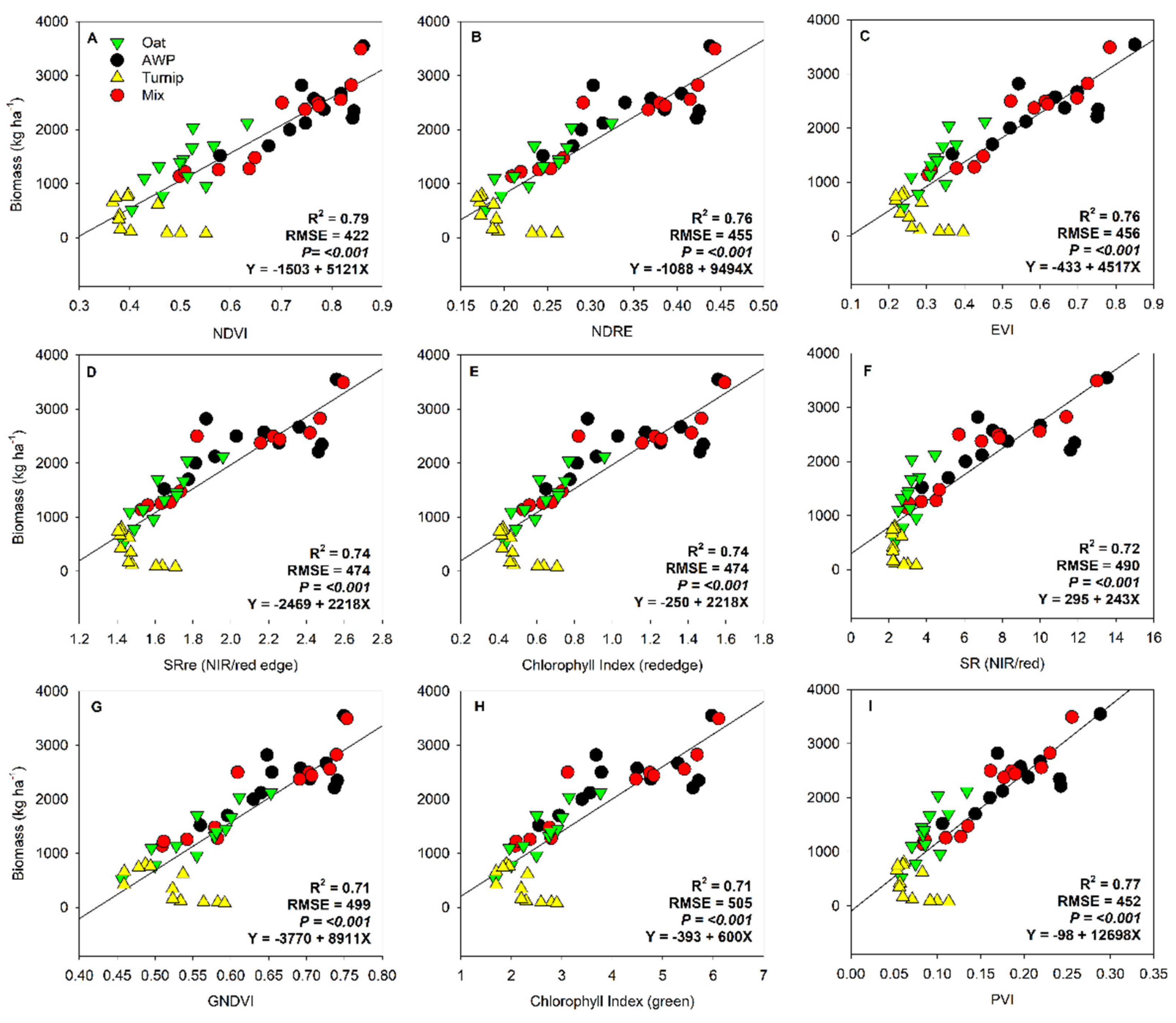
| NDVI | 0.92 | 0.88 |
| GNDVI | 0.91 | 0.91 |
| NDRE | 0.93 | 0.91 |
| SR | 0.93 | 0.84 |
| SR red edge | 0.93 | 0.90 |
| CI green | 0.92 | 0.89 |
| CI red edge | 0.93 | 0.90 |
| EVI | 0.93 | 0.86 |
| PVI | 0.93 | 0.86 |
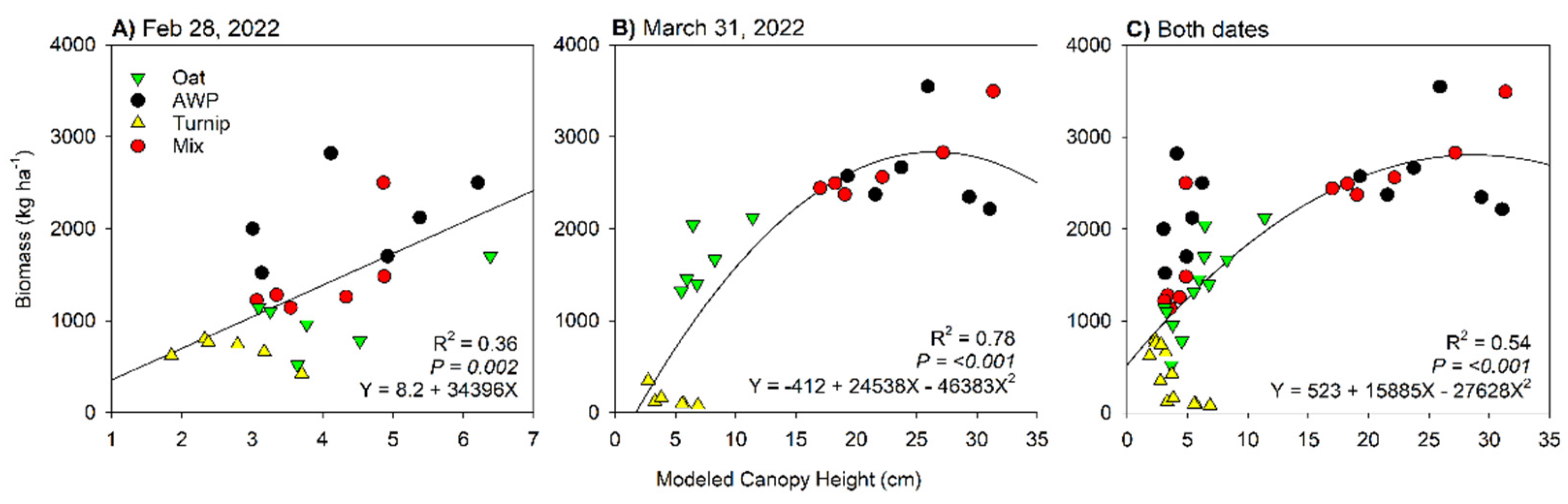
| CHM | 0.60 | 0.84 |
