Submitted:
21 March 2025
Posted:
28 March 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Related Work
2.1. Deep Learning in Medical Image Analysis
2.2. Natural Language Processing for Clinical Text Generation
2.3. Multimodal Learning for Radiology Report Synthesis
2.4. Challenges and Limitations in Existing Work
2.5. Summary of Related Work
3. Methodology
3.1. Search Strategy
- PubMed (for biomedical and clinical research)
- IEEE Xplore (for AI and engineering applications)
- ACM Digital Library (for computer science research)
- Scopus (for multidisciplinary coverage)
- Google Scholar (for additional gray literature)
- Deep learning (e.g., “deep learning,” “neural networks,” “CNN,” “RNN,” “transformers”)
- Radiology reports (e.g., “radiology report generation,” “medical report automation,” “radiology text generation”)
- Medical imaging (e.g., “X-ray,” “CT,” “MRI,” “medical image captioning”)
- Natural language processing (e.g., “NLP in healthcare,” “clinical text generation”)
(“deep learning” OR “neural networks” OR “CNN” OR “transformer”) AND (“radiology report” OR “medical report generation”) AND (“X-ray” OR “MRI” OR “CT”) AND (“natural language processing” OR “text generation”)
3.2. Inclusion and Exclusion Criteria
3.2.1. Inclusion Criteria
- Studies that apply deep learning models for automated radiology report generation [55].
- Research focusing on multimodal learning approaches combining medical imaging and natural language processing.
- Papers that provide experimental results and evaluations using benchmark datasets.
- Peer-reviewed journal articles, conference proceedings, and preprints with substantial contributions [56].
3.2.2. Exclusion Criteria
- Studies that focus solely on radiology image classification or segmentation without report generation.
- Papers that propose rule-based or template-based reporting systems without deep learning components.
- Review articles, opinion pieces, or studies lacking experimental validation [57].
- Articles written in languages other than English (due to accessibility constraints).
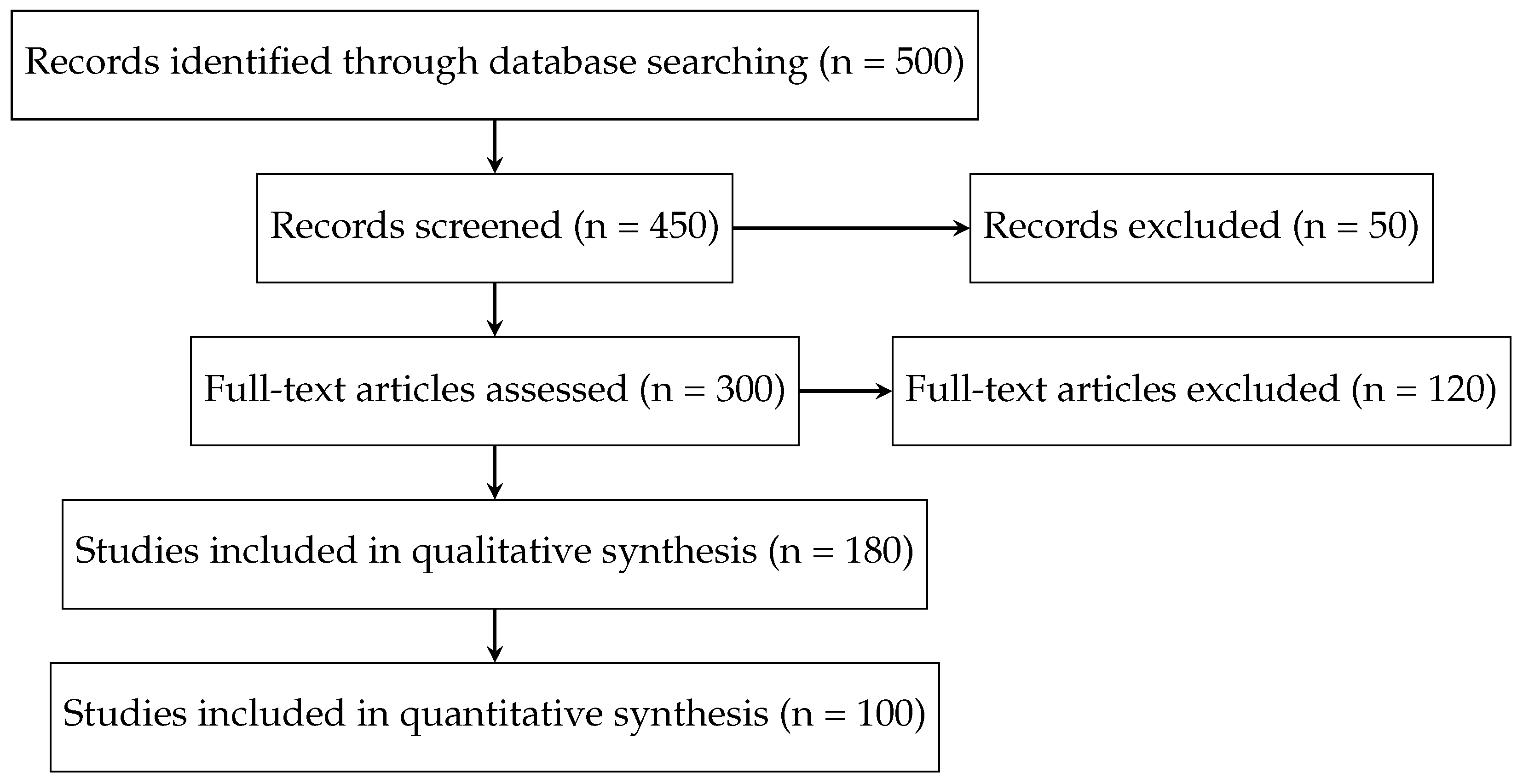
3.3. Study Selection Process
- (1)
- Title and Abstract Screening: Two independent reviewers screened the retrieved articles based on their titles and abstracts to remove irrelevant studies [58].
- (2)
- Full-Text Review: The remaining articles were reviewed in full to assess their relevance and methodological rigor [59].
- (3)
- Final Inclusion: Any disagreements between reviewers were resolved through discussion or consultation with a third reviewer to ensure unbiased selection.
3.4. Data Extraction and Synthesis
- Study Information: Authors, publication year, venue (conference/journal) [60].
- Dataset Used: Publicly available datasets (e.g., MIMIC-CXR, IU X-ray) or institution-specific datasets.
- Deep Learning Model: Architecture details (e.g., CNN-RNN, transformer-based, multimodal models) [61].
- Training and Evaluation Metrics: Metrics such as BLEU, ROUGE, METEOR, and clinical accuracy scores [62].
