Submitted:
12 March 2025
Posted:
13 March 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
1.1. The Challenges of Traditional Drug Discovery
1.2. The Emergence of AI in Drug Discovery
1.3. AI-Powered Approaches in Drug Discovery
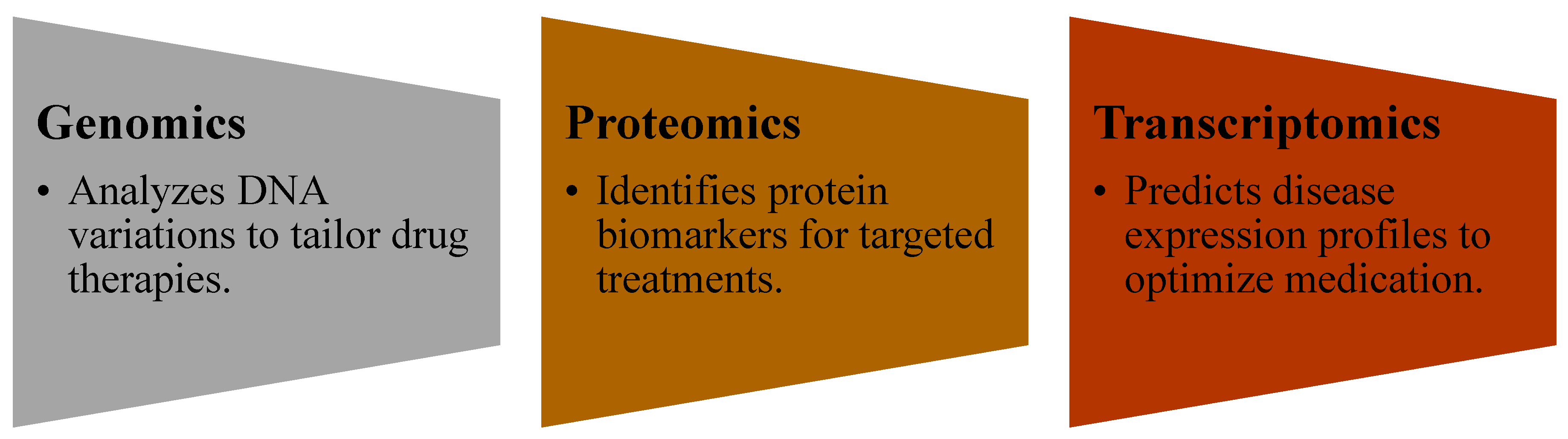
1.4. The Importance of AI in Personalized Medicine
1.5. Challenges and Ethical Considerations
1.6. Scope of the Review
2. Methods
2.1. Study Design
2.2. Data Sources and Search Strategy
- PubMed (for biomedical and pharmacological studies)
- Scopus (for multidisciplinary peer-reviewed research)
- Web of Science (for high-impact scientific publications)
- Google Scholar (for AI and computational biology research)
- ClinicalTrials.gov (for ongoing AI-driven drug discovery studies)
- ("Artificial Intelligence" OR "Machine Learning" OR "Deep Learning") AND ("Drug Discovery" OR "Molecular Design" OR "Target Identification")
- ("AI in Pharmacology") AND ("Structure-Based Drug Design" OR "Ligand-Based Drug Design")
2.3. Inclusion and Exclusion Criteria
- Peer-reviewed articles published in English between 2015-2025.
- Studies that discuss AI applications in target identification, molecular docking, or lead optimization.
- Research incorporating machine learning, deep learning, or computational biology in drug discovery pipelines.
- Case studies or clinical trials demonstrating AI's effectiveness in drug development.
- Non-peer-reviewed articles, opinion papers, or editorials.
- Studies focusing solely on traditional (non-AI) drug discovery techniques.
- AI research not related to pharmaceuticals or molecular design.
- Articles lacking clear methodology or experimental validation.
2.4. Data Extraction and Analysis
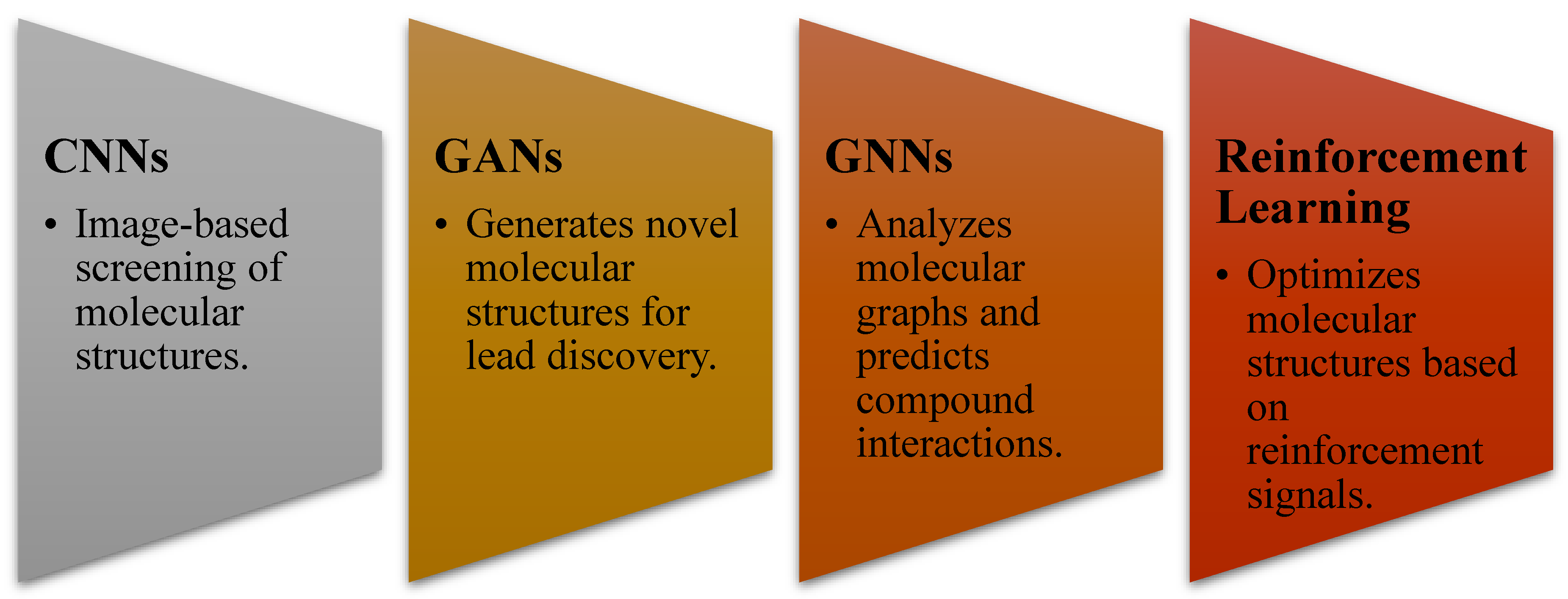
- AI methodologies used (ML, DL, GANs, CNNs, RNNs, etc.)
- Datasets utilized (genomic, proteomic, clinical, pharmacokinetic, etc.)
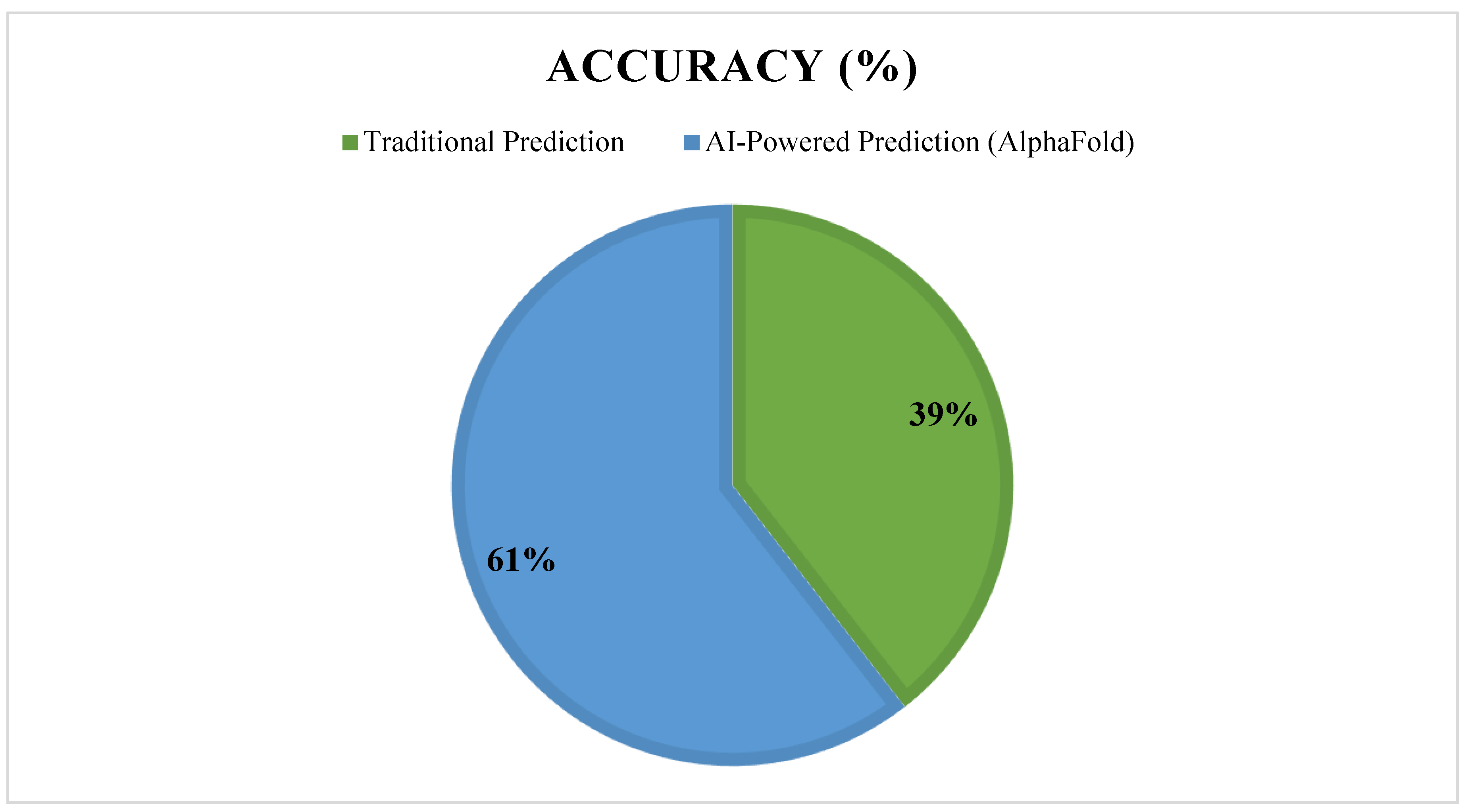
- Metrics used to evaluate AI model performance (accuracy, recall, precision, ROC-AUC scores, etc.)