| Variable | Coefficient | P-value | Adjusted R2 | RMSE |
| February 28, 2022 | ||||
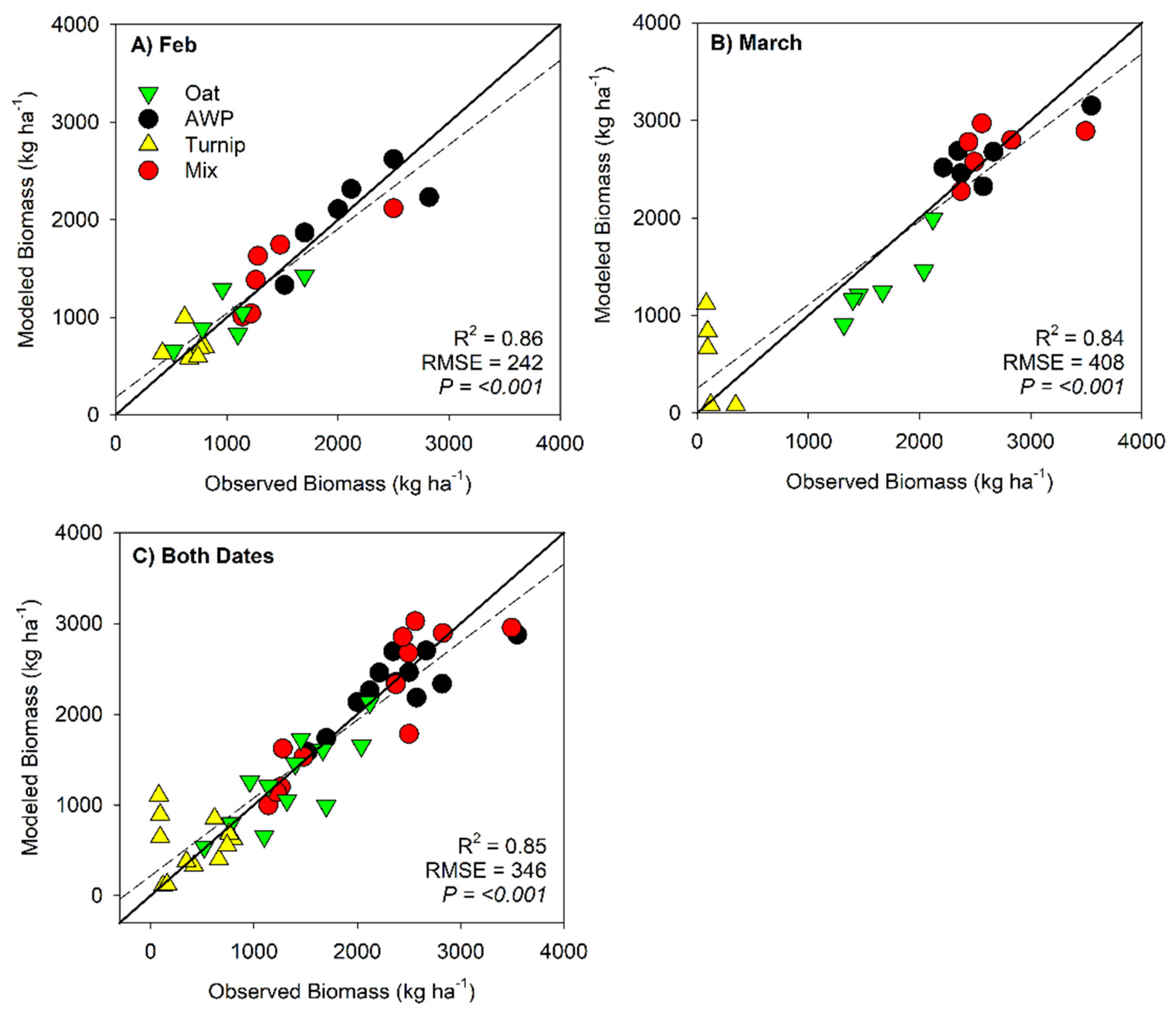
| Intercept | -169.1 | <0.001 | 0.86 | 242.3 |
| PVI | 14174 | |||
| March 31, 2022 | ||||
| Intercept | -3297.5 | <0.001 | 0.84 | 408.3 |
| NDRE | 18671.1 | |||
| CHM | -6679.7 | |||
| Both dates | ||||