- Key Findings and Limitations: Summary of results, major contributions, and study limitations.
3.5. Quality Assessment
- Reproducibility: Whether the dataset and code were made publicly available [66].
- Model Robustness: Use of cross-validation, external validation, and error analysis [67].
- Clinical Relevance: Whether the study involved domain experts (radiologists) for qualitative evaluation [68].
- Bias and Limitations: Identification of biases in dataset selection, training methodology, and evaluation [69].
3.6. Limitations of the Review Process
3.7. Summary of Methodology
4. Findings and Analysis
4.1. Overview of Included Studies
4.2. Commonly Used Datasets
- IU X-ray : A smaller dataset comprising approximately 7,000 chest X-rays with structured reports. This dataset is frequently used for model evaluation and benchmarking.
- CheXpert : While primarily used for classification tasks, some studies utilize its reports for supervised training of NLP-based models [80].
4.3. Deep Learning Architectures for Radiology Report Generation
4.3.1. CNN-RNN Architectures
- proposed a CNN-LSTM model with an attention mechanism to generate reports from X-ray images [83].
- introduced a hierarchical LSTM structure that improved coherence in generated reports.
4.3.2. Transformer-Based Models
- applied a Transformer-based model to generate radiology reports, demonstrating improved linguistic fluency and clinical accuracy [85].
- incorporated a medical knowledge graph into a Transformer-based architecture, enhancing factual correctness [86].
4.3.3. Multimodal Vision-Language Models
- introduced a knowledge-guided attention mechanism to enhance multimodal alignment.
- proposed a cross-modal learning approach using contrastive loss to better link textual and visual features [88].
4.4. Evaluation Metrics
- BLEU : Measures n-gram overlap between generated and reference reports [90].
- ROUGE : Evaluates recall-based text overlap.
- METEOR : Incorporates synonym matching and stemming for a more nuanced assessment [91].
- Clinical Accuracy Metrics: Some studies introduced domain-specific metrics such as RadGraph , which evaluates the factual correctness of generated reports.
4.5. Trends and Open Challenges
- Shift toward Transformer Models: Transformer-based architectures have increasingly replaced traditional CNN-RNN frameworks due to their superior language modeling capabilities [93].
- Integration of Medical Knowledge: Recent studies have incorporated medical ontologies and knowledge graphs to improve clinical accuracy [94].
- Challenges in Data and Evaluation: Despite the availability of large datasets, issues such as dataset bias, annotation inconsistencies, and the lack of standardized evaluation metrics hinder model generalization.
4.6. Summary of Findings
5. Discussion and Future Directions
5.1. Challenges and Limitations
5.1.1. Data-Related Challenges
- Class Imbalance and Bias: Medical datasets often exhibit class imbalances, with underrepresentation of rare conditions [102]. Models trained on such datasets may fail to generalize well to rare diseases, leading to biased predictions.
- Variability in Radiology Reports: Radiology reports are highly variable in structure and style, depending on institutional guidelines, radiologist preferences, and regional practices [103]. This variability poses challenges in training models that generalize across diverse settings.
5.1.2. Model Limitations
- Inability to Capture Fine-Grained Medical Details: Current deep learning models, particularly those based on CNN-RNN architectures, often struggle to capture intricate medical details, leading to incomplete or incorrect report generation [104].
- Hallucination in Text Generation: Transformer-based models, while effective for NLP tasks, have been observed to generate plausible but incorrect medical statements (hallucinations) [105]. Ensuring factual accuracy remains a critical challenge.
- Limited Explainability and Interpretability: Deep learning models often function as black boxes, making it difficult for radiologists to understand the rationale behind generated reports [106]. This lack of interpretability reduces clinical trust and adoption.
5.1.3. Clinical Integration Challenges
- Regulatory and Ethical Concerns: Deploying AI-driven radiology reporting systems in clinical practice requires adherence to strict regulatory guidelines (e.g., FDA approval). Ethical concerns related to accountability, liability, and patient safety also need careful consideration [109].
- Resistance to Adoption: Many radiologists remain skeptical about AI-generated reports due to concerns over accuracy and reliability. Seamless integration with existing radiology workflows and decision-support mechanisms is essential for practical adoption [110].
5.1.4. Evaluation Limitations
- Over-Reliance on NLP Metrics: Commonly used NLP evaluation metrics (BLEU, ROUGE, METEOR) do not fully capture the clinical correctness of generated reports. There is a need for domain-specific evaluation metrics that better reflect diagnostic accuracy.
- Lack of Standardized Benchmarks: There is no universally accepted benchmark dataset or evaluation framework for radiology report generation, making it difficult to compare models across studies.
- Limited Expert-Based Evaluation: Few studies involve radiologists in evaluating generated reports, which is crucial for assessing clinical relevance and diagnostic correctness [111].
5.2. Future Research Directions
5.2.1. Advancing Data Collection and Curation
- Expansion of Publicly Available Datasets: Efforts should be made to develop large-scale, well-annotated, and diverse datasets that cover a broad spectrum of diseases, imaging modalities, and demographic variations [112].
- Federated Learning for Privacy-Preserving AI: Federated learning enables training AI models across multiple institutions without sharing raw data, mitigating privacy concerns while improving model robustness.
- Standardization of Radiology Reports: Encouraging the use of structured reporting templates in radiology could reduce variability and improve the consistency of training data [113].
5.2.2. Improving Model Architectures
- Hybrid Deep Learning Models: Combining CNNs, Transformers, and graph-based learning approaches could enhance the model’s ability to capture both visual and textual relationships in medical images [114].
- Knowledge-Enhanced NLP Models: Incorporating medical knowledge graphs and ontologies (e.g., UMLS ) can improve the factual accuracy and interpretability of generated reports [115].
- Self-Supervised and Few-Shot Learning: Techniques such as contrastive learning and few-shot learning could enable models to learn from limited labeled data while improving generalization [116].
5.2.3. Enhancing Clinical Integration and Evaluation
- Clinician-in-the-Loop AI Systems: Future models should focus on AI-assisted reporting rather than full automation, allowing radiologists to edit, refine, and validate generated reports[117].
- Development of Clinically Meaningful Evaluation Metrics: Creating new evaluation frameworks that incorporate clinical accuracy, disease detection performance, and expert validation will be critical for reliable assessments.
- Real-World Clinical Trials: Conducting multi-institutional clinical trials to assess model performance in real-world radiology workflows is essential for validating AI-generated reports [118].
5.2.4. Regulatory and Ethical Considerations
- Establishing AI Governance Frameworks: Developing clear guidelines on AI-driven medical report generation, addressing issues of bias, transparency, and accountability.