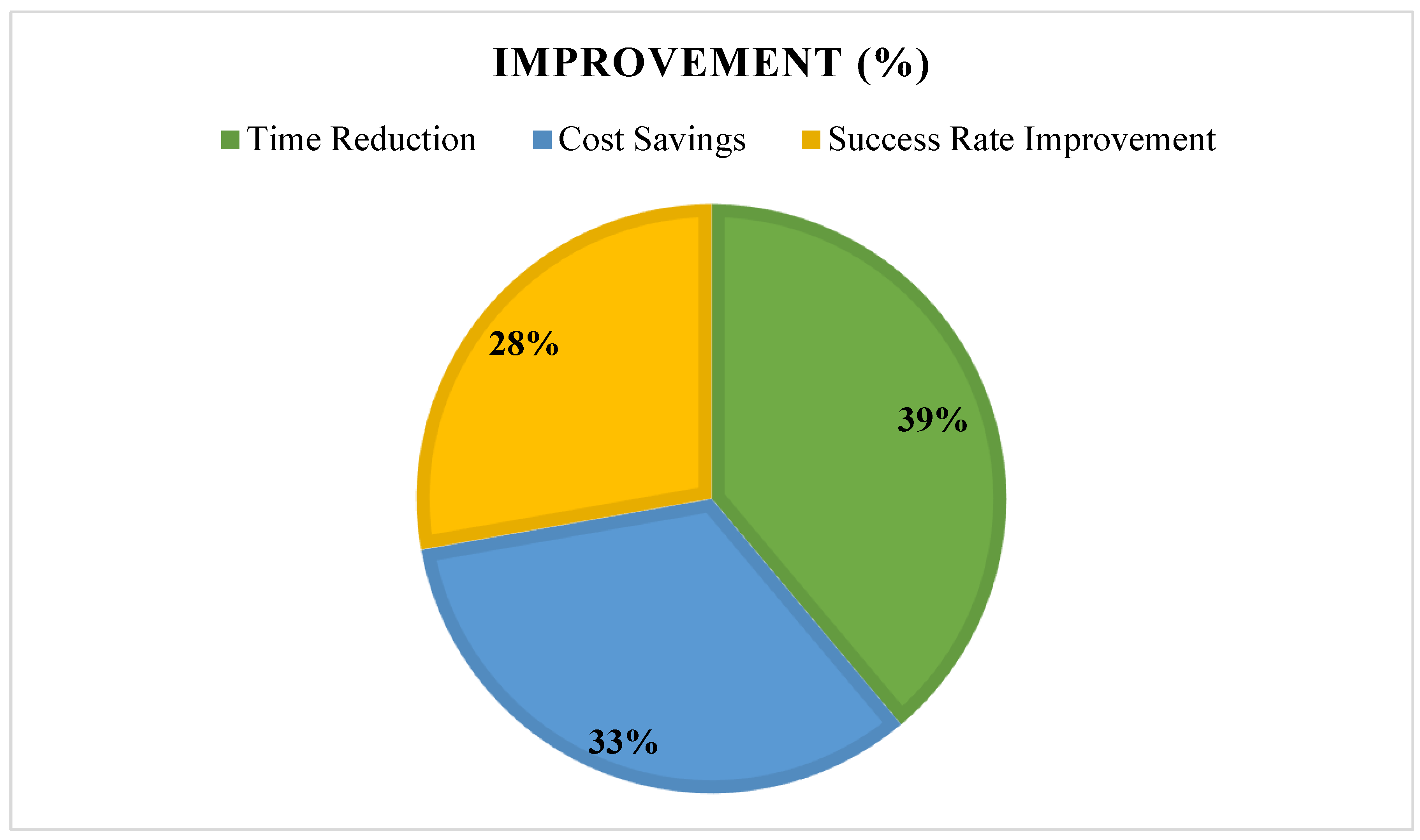
- Clinical relevance of AI-driven findings (success in preclinical/clinical trials), see see Figure 11.
2.5. Quality Assessment and Bias Control
3. Results
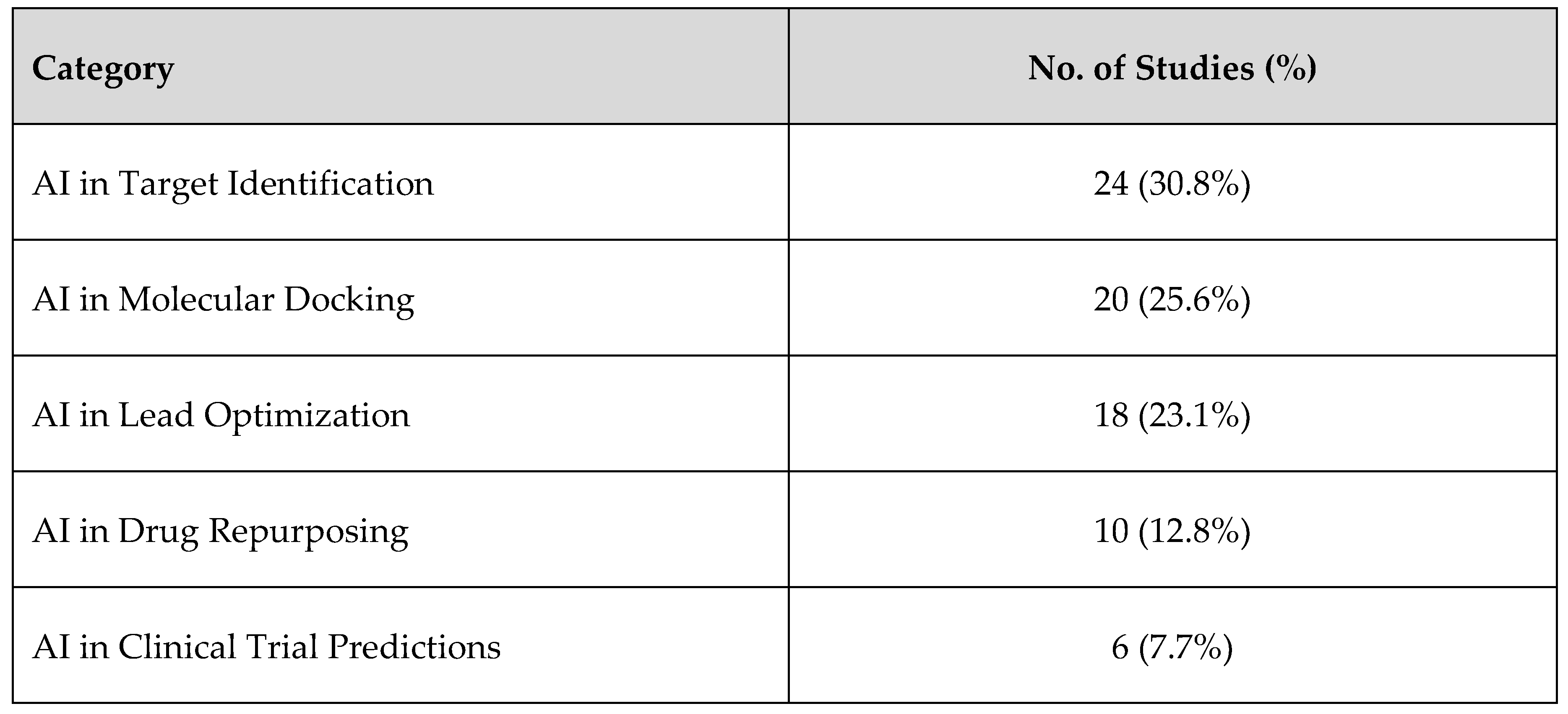
3.1. Overview of Selected Studies
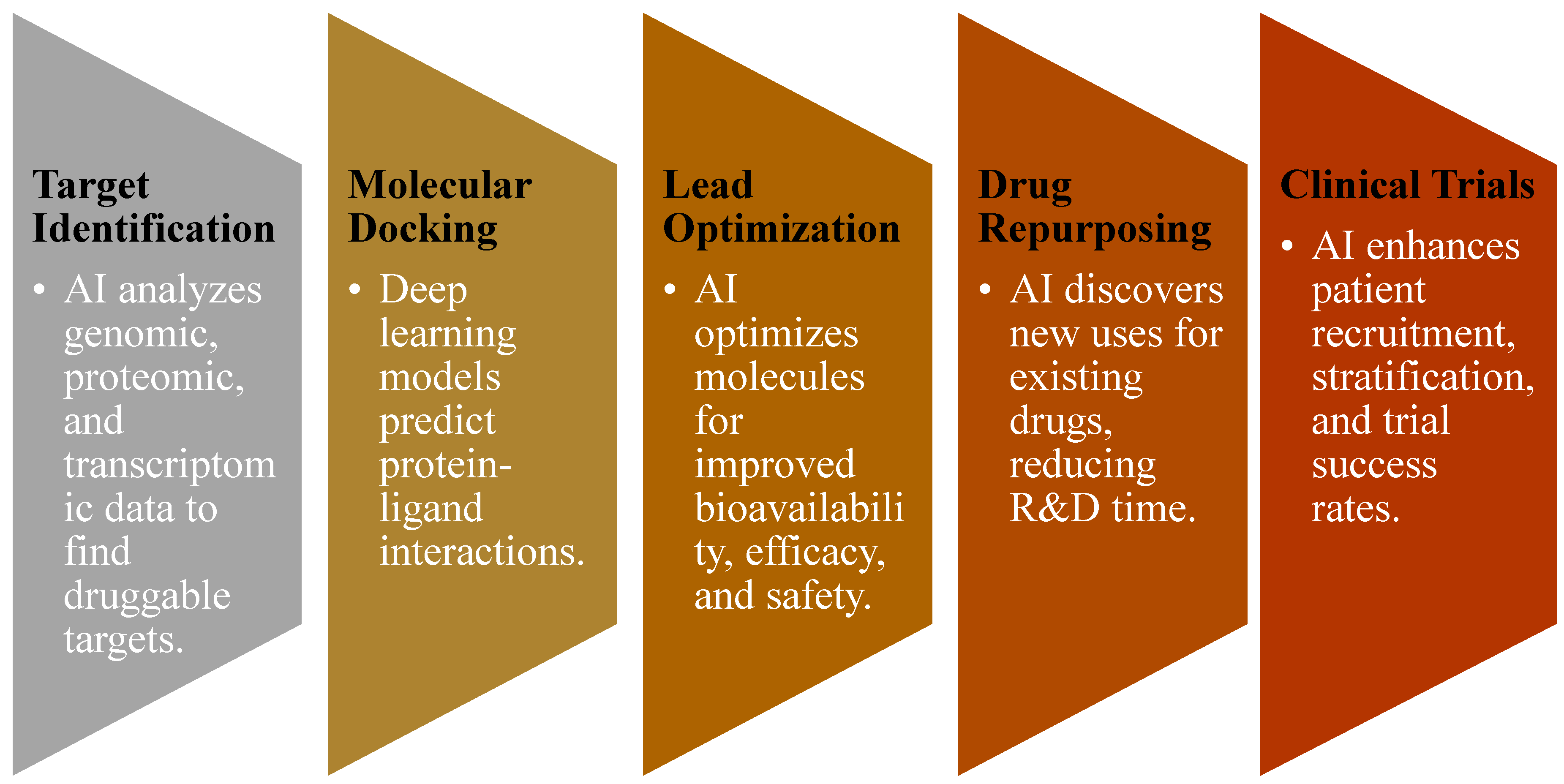
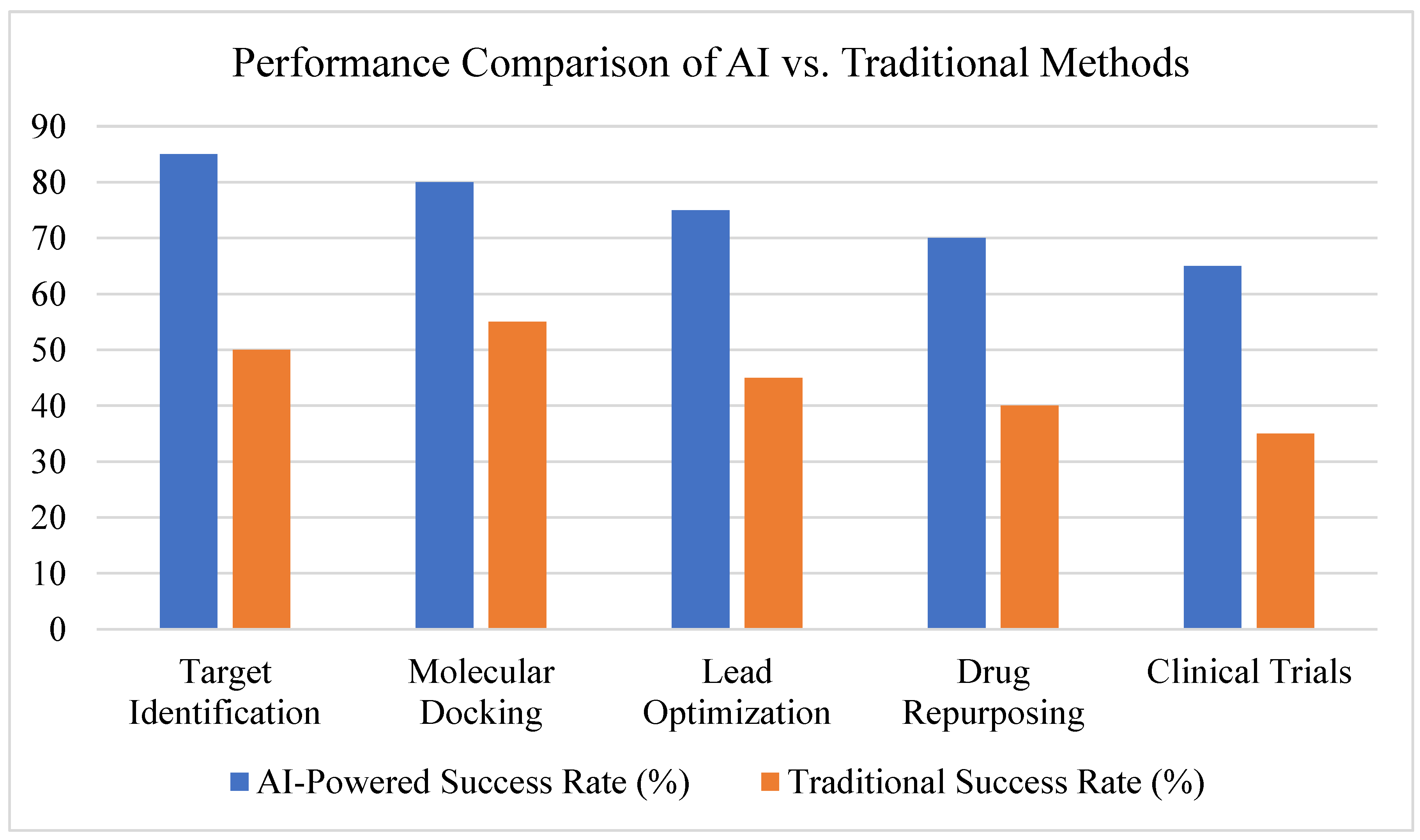
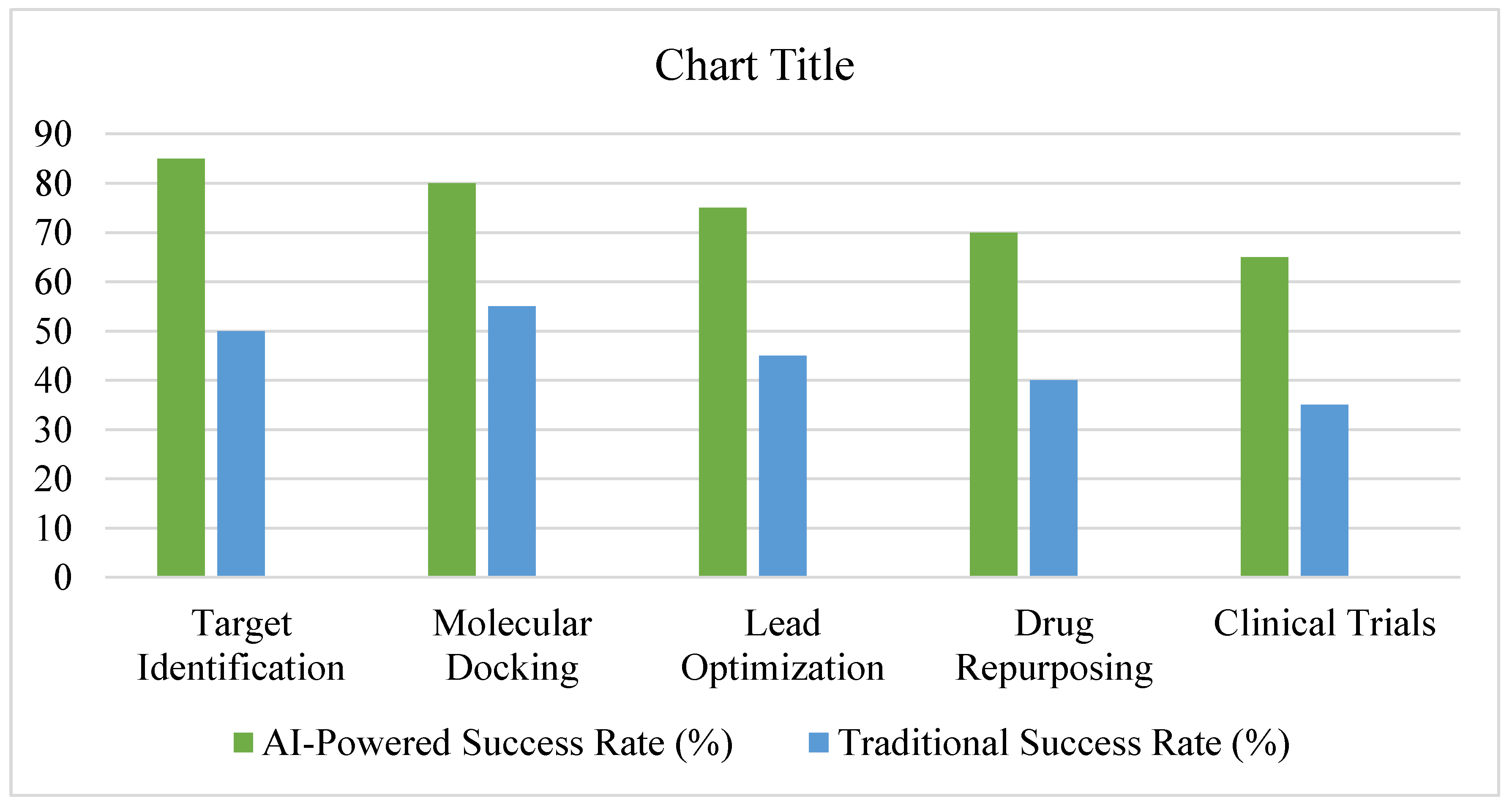