| Intercept | -694.7 | <0.001 | 0.85 | 345.8 |
| Green | -33464.3 | |||
| CHM | -6760.3 | |||
| SRre | 3160.1 | |||
| Variable | Coefficient | P-value | Adjusted R2 | RMSE |
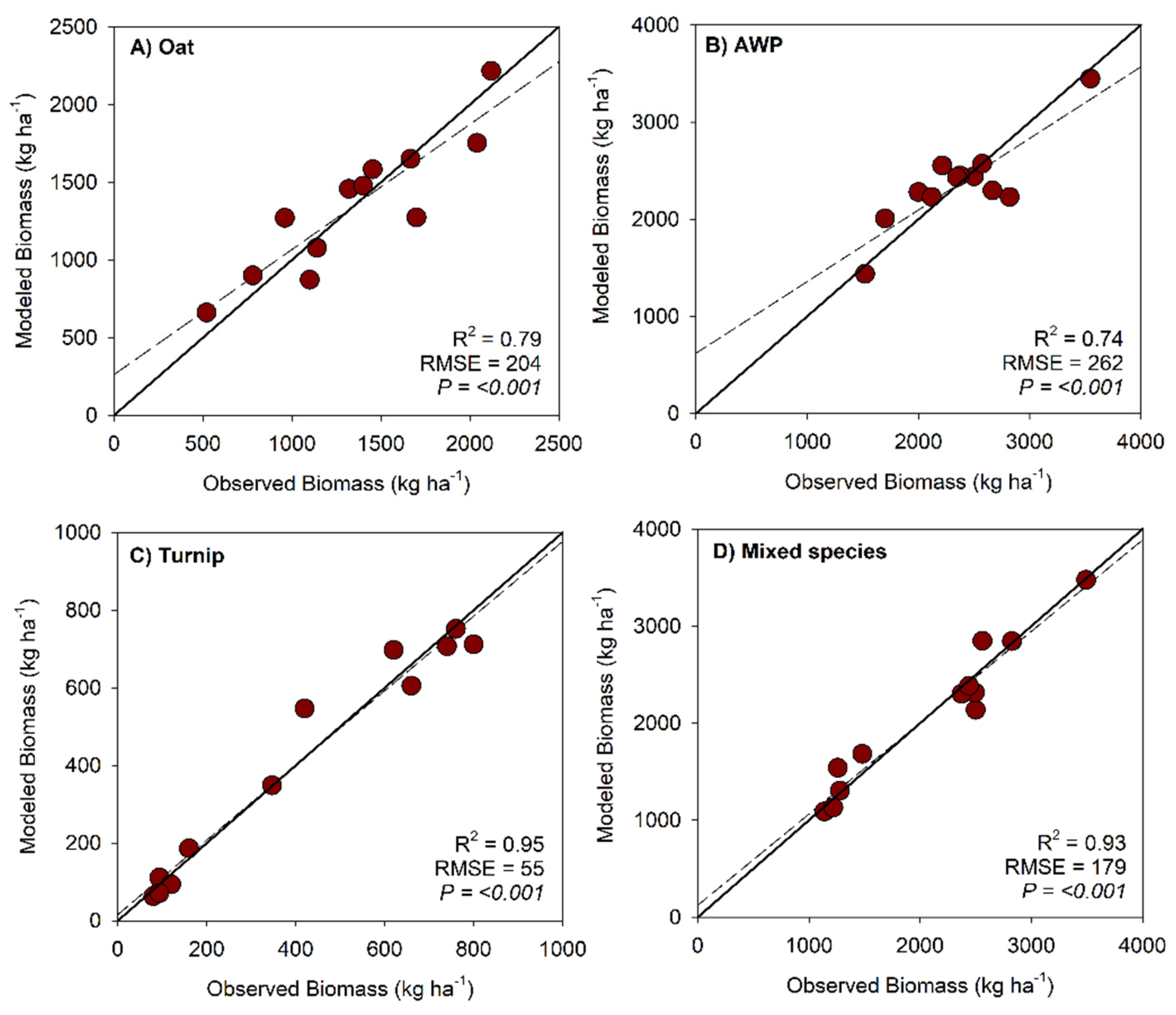
| Oat | ||||
| Intercept | -500.9 | <0.001 | 0.86 | 242.4 |
| EVI | 5018.3 | |||
| Austrian winter pea | ||||
| Intercept | -5530.1 | <0.001 | 0.71 | 261.7 |
| Red edge | 39904.1 | |||
| Turnip | ||||
| Intercept | 952.1 | <0.001 | 0.95 | 55.1 |
| NIR | -7690.8 | |||
| GNDVI | 3910.9 | |||
| CHM | -5773.6 | |||
| Mixed species | ||||
| Intercept | -6421.7 | <0.001 | 0.93 | 179.4 |
| NIR | 141191.7 | |||
| Blue | 81897.9 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