- Ensuring Fairness and Bias Mitigation: Future research should focus on fairness-aware AI models that minimize biases related to gender, ethnicity, and socioeconomic factors in medical AI applications [119].
- Explainable AI for Clinical Trust: Implementing explainable AI (XAI) techniques, such as attention heatmaps and counterfactual explanations, could help radiologists better understand and trust AI-generated reports.
5.3. Summary of Discussion
6. Conclusion
6.1. Summary of Key Findings
- Shift Toward Transformer-Based Architectures: Traditional CNN-RNN frameworks have been progressively replaced by Transformer-based models, which offer improved language modeling and contextual understanding.
- Multimodal Learning as a Key Trend: The integration of medical image features with textual components (e.g., medical knowledge graphs, structured reports) has improved the factual consistency of generated reports.
- Challenges in Clinical Accuracy and Evaluation: Despite improvements in NLP metrics (e.g., BLEU, ROUGE, METEOR), these metrics do not fully capture the clinical correctness of reports, highlighting the need for expert-involved evaluations.
- Limited Dataset Availability and Bias Concerns: While datasets like MIMIC-CXR and IU X-ray have driven progress, issues such as dataset bias, imbalance, and privacy restrictions remain major barriers to real-world deployment.
- Need for Clinician-AI Collaboration: Fully automated report generation remains an ambitious goal; future systems should focus on AI-assisted reporting, where deep learning models support, rather than replace, radiologists.
6.2. Broader Impact and Clinical Implications
- Regulatory Approval: AI-driven medical applications require validation through rigorous clinical trials and compliance with regulatory standards (e.g., FDA, CE certification).
- Ethical Considerations: Ensuring fairness, transparency, and accountability in AI-generated reports is crucial to prevent biases that may negatively impact patient care.
- Seamless Integration into Radiology Workflows: AI models should complement radiologists’ decision-making rather than operate in isolation, allowing for real-time editing, feedback, and verification.
6.3. Future Outlook
- The creation of large-scale, diverse, and publicly available datasets with structured annotations to improve model generalization.
- The adoption of self-supervised learning and knowledge-enhanced AI to mitigate data scarcity issues and improve factual correctness in generated reports.
- The development of clinically meaningful evaluation metrics that assess diagnostic accuracy rather than mere text similarity.
- The implementation of explainable AI techniques to enhance transparency and trust among clinicians and regulatory bodies.
- The conduction of real-world clinical trials to evaluate the effectiveness of AI-assisted radiology reporting in hospital settings.
6.4. Final Remarks
References
- Smit, A.; Jain, S.; Rajpurkar, P.; Pareek, A.; Ng, A.; Lungren, M. Combining Automatic Labelers and Expert Annotations for Accurate Radiology Report Labeling Using BERT. In Proceedings of the EMNLP 2020, Online, 2020; pp. 1500–1519.
- Moradi, M.; Wong, K.C.L.; Syeda-Mahmood, T.; Wu, J.T. Identifying disease-free chest x-ray images with deep transfer learning. In Proceedings of the Medical Imaging 2019: Computer-Aided Diagnosis. SPIE, 2019, p. 24. [CrossRef]
- Rajaraman, S.; Sornapudi, S.; Kohli, M.; Antani, S. Assessment of an ensemble of machine learning models toward abnormality detection in chest radiographs. In Proceedings of the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC). IEEE, 2019, pp. 3689–3692. [CrossRef]
- Xue, Z.; Long, R.; Jaeger, S.; Folio, L.; George Thoma, R.; Antani, a.S. Extraction of Aortic Knuckle Contour in Chest Radiographs Using Deep Learning. Proceedings of the 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC). IEEE, 2018; 5890–5893. [Google Scholar] [CrossRef]
- Pan, I.; Agarwal, S.; Merck, D. Generalizable Inter-Institutional Classification of Abnormal Chest Radiographs Using Efficient Convolutional Neural Networks. Journal of Digital Imaging 2019, 32, 888–896. [Google Scholar] [CrossRef] [PubMed]
- Gyawali, P.K.; Ghimire, S.; Bajracharya, P.; Li, Z.; Wang, L. Semi-supervised Medical Image Classification with Global Latent Mixing. In Medical Image Computing and Computer Assisted Intervention – MICCAI 2020; Springer, 2020; Vol. 12261, pp. 604–613. [CrossRef]
- Majkowska, A.; Mittal, S.; Steiner, D.F.; Reicher, J.J.; McKinney, S.M.; Duggan, G.E.; Eswaran, K.; Cameron Chen, P.H.; Liu, Y.; Kalidindi, S.R.; et al. Chest Radiograph Interpretation with Deep Learning Models: Assessment with Radiologist-adjudicated Reference Standards and Population-adjusted Evaluation. Radiology 2019, 294, 421–431, Publisher: Radiological Society of North America. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Jia, H.; Lu, L.; Xia, Y. Thorax-Net: An Attention Regularized Deep Neural Network for Classification of Thoracic Diseases on Chest Radiography. IEEE Journal of Biomedical and Health Informatics 2020, 24, 475–485. [Google Scholar] [CrossRef] [PubMed]
- Dallal, A.H.; Agarwal, C.; Arbabshirani, M.R.; Patel, A.; Moore, G. Automatic estimation of heart boundaries and cardiothoracic ratio from chest x-ray images. Proceedings of the Medical Imaging 2017: Computer-Aided Diagnosis. SPIE, 2017; 101340K. [Google Scholar] [CrossRef]
- Yahyatabar, M.; Jouvet, P.; Cheriet, F. Dense-Unet: a light model for lung fields segmentation in Chest X-Ray images. In Proceedings of the 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC). IEEE, 2020, pp. 1242–1245. [CrossRef]
- Hermoza, R.; Maicas, G.; Nascimento, J.C.; Carneiro, G. Region Proposals for Saliency Map Refinement for Weakly-Supervised Disease Localisation and Classification. In Medical Image Computing and Computer Assisted Intervention – MICCAI 2020; Springer, 2020; Vol. 12266, pp. 539–549. [CrossRef]
- Cruz, B.G.S.; Bossa, M.N.; Sölter, J.; Husch, A.D. Public Covid-19 X-ray datasets and their impact on model bias - a systematic review of a significant problem. medRxiv 2021. [Google Scholar] [CrossRef]
- van Ginneken, B. Fifty years of computer analysis in chest imaging: rule-based, machine learning, deep learning. Radiological Physics and Technology 2017, 10, 23–32. [Google Scholar] [CrossRef]
- Zhao, Z.Q.; Zheng, P.; Xu, S.T.; Wu, X. Object Detection With Deep Learning: A Review. IEEE Transactions on Neural Networks and Learning Systems 2019, 30, 3212–3232. [Google Scholar] [CrossRef]
- Yuan, J.; Liao, H.; Luo, R.; Luo, J. Automatic Radiology Report Generation Based on Multi-view Image Fusion and Medical Concept Enrichment. In Medical Image Computing and Computer Assisted Intervention – MICCAI 2019; Springer, 2019; Vol. 11769, pp. 721–729. [CrossRef]
- von Berg, J.; Krönke, S.; Gooßen, A.; Bystrov, D.; Brück, M.; Harder, T.; Wieberneit, N.; Young, S. Robust chest x-ray quality assessment using convolutional neural networks and atlas regularization. In Proceedings of the Medical Imaging 2020: Image Processing. SPIE, 2020, p. 56. [CrossRef]
- Chauhan, G.; Liao, R.; Wells, W.; Andreas, J.; Wang, X.; Berkowitz, S.; Horng, S.; Szolovits, P.; Golland, P. Joint Modeling of Chest Radiographs and Radiology Reports for Pulmonary Edema Assessment. In Medical Image Computing and Computer Assisted Intervention – MICCAI 2020; Springer, 2020; Vol. 12262, pp. 529–539. [CrossRef]
- Feng, Y.; Teh, H.S.; Cai, Y. Deep Learning for Chest Radiology: A Review. Current Radiology Reports 2019, 7. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; v. d. Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2017, pp. 2261–2269. [CrossRef]
- Ypsilantis, P.; Montana, G. Learning What to Look in Chest X-rays with A Recurrent Visual Attention Model. CoRR 2017, abs/1701.06452, [1701.06452].