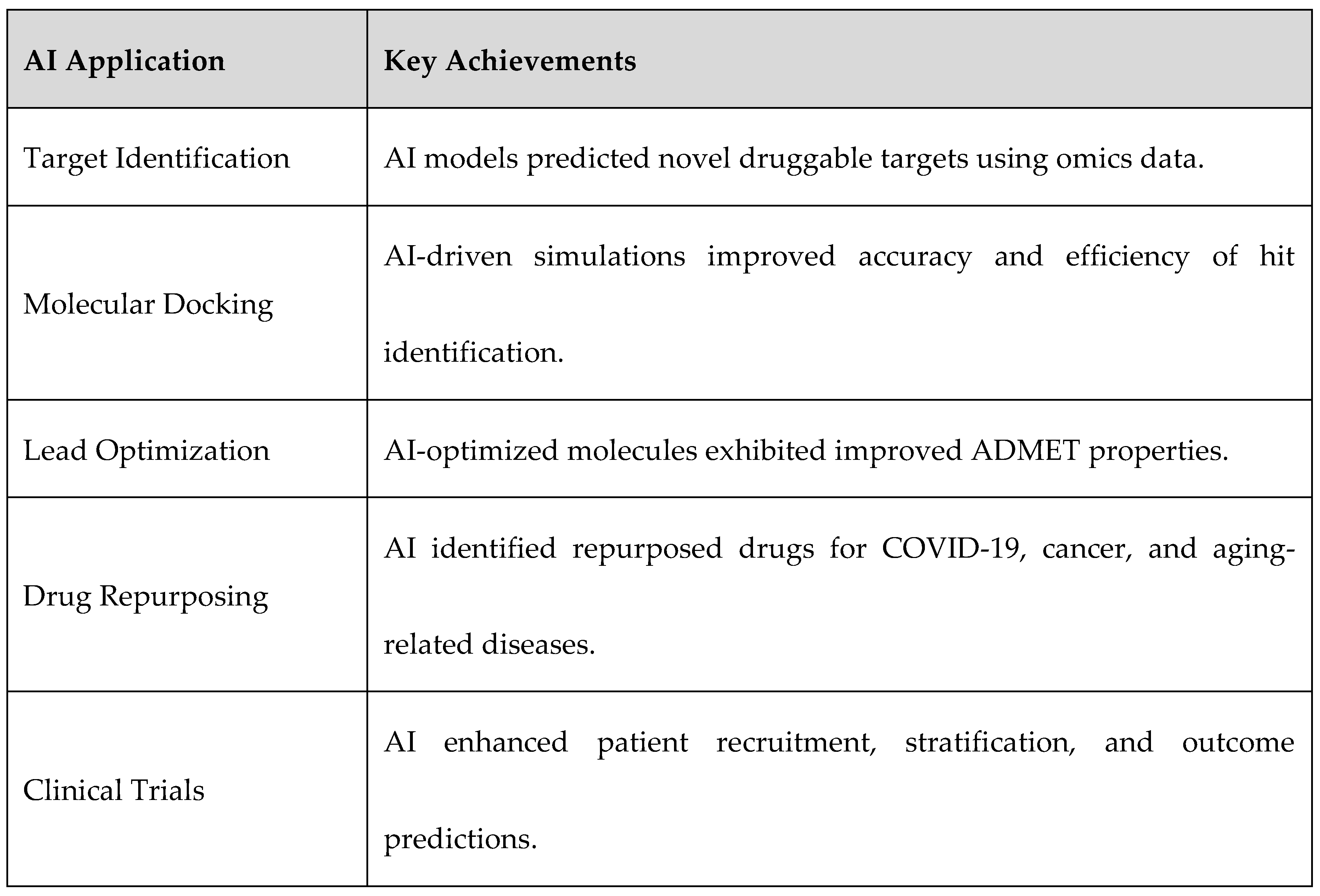
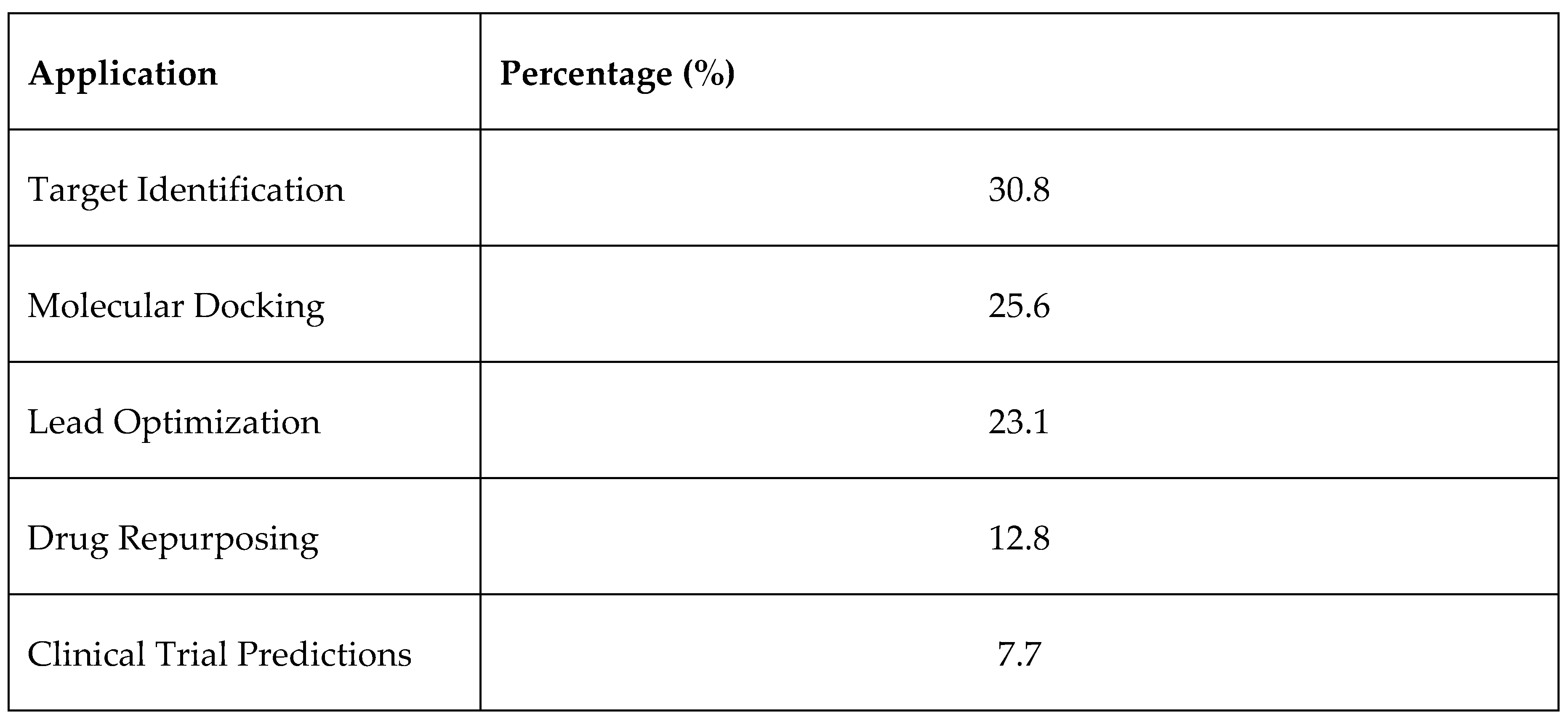
3.2. AI in Target Identification
- DeepMind’s AlphaFold: Revolutionized protein structure prediction, aiding in novel target identification for drug discovery [24].
- BenevolentAI: Utilized AI to identify Janus kinase inhibitors (JAK) as potential COVID-19 treatments, demonstrating AI's capability in target discovery [25].
- AI-Driven CRISPR Screening: AI-assisted genome-wide CRISPR screening has led to the identification of essential oncogenes and tumor suppressor targets [26].
- Reduces time required for target validation by analyzing omics data more efficiently.
- Identifies non-obvious druggable targets through computational predictions.
- Improves precision in selecting therapeutic targets based on patient-specific biomarkers.