- Song, J.; Meng, C.; Ermon, S. Denoising Diffusion Implicit Models, 2022, [arXiv:cs.LG/2010.02502].
- Milletari, F.; Rieke, N.; Baust, M.; Esposito, M.; Navab, N. CFCM: Segmentation via Coarse to Fine Context Memory. In Medical Image Computing and Computer Assisted Intervention – MICCAI 2018; Springer, 2018; Vol. 11073, pp. 667–674. [CrossRef]
- Wang, L.; Ning, M.; Lu, D.; Wei, D.; Zheng, Y.; Chen, J. An Inclusive Task-Aware Framework for Radiology Report Generation. In Proceedings of the MICCAI 2022, 2022, Vol. 13438, pp. 568–577.
- RSNA. RSNA Pneumonia Detection Challenge, 2018. Library Catalog: www.kaggle.com.
- Bayat, A.; Sekuboyina, A.; Paetzold, J.C.; Payer, C.; Stern, D.; Urschler, M.; Kirschke, J.S.; Menze, B.H. Inferring the 3D Standing Spine Posture from 2D Radiographs. In Medical Image Computing and Computer Assisted Intervention – MICCAI 2020; Springer, 2020; Vol. 12266, pp. 775–784. [CrossRef]
- Wang, J.; Bhalerao, A.; He, Y. Cross-Modal Prototype Driven Network for Radiology Report Generation. In Proceedings of the ECCV, 2022, Vol. 13695, pp. 563–579.
- Cornia, M.; Stefanini, M.; Baraldi, L.; Cucchiara, R. Meshed-memory Transformer for Image Captioning. Proceedings of the CVPR, 2020; 10578–10587. [Google Scholar]
- Jarrett, K.; Kavukcuoglu, K.; Ranzato, M.; LeCun, Y. What is the best multi-stage architecture for object recognition? Proceedings of the Proceedings of the IEEE International Conference on Computer Vision. IEEE, 2009; 2146–2153. [Google Scholar] [CrossRef]
- Hata, A.; Yanagawa, M.; Yoshida, Y.; Miyata, T.; Tsubamoto, M.; Honda, O.; Tomiyama, N. Combination of Deep Learning–Based Denoising and Iterative Reconstruction for Ultra-Low-Dose CT of the Chest: Image Quality and Lung-RADS Evaluation. Am. J. Roentgenol. 2020, 215, 1321–1328. [Google Scholar] [CrossRef]
- Rubin, J.; Sanghavi, D.; Zhao, C.; Lee, K.; Qadir, A.; Xu-Wilson, M. Large Scale Automated Reading of Frontal and Lateral Chest X-Rays using Dual Convolutional Neural Networks 2018. [1804.07839].
- Bengio, Y.; Louradour, J.; Collobert, R.; Weston, J. Curriculum Learning. In Proceedings of the ICML, New York, NY, USA, 2009; p. 41–48.
- Sim, Y.; Chung, M.J.; Kotter, E.; Yune, S.; Kim, M.; Do, S.; Han, K.; Kim, H.; Yang, S.; Lee, D.J.; et al. Deep Convolutional Neural Network–based Software Improves Radiologist Detection of Malignant Lung Nodules on Chest Radiographs. Radiology 2019, 294, 199–209. [Google Scholar] [CrossRef]
- Crosby, J.; Chen, S.; Li, F.; MacMahon, H.; Giger, M. Network output visualization to uncover limitations of deep learning detection of pneumothorax. In Proceedings of the Medical Imaging 2020: Image Perception, Observer Performance, and Technology Assessment. SPIE, 2020, p. 22. [CrossRef]
- Thawakar, O.; Shaker, A.; Mullappilly, S.S.; Cholakkal, H.; Anwer, R.M.; Khan, S.H.; Laaksonen, J.; Khan, F.S. XrayGPT: Chest Radiographs Summarization using Medical Vision-Language Models. CoRR, 2306. [Google Scholar]
- Kim, Y.G.; Cho, Y.; Wu, C.J.; Park, S.; Jung, K.H.; Seo, J.B.; Lee, H.J.; Hwang, H.J.; Lee, S.M.; Kim, N. Short-term Reproducibility of Pulmonary Nodule and Mass Detection in Chest Radiographs: Comparison among Radiologists and Four Different Computer-Aided Detections with Convolutional Neural Net. Scientific Reports 2019, 9, 18738. [Google Scholar] [CrossRef] [PubMed]
- Xue, Z.; Antani, S.; Long, R.; Thoma, G.R. Using deep learning for detecting gender in adult chest radiographs. Proceedings of the Medical Imaging 2018: Imaging Informatics for Healthcare, Research, and Applications. SPIE, 2018; 10. [Google Scholar] [CrossRef]
- Neumann, M.; King, D.; Beltagy, I.; Ammar, W. ScispaCy: Fast and Robust Models for Biomedical Natural Language Processing. Proceedings of the BioNLP, Florence, Italy, 2019; 319–327. [Google Scholar]
- Schroeder, J.D.; Bigolin Lanfredi, R.; Li, T.; Chan, J.; Vachet, C.; Paine, R.; Srikumar, V.; Tasdizen, T. Prediction of Obstructive Lung Disease from Chest Radiographs via Deep Learning Trained on Pulmonary Function Data. International Journal of Chronic Obstructive Pulmonary Disease 2021, Volume 15, 3455–3466. [Google Scholar] [CrossRef]
- Brestel, C.; Shadmi, R.; Tamir, I.; Cohen-Sfaty, M.; Elnekave, E. RadBot-CXR: Classification of Four Clinical Finding Categories in Chest X-Ray Using Deep Learning. Proceedings of the International Conference on Medical Imaging with Deep Learning, 2018; 1–8. [Google Scholar]
- Yang, S.; Wu, X.; Ge, S.; Zhou, S.K.; Xiao, L. Knowledge Matters: Chest Radiology Report Generation with General and Specific Knowledge. Medical Image Anal. 2022, 80, 102510. [Google Scholar] [CrossRef]
- Demner-Fushman, D.; Kohli, M.D.; Rosenman, M.B.; Shooshan, S.E.; Rodriguez, L.; Antani, S.K.; Thoma, G.R.; McDonald, C.J. Preparing a collection of radiology examinations for distribution and retrieval. Journal of the American Medical Informatics Association 2016, 23, 304–310. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Schwab, E.; Rubin, J.; Klassen, P.; Liao, R.; Berkowitz, S.; Golland, P.; Horng, S.; Dalal, S. Pulmonary Edema Severity Estimation in Chest Radiographs Using Deep Learning. Proceedings of the International Conference on Medical Imaging with Deep Learning–Extended Abstract Track, 2019; 1. [Google Scholar]
- Guan, Q.; Huang, Y.; Zhong, Z.; Zheng, Z.; Zheng, L.; Yang, Y. Diagnose like a Radiologist: Attention Guided Convolutional Neural Network for Thorax Disease Classification. ArXiv 2018, abs/1801.09927.
- Cha, M.J.; Chung, M.J.; Lee, J.H.; Lee, K.S. Performance of Deep Learning Model in Detecting Operable Lung Cancer With Chest Radiographs. Journal of Thoracic Imaging 2019, 34, 86–91. [Google Scholar] [CrossRef]
- Bar, Y.; Diamant, I.; Wolf, L.; Greenspan, H. Deep learning with non-medical training used for chest pathology identification. In Proceedings of the Medical Imaging 2015: Computer-Aided Diagnosis. SPIE, mar 2015, Vol. 9414, p. 94140V. [CrossRef]
- Pham, V.T.; Tran, C.M.; Zheng, S.; Vu, T.M.; Nath, S. Chest x-ray abnormalities localization via ensemble of deep convolutional neural networks. Proceedings of the 2021 International Conference on Advanced Technologies for Communications (ATC). IEEE, 2021; 125–130. [Google Scholar]
- Shi, Z.; Liu, H.; Zhu, X. Enhancing Descriptive Image Captioning with Natural Language Inference. Proceedings of the ACL 2021, Online, 2021; 269–277. [Google Scholar]
- Sabottke, C.F.; Breaux, M.A.; Spieler, B.M. Estimation of age in unidentified patients via chest radiography using convolutional neural network regression. Emergency Radiology 2020, 27, 463–468. [Google Scholar] [CrossRef]
- Pavlopoulos, J.; Kougia, V.; Androutsopoulos, I. A Survey on Biomedical Image Captioning. In Proceedings of the Proceedings of the Second Workshop on Shortcomings in Vision and Language, Minneapolis, Minnesota, 2019; pp. 26–36.
- von Berg, J.; Young, S.; Carolus, H.; Wolz, R.; Saalbach, A.; Hidalgo, A.; Giménez, A.; Franquet, T. A novel bone suppression method that improves lung nodule detection. International Journal of Computer Assisted Radiology and Surgery 2015, 11, 641–655. [Google Scholar] [CrossRef] [PubMed]
- Dong, N.; Xu, M.; Liang, X.; Jiang, Y.; Dai, W.; Xing, E. Neural Architecture Search for Adversarial Medical Image Segmentation. In Medical Image Computing and Computer Assisted Intervention – MICCAI 2019; Springer, 2019; Vol. 11769, pp. 828–836. [CrossRef]
- Salehinejad, H.; Colak, E.; Dowdell, T.; Barfett, J.; Valaee, S. Synthesizing Chest X-Ray Pathology for Training Deep Convolutional Neural Networks. IEEE Transactions on Medical Imaging 2019, 38, 1197–1206. [Google Scholar] [CrossRef]
- Park, S.; Lee, S.M.; Kim, N.; Choe, J.; Cho, Y.; Do, K.H.; Seo, J.B. Application of deep learning–based computer-aided detection system: detecting pneumothorax on chest radiograph after biopsy. European Radiology 2019, 29, 5341–5348. [Google Scholar] [CrossRef]
- Liu, H.; Wang, L.; Nan, Y.; Jin, F.; Wang, Q.; Pu, J. SDFN: Segmentation-based deep fusion network for thoracic disease classification in chest X-ray images. Computerized Medical Imaging and Graphics 2019, 75, 66–73. [Google Scholar] [CrossRef]
- Qin, Z.Z.; Sander, M.S.; Rai, B.; Titahong, C.N.; Sudrungrot, S.; Laah, S.N.; Adhikari, L.M.; Carter, E.J.; Puri, L.; Codlin, A.J.; et al. Using artificial intelligence to read chest radiographs for tuberculosis detection: A multi-site evaluation of the diagnostic accuracy of three deep learning systems. Scientific Reports 2019, 9, 15000. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.; Song, H.; Shin, Y.J.; Kim, J.; Kim, J.; Lee, K.W.; Lee, S.S.; Lee, W.; Lee, S.; Lee, K.H. Deep Learning–based Automatic Detection Algorithm for Reducing Overlooked Lung Cancers on Chest Radiographs. Radiology 2020, 296, 652–661. [Google Scholar] [CrossRef]
- Owais, M.; Arsalan, M.; Mahmood, T.; Kim, Y.H.; Park, K.R. Comprehensive Computer-Aided Decision Support Framework to Diagnose Tuberculosis From Chest X-Ray Images: Data Mining Study. JMIR Medical Informatics 2020, 8, e21790. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, J.R.; Armando Cardona Cardenas, D.; Moreno, R.A.; de Fatima de Sa Rebelo, M.; Krieger, J.E.; Antonio Gutierrez, M. Multi-View Ensemble Convolutional Neural Network to Improve Classification of Pneumonia in Low Contrast Chest X-Ray Images. In Proceedings of the 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC). IEEE, 2020, pp. 1238–1241. [CrossRef]