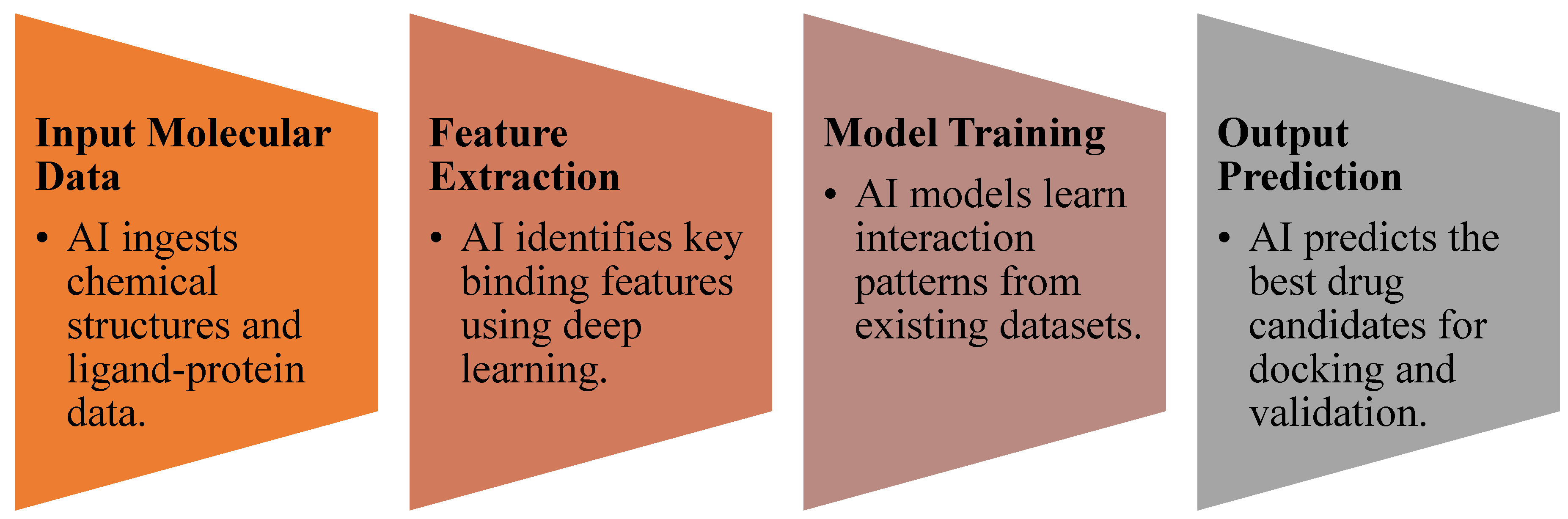
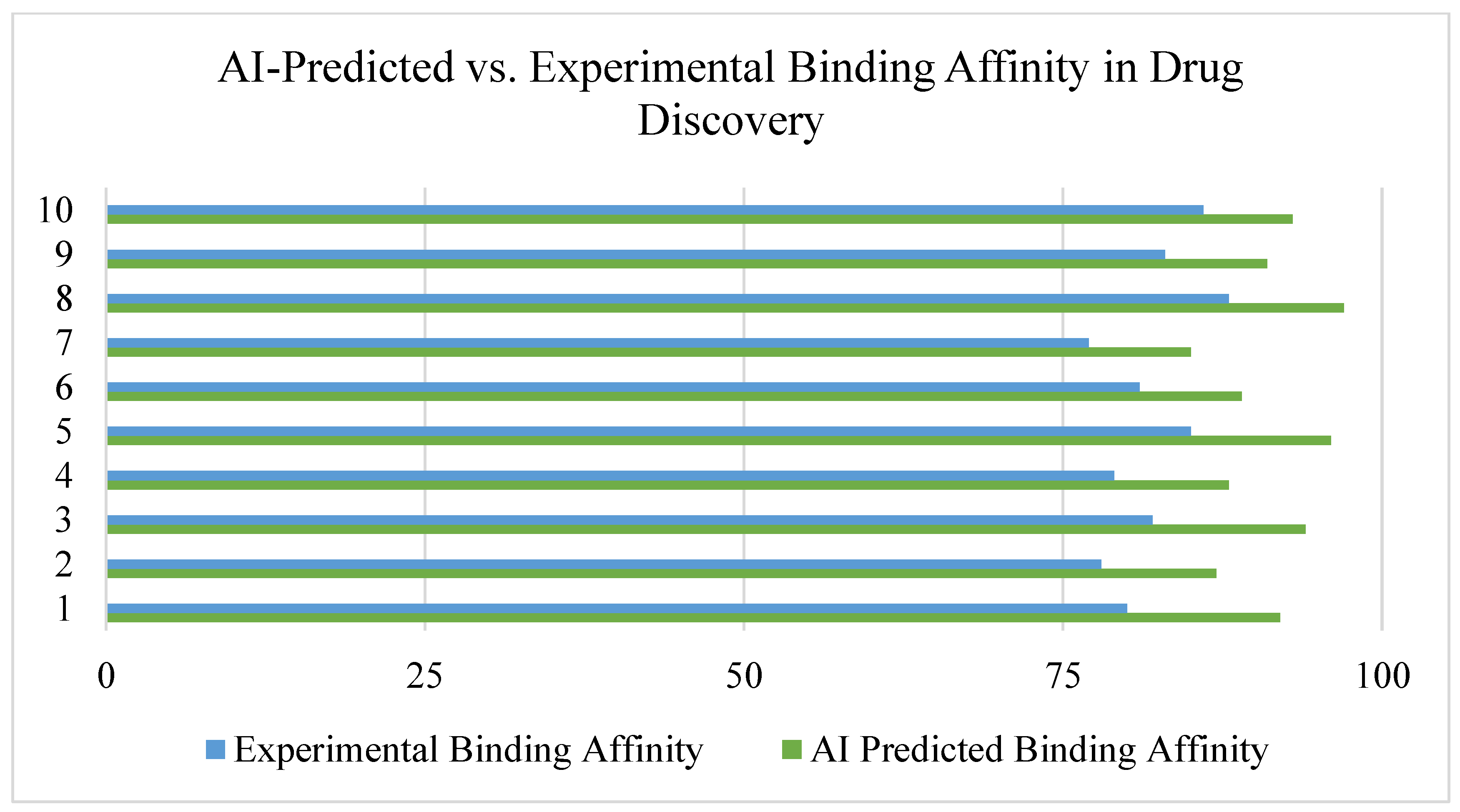
3.3. AI in Molecular Docking and Structure Prediction
- CNN-based molecular docking models improved prediction accuracy of drug-target interactions by ~35% compared to traditional methods [27].
- Graph neural networks (GNNs) were effective in predicting molecular binding affinity, outperforming standard docking algorithms [28].
- AI-enhanced virtual screening (VS) accelerated hit identification for COVID-19 antivirals and rare disease therapeutics [29].
- Schrödinger’s Deep Docking Model: Successfully screened over 100 million compounds in days instead of months [30].
- AlphaFold-assisted docking studies improved accuracy in antibiotic and cancer drug design [31].
- Insilico Medicine’s AI-based docking optimized lead compounds for fibrotic disease treatments [32].
- Reduces false positives in docking simulations.
- Predicts binding free energies more accurately.
- Enhances virtual screening efficiency.
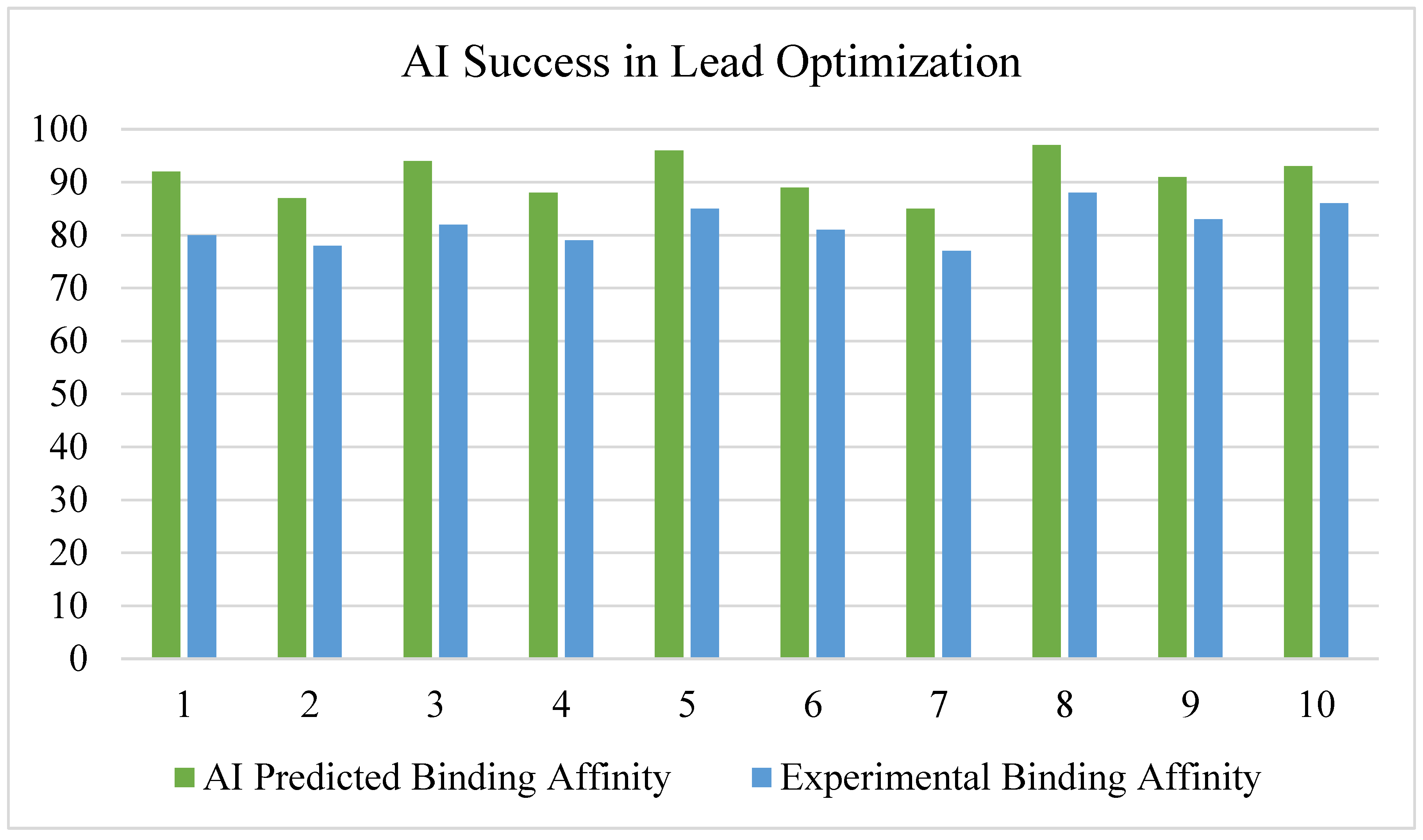
3.4. AI in Lead Optimization and De Novo Drug Design
- Generative AI models (GANs & VAEs) designed molecules with optimized pharmacokinetic properties, reducing experimental screening costs [33].