- Yang, W.; Chen, Y.; Liu, Y.; Zhong, L.; Qin, G.; Lu, Z.; Feng, Q.; Chen, W. Cascade of multi-scale convolutional neural networks for bone suppression of chest radiographs in gradient domain. Medical Image Analysis 2017, 35, 421–433. [Google Scholar] [CrossRef] [PubMed]
- López-Cabrera, J.D.; Orozco-Morales, R.; Portal-Diaz, J.A.; Lovelle-Enríquez, O.; Pérez-Díaz, M. Current limitations to identify COVID-19 using artificial intelligence with chest X-ray imaging. Health and Technology 2021, 11, 411–424. [Google Scholar] [CrossRef]
- Sethy, P.K.; Behera, S.K.; Anitha, K.; Pandey, C.; Khan, M. Computer aid screening of COVID-19 using X-ray and CT scan images: An inner comparison. Journal of X-Ray Science and Technology 2021, pp. 1–14. [CrossRef]
- Yan, B.; Pei, M.; Zhao, M.; Shan, C.; Tian, Z. Prior Guided Transformer for Accurate Radiology Reports Generation. IEEE Journal of Biomedical and Health Informatics 2022, 26, 5631–5640. [Google Scholar] [CrossRef]
- Zucker, E.J.; Barnes, Z.A.; Lungren, M.P.; Shpanskaya, Y.; Seekins, J.M.; Halabi, S.S.; Larson, D.B. Deep learning to automate Brasfield chest radiographic scoring for cystic fibrosis. Journal of Cystic Fibrosis 2020, 19, 131–138. [Google Scholar] [CrossRef]
- Miura, Y.; Zhang, Y.; Tsai, E.; Langlotz, C.; Jurafsky, D. Improving Factual Completeness and Consistency of Image-to-text Radiology Report Generation. Proceedings of the NAACL 2021, Online, 2021; 5288–5304. [Google Scholar]
- Singh, R.K.; Pandey, R.; Babu, R.N. COVIDScreen: explainable deep learning framework for differential diagnosis of COVID-19 using chest X-rays. Neural Computing and Applications 2021. [Google Scholar] [CrossRef]
- Lee, D.; Kim, H.; Choi, B.; Kim, H.J. Development of a deep neural network for generating synthetic dual-energy chest x-ray images with single x-ray exposure. Physics in Medicine & Biology 2019, 64, 115017. [Google Scholar] [CrossRef]
- Adams, S.J.; Henderson, R.D.E.; Yi, X.; Babyn, P. Artificial Intelligence Solutions for Analysis of X-ray Images. Can. Assoc. Radiol. J. 2021, 72, 60–72. [Google Scholar] [CrossRef]
- Bonheur, S.; Štern, D.; Payer, C.; Pienn, M.; Olschewski, H.; Urschler, M. Matwo-CapsNet: A Multi-label Semantic Segmentation Capsules Network. In Medical Image Computing and Computer Assisted Intervention – MICCAI 2019; Springer, 2019; Vol. 11768, pp. 664–672. [CrossRef]
- Yu, X.; Wang, S.H.; Zhang, Y.D. CGNet: A graph-knowledge embedded convolutional neural network for detection of pneumonia. Information Processing & Management 2021, 58, 102411. [Google Scholar] [CrossRef]
- Yang, L.; Wang, Z.; Zhou, L. MedXChat: Bridging CXR Modalities with a Unified Multimodal Large Model, 2023, [arXiv:cs.CV/2312.02233].
- Kholiavchenko, M.; Sirazitdinov, I.; Kubrak, K.; Badrutdinova, R.; Kuleev, R.; Yuan, Y.; Vrtovec, T.; Ibragimov, B. Contour-aware multi-label chest X-ray organ segmentation. International Journal of Computer Assisted Radiology and Surgery 2020, 15, 425–436. [Google Scholar] [CrossRef] [PubMed]
- Pandit, M.; Banday, S.; Naaz, R.; Chishti, M. Automatic detection of COVID-19 from chest radiographs using deep learning. Radiography 2020, p. S1078817420302285. [CrossRef]
- Yao, L.; Prosky, J.; Poblenz, E.; Covington, B.; Lyman, K. Weakly Supervised Medical Diagnosis and Localization from Multiple Resolutions 2018. [1803.07703].
- Nour, M.; Cömert, Z.; Polat, K. A Novel Medical Diagnosis model for COVID-19 infection detection based on Deep Features and Bayesian Optimization. Applied Soft Computing 2020, 97, 106580. [Google Scholar] [CrossRef] [PubMed]
- Olatunji, T.; Yao, L.; Covington, B.; Upton, A. Caveats in Generating Medical Imaging Labels from Radiology Reports with Natural Language Processing. In Proceedings of the International Conference on Medical Imaging with Deep Learning – Extended Abstract Track, 08–10 Jul 2019, pp. 1–4.
- Lu, M.T.; Raghu, V.K.; Mayrhofer, T.; Aerts, H.J.; Hoffmann, U. Deep Learning Using Chest Radiographs to Identify High-Risk Smokers for Lung Cancer Screening Computed Tomography: Development and Validation of a Prediction Model. Annals of Internal Medicine 2020, 173, 704–713. [Google Scholar] [CrossRef]
- Jones, D.R.; Schonlau, M.; Welch, W.J. Efficient Global Optimization of Expensive Black-Box Functions. J. Global Optimization 1998, 13, 455–492. [Google Scholar] [CrossRef]
- Shiraishi, J.; Katsuragawa, S.; Ikezoe, J.; Matsumoto, T.; Kobayashi, T.; Komatsu, K.i.; Matsui, M.; Fujita, H.; Kodera, Y.; Doi, K. Development of a digital image database for chest radiographs with and without a lung nodule: receiver operating characteristic analysis of radiologists’ detection of pulmonary nodules. American Journal of Roentgenology 2000, 174, 71–74. [Google Scholar] [CrossRef]
- Yue, Z.; Ma, L.; Zhang, R. Comparison and Validation of Deep Learning Models for the Diagnosis of Pneumonia. Computational Intelligence and Neuroscience 2020, 2020, 1–8. [Google Scholar] [CrossRef]
- Wong, K.C.L.; Moradi, M.; Wu, J.; Pillai, A.; Sharma, A.; Gur, Y.; Ahmad, H.; Chowdary, M.S.; Chiranjeevi, J.; Reddy Polaka, K.K.; et al. A Robust Network Architecture to Detect Normal Chest X-Ray Radiographs. Proceedings of the 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI). IEEE, 2020; 1851–1855. [Google Scholar] [CrossRef]
- Madaan, V.; Roy, A.; Gupta, C.; Agrawal, P.; Sharma, A.; Bologa, C.; Prodan, R. XCOVNet: Chest X-ray Image Classification for COVID-19 Early Detection Using Convolutional Neural Networks. New Generation Computing 2021. [Google Scholar] [CrossRef]
- Bortsova, G.; Dubost, F.; Hogeweg, L.; Katramados, I.; de Bruijne, M. Semi-supervised Medical Image Segmentation via Learning Consistency Under Transformations. In Medical Image Computing and Computer Assisted Intervention – MICCAI 2019; Springer, 2019; Vol. 11769, pp. 810–818. [CrossRef]
- Heo, S.J.; Kim, Y.; Yun, S.; Lim, S.S.; Kim, J.; Nam, C.M.; Park, E.C.; Jung, I.; Yoon, J.H. Deep Learning Algorithms with Demographic Information Help to Detect Tuberculosis in Chest Radiographs in Annual Workers’ Health Examination Data. International Journal of Environmental Research and Public Health 2019, 16, 250. [Google Scholar] [CrossRef]
- Jiang, W.; Ma, L.; Jiang, Y.G.; Liu, W.; Zhang, T. Recurrent Fusion Network for Image Captioning. Proceedings of the ECCV 2018, Berlin, Heidelberg, 2018; 510–526. [Google Scholar]
- Arsalan, M.; Owais, M.; Mahmood, T.; Choi, J.; Park, K.R. Artificial Intelligence-Based Diagnosis of Cardiac and Related Diseases. Journal of Clinical Medicine 2020, 9, 871. [Google Scholar] [CrossRef]
- Chokshi, F.H.; Flanders, A.E.; Prevedello, L.M.; Langlotz, C.P. Fostering a Healthy AI Ecosystem for Radiology: Conclusions of the 2018 RSNA Summit on AI in Radiology. Radiology: Artificial Intelligence 2019, 1, 190021. [Google Scholar] [CrossRef] [PubMed]
- Márquez-Neila, P.; Sznitman, R. Image Data Validation for Medical Systems. In Medical Image Computing and Computer Assisted Intervention – MICCAI 2019; Springer, 2019; Vol. 11767, pp. 329–337. [CrossRef]
- Anavi, Y.; Kogan, I.; Gelbart, E.; Geva, O.; Greenspan, H. Visualizing and enhancing a deep learning framework using patients age and gender for chest x-ray image retrieval. In Proceedings of the Medical Imaging 2016: Computer-Aided Diagnosis. SPIE, jul 2016, Vol. 9785, p. 978510. [CrossRef]
- Liu, H.; Li, C.; Wu, Q.; Lee, Y.J. Visual Instruction Tuning. CoRR, 2304. [Google Scholar]
- Candemir, S.; Jaeger, S.; Lin, W.; Xue, Z.; Antani, S.K.; Thoma, G.R. Automatic heart localization and radiographic index computation in chest x-rays. In Proceedings of the Medical Imaging 2016: Computer-Aided Diagnosis. SPIE, 2016, Vol. 9785, p. 978517. [CrossRef]
- Liu, F.; Wu, X.; Ge, S.; Fan, W.; Zou, Y. Exploring and Distilling Posterior and Prior Knowledge for Radiology Report Generation. In Proceedings of the CVPR 2021, 2021, pp. 13753–13762. [Google Scholar]
- Daniels, Z.A.; Metaxas, D.N. Exploiting Visual and Report-Based Information for Chest X-RAY Analysis by Jointly Learning Visual Classifiers and Topic Models. Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019). IEEE, 2019; 1270–1274. [Google Scholar] [CrossRef]
- Bergstra, J.S.; Bardenet, R.; Bengio, Y.; Kégl, B. Algorithms for Hyper-Parameter Optimization. Proceedings of the Advances in Neural Information Processing Systems, 2011; 2546–2554. [Google Scholar]
- Rombach, R.; Blattmann, A.; Lorenz, D.; Esser, P.; Ommer, B. High-resolution Image Synthesis with Latent Diffusion Models. Proceedings of the CVPR, 2022; 10684–10695. [Google Scholar]
- Polat, C.; Karaman, O.; Karaman, C.; Korkmaz, G.; Balcı, M.C.; Kelek, S.E. COVID-19 diagnosis from chest X-ray images using transfer learning: Enhanced performance by debiasing dataloader. Journal of X-Ray Science and Technology 2021, 29, 19–36. [Google Scholar] [CrossRef]
- Candemir, S.; Rajaraman, S.; Thoma, G.; Antani, S. Deep Learning for Grading Cardiomegaly Severity in Chest X-Rays: An Investigation. Proceedings of the Life Sciences Conference, 2018; 109–113. [Google Scholar] [CrossRef]
- You, D.; Liu, F.; Ge, S.; Xie, X.; Zhang, J.; Wu, X. AlignTransformer: Hierarchical Alignment of Visual Regions and Disease Tags for Medical Report Generation. In Proceedings of the MICCAI 2021, 2021, Vol. 12903, pp. 72–82.
- Arjovsky, M.; Chintala, S.; Bottou, L. Wasserstein Generative Adversarial Networks. In Proceedings of the Proceedings of the 34th International Conference on Machine Learning. PMLR, 06–11 Aug 2017, Vol. 70, Proceedings of Machine Learning Research, pp. 214–223.
- Kruger, R.P.; Townes, J.R.; Hall, D.L.; Dwyer, S.J.; Lodwick, G.S. Automated Radiographic Diagnosis via Feature Extraction and Classification of Cardiac Size and Shape Descriptors. IEEE Transactions on Biomedical Engineering 1972, BME-19, 174–186. [CrossRef]
- Gu, X.; Pan, L.; Liang, H.; Yang, R. Classification of Bacterial and Viral Childhood Pneumonia Using Deep Learning in Chest Radiography. In Proceedings of the Proceedings of the 3rd International Conference on Multimedia and Image Processing - ICMIP 2018. ACM Press, 2018. [CrossRef]
- Tien, H.J.; Yang, H.C.; Shueng, P.W.; Chen, J.C. Cone-beam CT image quality improvement using Cycle-Deblur consistent adversarial networks (Cycle-Deblur GAN) for chest CT imaging in breast cancer patients. Sci. Rep. 2021, 11, 1133. [Google Scholar] [CrossRef] [PubMed]
- Bigolin Lanfredi, R.; Schroeder, J.D.; Vachet, C.; Tasdizen, T. Adversarial Regression Training for Visualizing the Progression of Chronic Obstructive Pulmonary Disease with Chest X-Rays. In Medical Image Computing and Computer Assisted Intervention – MICCAI 2019; Springer, 2019; Vol. 11769, pp. 685–693. [CrossRef]
- Irvin, J.; Rajpurkar, P.; Ko, M.; Yu, Y.; Ciurea-Ilcus, S.; Chute, C.; Marklund, H.; Haghgoo, B.; Ball, R.L.; Shpanskaya, K.S.; et al. CheXpert: A Large Chest Radiograph Dataset with Uncertainty Labels and Expert Comparison. In Proceedings of the AAAI 2019, 2019, pp. 590–597. [Google Scholar] [CrossRef]
- Portela, R.D.S.; Pereira, J.R.G.; Costa, M.G.F.; Filho, C.F.F.C. Lung Region Segmentation in Chest X-Ray Images using Deep Convolutional Neural Networks. In Proceedings of the 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC). IEEE, 2020, pp. 1246–1249. [CrossRef]
- Li, J.; Li, D.; Savarese, S.; Hoi, S.C.H. BLIP-2: Bootstrapping Language-Image Pre-training with Frozen Image Encoders and Large Language Models. In Proceedings of the ICML, 2023, Vol. 202, pp. 19730–19742.