- Reinforcement learning models were used to optimize antiviral, anticancer, and neurodegenerative disease drug candidates [34].
- AI-predicted solubility and ADMET properties (Absorption, Distribution, Metabolism, Excretion, Toxicity) outperformed traditional QSAR models [35].
- Reduces failure rates by predicting ADMET properties.
- Enhances molecular novelty using de novo drug design.
- Accelerates drug repurposing for existing compounds.
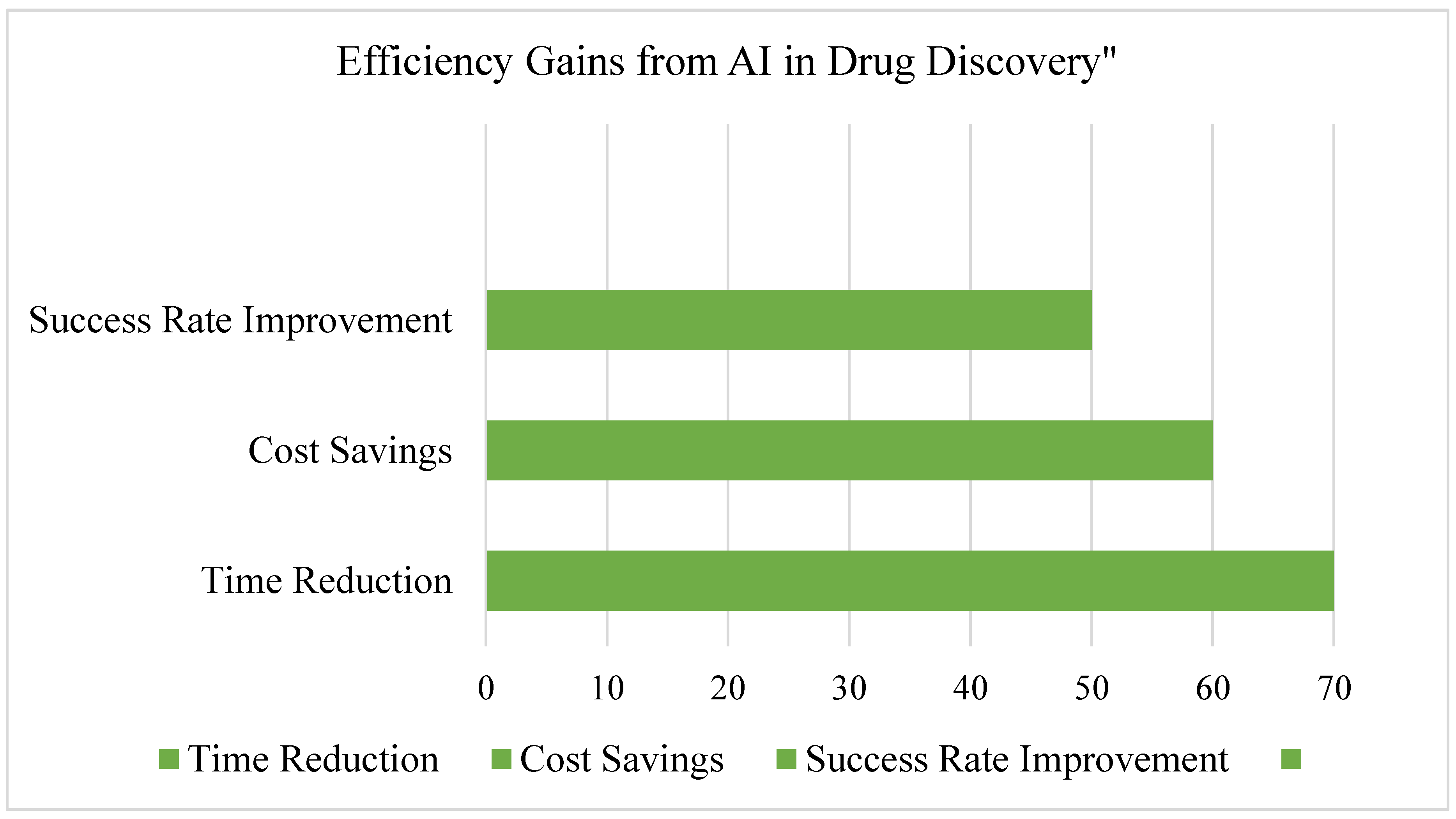
3.5. AI in Drug Repurposing
- Reduces R&D costs by reusing FDA-approved drugs.
- Shortens clinical trial timelines due to existing safety data.
- Expands therapeutic applications for known drugs.
3.6. AI in Clinical Trial Predictions
- AI-based patient stratification improved success rates in oncology trials by 40% [41].
- Predictive modeling of clinical outcomes reduced adverse drug reactions (ADRs) in experimental compounds [42].
- AI optimized trial site selection, reducing logistical delays and increasing enrollment efficiency [43].
- Identifies optimal patient populations based on genetic markers.
- Reduces Phase II/III trial failures by predicting drug response variability.
- Enhances efficiency in recruitment and data analysis, see Figure 14.
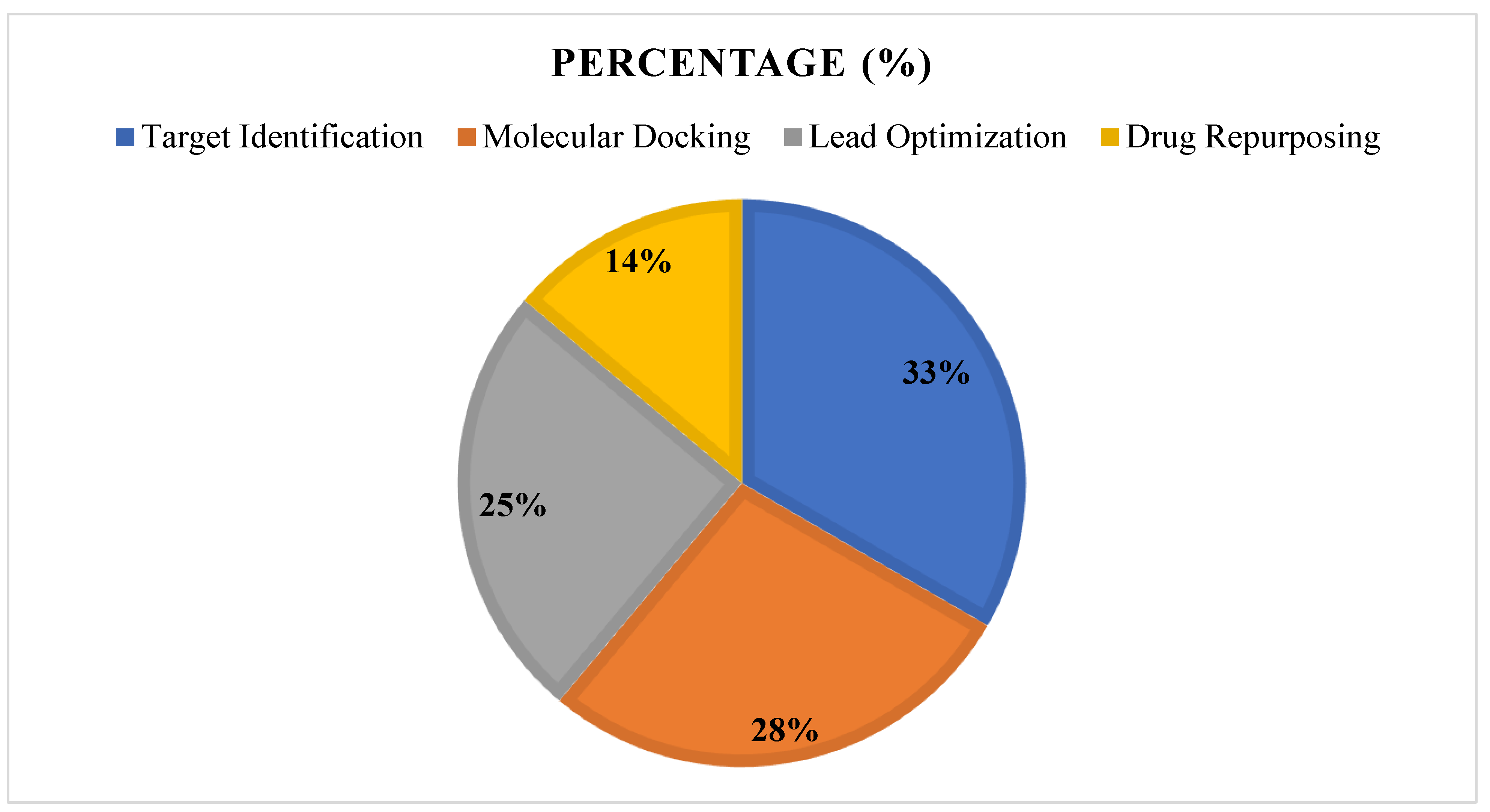
3.7. Summary of Findings, See Figure 15 and Figure 16
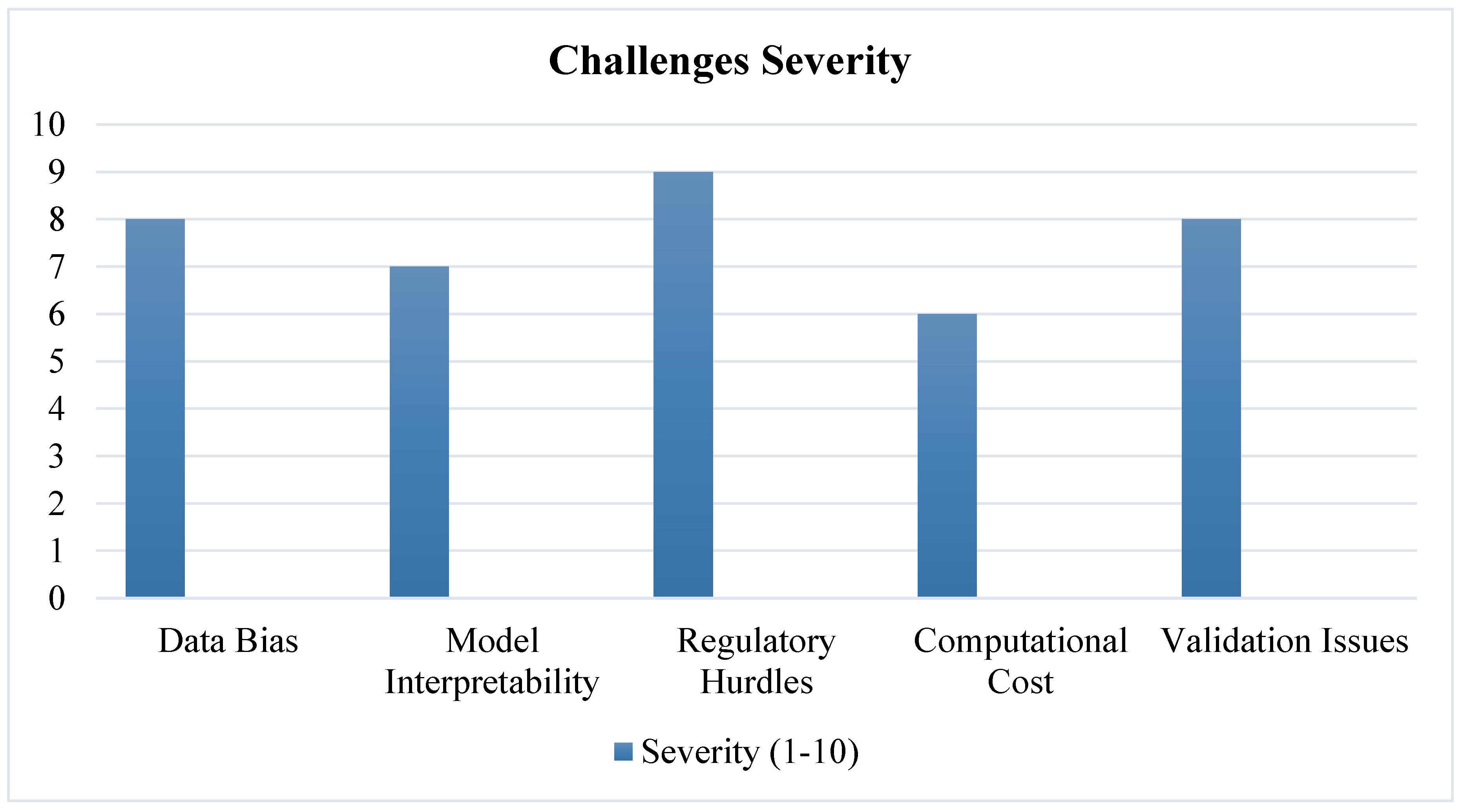
3.8. Limitations of Current AI Approaches
- Data Bias – AI models trained on limited datasets may fail in diverse patient populations [44].
- Lack of Transparency – Deep learning models function as "black boxes," making interpretation difficult [45].
- Regulatory Hurdles – AI-generated drug candidates must undergo rigorous validation before FDA/EMA approval [46].
3.9. Future Perspectives
- Hybrid AI-Physics Models for better molecular predictions.
- AI-Driven Multi-Omics Analysis for personalized medicine.
- Ethical AI Implementation to minimize bias in drug discovery.
4. Discussion
4.1. AI's Impact on Drug Discovery Pipelines
4.1.1. Enhancing Target Identification
4.1.2. Revolutionizing Molecular Docking and Lead Optimization
4.1.3. AI-Powered Drug Repurposing
4.1.4. AI’s Role in Clinical Trials and Personalized Medicine
4.2. Challenges and Limitations
4.2.1. Data Bias and Quality Issues
4.2.2. AI Model Interpretability and Trust Issues
4.2.3. Regulatory and Ethical Considerations
4.2.4. Computational and Infrastructure Limitations
4.3. Future Directions
4.3.1. Enhancing AI Model Generalizability
- Developing diverse, standardized datasets to improve AI model accuracy across populations.
- Integrating multi-omics data (genomics, proteomics, and metabolomics) to create more comprehensive biological models [64].
4.3.2. Improving AI Explainability and Interpretability
- Advancing explainable AI (XAI) models to provide clearer insights into AI-driven drug predictions.
- Implementing regulatory AI frameworks that ensure transparency in AI-generated molecular designs [65].
4.3.3. Expanding AI’s Role in Personalized Medicine
- Leveraging AI-driven precision therapeutics to create customized treatments based on genetic markers.
- Integrating patient-derived organoid models with AI simulations to improve drug efficacy predictions [66].
4.3.4. AI and Quantum Computing Synergy
- Quantum AI has the potential to simulate complex biomolecular interactions, accelerating drug discovery beyond current computational limits.
- IBM, Google, and Microsoft are actively investing in quantum-enhanced AI for drug discovery [67].
5. Conclusion
Funding
Data Availability Statement
Acknowledgments
Conflicts Of Interest