- Yang, Y.; Lure, F.Y.; Miao, H.; Zhang, Z.; Jaeger, S.; Liu, J.; Guo, L. Using artificial intelligence to assist radiologists in distinguishing COVID-19 from other pulmonary infections. Journal of X-Ray Science and Technology 2021, 29, 1–17. [Google Scholar] [CrossRef]
- Liang, C.H.; Liu, Y.C.; Wu, M.T.; Garcia-Castro, F.; Alberich-Bayarri, A.; Wu, F.Z. Identifying pulmonary nodules or masses on chest radiography using deep learning: external validation and strategies to improve clinical practice. Clinical Radiology 2020, 75, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Toriwaki, J.I.; Suenaga, Y.; Negoro, T.; Fukumura, T. Pattern recognition of chest X-ray images. Computer Graphics and Image Processing 1973, 2, 252–271. [Google Scholar] [CrossRef]
- Gozes, O.; Greenspan, H. Lung Structures Enhancement in Chest Radiographs via CT Based FCNN Training. In Image Analysis for Moving Organ, Breast, and Thoracic Images; Springer, 2018; pp. 147–158. [CrossRef]
- Saednia, K.; Jalalifar, A.; Ebrahimi, S.; Sadeghi-Naini, A. An Attention-Guided Deep Neural Network for Annotating Abnormalities in Chest X-ray Images: Visualization of Network Decision Basis *. In Proceedings of the 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC). IEEE, 2020, pp. 1258–1261. [CrossRef]
- Singh, R.; Kalra, M.K.; Nitiwarangkul, C.; Patti, J.A.; Homayounieh, F.; Padole, A.; Rao, P.; Putha, P.; Muse, V.V.; Sharma, A.; et al. Deep learning in chest radiography: Detection of findings and presence of change. PloS One 2018, 13, e0204155. [Google Scholar] [CrossRef]
- Bustos, A.; Pertusa, A.; Salinas, J.M.; de la Iglesia-Vayá, M. PadChest: A Large Chest X-Ray Image Dataset with Multi-Label Annotated Reports. Medical Image Analysis 2020, 66, 101797. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Guo, H.; Yi, K.; Li, B.; Elhoseiny, M. VisualGPT: Data-Efficient Adaptation of Pretrained Language Models for Image Captioning. Proceedings of the CVPR, 2022; 18009–18019. [Google Scholar]
- Chlebus, G.; Schenk, A.; Hendrik Moltz, J.; van Ginneken, B.; Meine, H.; Karl Hahn, H. Automatic liver tumor segmentation in CT with fully convolutional neural networks and object-based postprocessing. Scientific Reports 2018, 8, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Ghesu, F.C.; Georgescu, B.; Gibson, E.; Guendel, S.; Kalra, M.K.; Singh, R.; Digumarthy, S.R.; Grbic, S.; Comaniciu, D. Quantifying and Leveraging Classification Uncertainty for Chest Radiograph Assessment. In Medical Image Computing and Computer Assisted Intervention – MICCAI 2019; Springer, 2019; Vol. 11769, pp. 676–684. [CrossRef]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. Proceedings of the IEEE conference on computer vision and pattern recognition, 2015; 1–9. [Google Scholar]
- Pham, V.T.; Nguyen, T.P. Identification and localization covid-19 abnormalities on chest radiographs. Proceedings of the The International Conference on Artificial Intelligence and Computer Vision. Springer, 2023; 251–261. [Google Scholar]
- van Leeuwen, K.G.; Schalekamp, S.; Rutten, M.J.; van Ginneken, B.; de Rooij, M. Artificial intelligence in Radiology; 100 commercially available products and their scientific evidence, 2021. European Radiology (in press).
- Kim, D.W.; Jang, H.Y.; Kim, K.W.; Shin, Y.; Park, S.H. Design Characteristics of Studies Reporting the Performance of Artificial Intelligence Algorithms for Diagnostic Analysis of Medical Images: Results from Recently Published Papers. Korean Journal of Radiology 2019, 20, 405. [Google Scholar] [CrossRef] [PubMed]
- Anavi, Y.; Kogan, I.; Gelbart, E.; Geva, O.; Greenspan, H. A comparative study for chest radiograph image retrieval using binary texture and deep learning classification. International Conference of the IEEE Engineering in Medicine and Biology Society 2015, 2015, 2940–2943. [Google Scholar] [CrossRef]
- Ouyang, X.; Xue, Z.; Zhan, Y.; Zhou, X.S.; Wang, Q.; Zhou, Y.; Wang, Q.; Cheng, J.Z. Weakly Supervised Segmentation Framework with Uncertainty: A Study on Pneumothorax Segmentation in Chest X-ray. In Medical Image Computing and Computer Assisted Intervention – MICCAI 2019; Springer, 2019; Vol. 11769, pp. 613–621. [CrossRef]
- Kallianos, K.; Mongan, J.; Antani, S.; Henry, T.; Taylor, A.; Abuya, J.; Kohli, M. How far have we come? Artificial intelligence for chest radiograph interpretation. Clinical Radiology 2019, 74, 338–345. [Google Scholar] [CrossRef]
| Study | Dataset | Model Architecture | Evaluation Metrics |
|---|---|---|---|
| Author et al. (Year) | MIMIC-CXR | CNN-RNN | BLEU, ROUGE |
| Author et al [76]. (Year) | IU X-ray | Transformer-based | METEOR, Clinical Accuracy |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