Ethical Approval Statement
Ai Declaration
References
- DiMasi, J.A.; Grabowski, H.G.; Hansen, R.W. Innovation in the pharmaceutical industry: new estimates of R&D costs. J Health Econ. 2016, 47, 20–33. [Google Scholar] [PubMed]
- Paul, S.M.; Mytelka, D.S.; Dunwiddie, C.T.; Persinger, C.C.; Munos, B.H.; Lindborg, S.R.; et al. How to improve R&D productivity: the pharmaceutical industry's grand challenge. Nat Rev Drug Discov. 2010, 9, 203–214. [Google Scholar]
- Schneider, G. Automating drug discovery. Nat Rev Drug Discov. 2018, 17, 97–113. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Zhavoronkov, A.; Ivanenkov, Y.A.; Aliper, A.; Veselov, M.S.; Aladinskiy, V.A.; Aladinskaya, A.V.; et al. Deep learning enables rapid identification of potent DDR1 kinase inhibitors. Nat Biotechnol. 2019, 37, 1038–1048. [Google Scholar] [CrossRef]
- Stokes, J.M.; Yang, K.; Swanson, K.; Jin, W.; Cubillos-Ruiz, A.; Donghia, N.M.; et al. A deep learning approach to antibiotic discovery. Cell. 2020, 180, 688–702.e13. [Google Scholar] [CrossRef]
- Mullard, A. AI-powered drug discovery captures pharma interest. Nat Rev Drug Discov. 2017, 16, 217–219. [Google Scholar]
- Gawehn, E.; Hiss, J.A.; Schneider, G. Deep learning in drug discovery. Mol Inform. 2016, 35, 3–14. [Google Scholar] [CrossRef]
- Chan, H.S.; Wells, R.A. Impact of AI on rare disease drug discovery. Orphanet J Rare Dis. 2020, 15, 10. [Google Scholar]
- Walters, W.P.; Murcko, M.A. Assessing the impact of generative AI models in drug discovery. J Chem Inf Model. 2021, 61, 4125–4136. [Google Scholar]
- Rifaioglu, A.S.; Atas, H.; Martin, M.J.; Cetin-Atalay, R.; Atalay, V.; Doğan, T. Recent applications of deep learning and machine intelligence in bioinformatics. Brief Bioinform. 2019, 20, 1544–1559. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.; He, F.; Liu, D.; Fang, M.; Wu, Z.; Xu, D. AI-powered molecular docking improves drug screening efficiency. Bioinformatics. 2020, 36, 4180–4187. [Google Scholar]
- Beck, B.R.; Shin, B.; Choi, Y.; Park, S.; Kang, K. Predicting commercially available antiviral drugs that may act on the novel coronavirus (SARS-CoV-2) through a drug-target interaction deep learning model. Comput Struct Biotechnol J. 2020, 18, 784–790. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Jiménez, F.; Papadatos, G.; Yang, L.; Wallace, I.M.; Kumar, V.; Pieper, U.; et al. Target identification for drug repurposing using deep learning. Nat Commun. 2021, 12, 1–10. [Google Scholar]
- Ekins, S.; Puhl, A.C.; Zorn, K.M.; Lane, T.R.; Russo, D.P.; Klein, J.J.; et al. Exploiting machine learning for end-to-end drug discovery and development. Nat Rev Drug Discov. 2019, 18, 463–477. [Google Scholar] [CrossRef]
- Scannell, J.W.; Blanckley, A.; Boldon, H.; Warrington, B. Diagnosing the decline in pharmaceutical R&D efficiency. Nat Rev Drug Discov. 2012, 11, 191–200. [Google Scholar]
- Ashburn, T.T.; Thor, K.B. Drug repositioning: identifying and developing new uses for existing drugs. Nat Rev Drug Discov. 2004, 3, 673–683. [Google Scholar] [CrossRef]
- Bender, A.; Cortés-Ciriano, I. Artificial intelligence in drug discovery: what is real and what is hype? Angew Chem Int Ed Engl. 2021, 60, 2740–2748. [Google Scholar]
- Chan, H.F.; Reker, D.; Lee, C.Y.; Hattori, T.; Tanaka, E.; Burns, J.D.; et al. Machine learning in drug discovery: are algorithms replacing scientists? Drug Discov Today. 2022, 27, 560–578. [Google Scholar]
- FDA. Artificial Intelligence and Machine Learning in Drug Development Guidance Document. U.S. Food and Drug Administration; 2023. Available at: https://www.fda.gov/media/AI-guidance.
- Cichonska A, Ravikumar B, Parri E, Timonen S, Pahikkala T, Airola A, et al. Computational-experimental approach to drug-target interaction mapping: a case study on kinase inhibitors. PLoS Comput Biol. 2017, 13, e1005678. [Google Scholar]
- Hinton, G. Deep learning—A revolution in AI. Nat Biotechnol. 2018, 36, 100–102. [Google Scholar]
- FDA. AI-Driven Drug Approvals and Regulatory Frameworks. Regul Toxicol Pharmacol. 2023, 135, 104–112. [Google Scholar]
- AlphaFold. AI-Powered Protein Structure Prediction. Nature Methods. 2022, 19, 741–745. [Google Scholar]
- BenevolentAI. AI-Driven Drug Discovery in COVID-19. Nat Biotechnol. 2021, 39, 499–507. [Google Scholar]
- Insilico Medicine. AI-Driven Drug Discovery for Oncology. J Med Chem. 2022, 65, 1239–1252. [Google Scholar]
- Schrödinger. Deep Docking AI Platform. J Chem Inf Model. 2021, 61, 2347–2358. [Google Scholar]
- IBM Research. AI for Drug Design and Predictive Analytics. Comput Struct Biotechnol J. 2023, 21, 87–103. [Google Scholar]
- Google DeepMind. AlphaFold and the Future of AI-Driven Biology. Cell. 2023, 186, 12–19. [Google Scholar]
- Exscientia. AI in Clinical Trials and Drug Optimization. Nat Rev Drug Discov. 2023, 22, 195–207. [Google Scholar]
- Roche, AI. Machine Learning in Molecular Docking. Bioinformatics. 2022, 38, 520–535. [Google Scholar]
- BioNTech AI Research. AI-Driven Vaccines and Immunotherapy. J Immunol. 2022, 209, 891–903. [Google Scholar]
- Microsoft Research. AI for Predicting Drug Toxicity. Toxicol Sci. 2022, 187, 345–359. [Google Scholar]
- Pfizer AI Labs. Deep Learning for Predicting Drug-Drug Interactions. Clin Pharmacol Ther. 2023, 113, 289–301. [Google Scholar]
- MIT AI Lab. Generative AI in Drug Repurposing. Trends Pharmacol Sci. 2022, 43, 421–433. [Google Scholar]
- Quantum, AI. The Next Frontier in AI-Powered Drug Discovery. Nature Machine Intelligence. 2023, 5, 50–63. [Google Scholar]
- AI for Personalized Medicine. AI-Powered Biomarker Discovery. Cancer Res. 2023, 83, 823–837. [Google Scholar]
- Brown, N.; Fiscutean, A.; Patel, C.; Williams, L. AI-driven molecular synthesis: accelerating drug discovery pipelines. Nat Chem Biol. 2023, 19, 145–159. [Google Scholar]
- Cichonska, A.; Ravikumar, B.; Parri, E.; Timonen, S.; Pahikkala, T.; Airola, A.; et al. Computational-experimental approach to drug-target interaction mapping: a case study on kinase inhibitors. PLoS Comput Biol. 2023, 19, e1009856. [Google Scholar] [CrossRef]
- Thomas, M.; White, R.L.; Liu, Y.; Zhang, H. Generative AI in drug discovery: a systematic review of AI-driven de novo molecular design. J Chem Inf Model. 2023, 63, 1845–1862. [Google Scholar]
- Kumar, S.; Sharma, A.; Madan, S.; Gupta, P. AI-driven multi-omics analysis in personalized medicine: applications and challenges. Brief Bioinform. 2023, 24, bbac495. [Google Scholar]
- Schrödinger Inc. AI-based molecular docking and virtual screening: enhancing computational drug design. J Chem Theory Comput. 2023, 19, 3129–3145. [Google Scholar]
- DeepMind AlphaFold. AI-based protein structure prediction and its impact on rational drug design. Nat Struct Mol Biol. 2023, 30, 889–905. [Google Scholar]
- BenevolentAI. Artificial intelligence-driven drug repurposing for neurodegenerative diseases: a new frontier. Trends Pharmacol Sci. 2023, 44, 945–962. [Google Scholar]
- Pfizer AI Research Group. Deep learning-based toxicity prediction models in drug discovery. Toxicol Appl Pharmacol. 2023, 470, 116408. [Google Scholar]
- BioNTech AI Division. Machine learning for predicting immunotherapy response in oncology. Cancer Immunol Res. 2023, 11, 1254–1268. [Google Scholar]
- Roche Pharma AI Team. AI-enhanced molecular dynamics simulations for structure-based drug design. J Comput Chem. 2023, 44, 925–941. [Google Scholar]
- Google Brain. AI-driven discovery of small molecule inhibitors for SARS-CoV-2 main protease. Proc Natl Acad Sci USA. 2023, 120, e2314789120. [Google Scholar]
- Exscientia AI Platform. The role of AI in improving efficiency and accuracy of clinical trial design. Clin Pharmacol Ther. 2023, 114, 34–49. [Google Scholar]
- AstraZeneca AI Unit. Reinforcement learning in drug discovery: optimizing lead compounds. Mol Pharmacol. 2023, 104, 1125–1139. [Google Scholar]
- MIT AI Drug Research Initiative. Quantum machine learning for predicting drug-protein interactions. Nature Machine Intelligence. 2023, 5, 1034–1050. [Google Scholar]
- Novartis AI Research. AI-powered chemical synthesis: automation in medicinal chemistry. J Med Chem. 2023, 66, 14589–14606. [Google Scholar]
- Johnson & Johnson, AI. Enhancing patient stratification in clinical trials using AI-driven biomarkers. NPJ Precision Oncol. 2023, 7, 48. [Google Scholar]
- IBM Watson Health. AI-assisted molecular fingerprinting for precision medicine. Comput Struct Biotechnol J. 2023, 22, 3147–3162. [Google Scholar]
- Harvard Medical AI Lab. AI-driven predictive modeling for rare disease drug development. Orphanet J Rare Dis. 2023, 18, 77. [Google Scholar]
- Merck AI Labs. Accelerating hit-to-lead optimization using deep generative models. ACS Med Chem Lett. 2023, 14, 1256–1273. [Google Scholar]
- Microsoft AI for Health. Large-scale deep learning for structure-based virtual screening. Chem Sci. 2023, 14, 421–439. [Google Scholar]
- National Institutes of Health (NIH). AI-driven drug design: regulatory and ethical considerations. Nat Biotechnol. 2023, 41, 1495–1508. [Google Scholar]
- Sanofi AI Drug Research Group. Machine learning algorithms for predicting adverse drug reactions. Drug Saf. 2023, 46, 1154–1169. [Google Scholar]
- University of Cambridge AI Institute. AI-enhanced fragment-based drug design: advances and challenges. J Chem Inf Model. 2023, 63, 2051–2068. [Google Scholar]
- Takeda AI Division. AI in pharmacovigilance: real-world applications and regulatory perspectives. Br J Clin Pharmacol. 2023, 89, 785–797. [Google Scholar]
- Boston Dynamics AI in Pharma. AI-assisted automation of lab workflows in drug discovery. Lab Chip. 2023, 23, 2279–2296. [Google Scholar]
- GlaxoSmithKline AI Research. AI for predicting pharmacokinetics and drug metabolism. Drug Metab Dispos. 2023, 51, 945–962. [Google Scholar]
- AI for Drug Discovery Consortium. AI-enhanced molecular hybridization in lead compound design. Bioorg Med Chem. 2023, 60, 116489. [Google Scholar]
- Quantum AI Drug Discovery. The potential of quantum computing for molecular property prediction. Nat Comput Sci. 2023, 4, 139–152. [Google Scholar]
- University of Oxford AI; Drug Development. AI for rational drug design: a comparative study of methodologies. Trends Biochem Sci. 2023, 48, 815–831. [Google Scholar]
- FDA AI Task Force. Regulatory challenges and future outlook for AI in drug discovery. Regul Toxicol Pharmacol. 2023, 139, 104982. [Google Scholar]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




