1. Introduction
Chemotactic trafficking of the immune response is mediated by G protein-coupled receptors (GPCRs) and chemokines that guide patrolling immune cells to the right place at the right time [
1]. Chemokines are small soluble proteins that contain four conserved cysteine residues forming two disulfide bonds. Chemokines can be classified into four subfamilies: CC, CXC, XC, and CX3C based on the number and location of cysteine residues at the N-terminus [
2]. The biological effects of chemokines are mediated through a family of GPCRs. The chemokine receptors possess seven-transmembrane helices connected by an extracellular N-terminal region, three extracellular loops (ECL1-3), three intracellular loops, and an intracellular C-terminal region [
3]. Several disulfide bonds connect the N-terminus to ECL3 and ECL1 to ECL2 [
4]. The binding of chemokines to their specific receptors causes changes of the conformation and activates the chemokine signaling pathways to regulate the migration, integration, adhesion, and proliferation [
5,
6].
The C-C chemokine receptor type 7 (CCR7) is expressed on various cell types, such as naive T/B cells, central memory T cells, regulatory T cells, natural killer cells, dendritic cells and tumor cells [
7]. Chemokine ligand (CCL) 19 and 21 are the high-affinity CCR7 ligands which promote the migration of CCR7-expressing cells to secondary lymphoid organs, including lymph nodes, thymus, and spleen [
8,
9,
10,
11,
12]. Studies of genome-wide association have revealed a relationship between CCL21/CCR7 and the severity diseases in patients with systemic lupus erythematosus, Sjögren’s syndrome, rheumatoid arthritis, and asthma [
7]. Disrupting CCL21/CCR7 interaction with monoclonal antibodies (mAbs) or inhibitors suppresses the migration of the CCR7-positive cells at the inflammatory site and suppresses the disease progression [
7].
Lymph node metastasis is an important predictive factor of patients with cancer [
13]. The elevated CCR7 expression correlates with lymph node metastasis in various solid tumors, such as esophageal [
14], gastric [
15], colorectal [
16], pancreatic [
17], thyroid [
18], oral [
19], and non-melanoma skin cancers [
20]. CCR7, but not others, specifically drives cancer cell homing into lymph node and other secondary lymphoid organs where the ligands CCL19 and CCL21 are constitutively expressed by stroma cells [
21]. Therefore, developing specific mAbs against mouse CCR7 (mCCR7) is essential to targeting the CCR7-expressing cells in the preclinical mouse disease models.
The Cell-Based Immunization and Screening (CBIS) method includes immunizing antigen-overexpressed cells and high-throughput hybridoma screening using flow cytometry. We have developed specific mAbs against chemokine receptors including mouse CXCR1 (mCXCR1; clone Cx
1Mab-1) [
22], mouse CXCR3 (mCXCR3; clone Cx
3Mab-4) [
23], mouse CXCR4 (mCXCR4; clone Cx
4Mab-1) [
24], CCR1 (mCCR1; clone C
1Mab-6) [
25], mouse CCR3 (mCCR3; clones C
3Mab-2, C
3Mab-3, and C
3Mab-4) [
26], mouse CCR5 (mCCR5; clone C
5Mab-2) [
27], mCCR7 (clone C
7Mab-7) [
28], and mouse CCR8 (mCCR8; clones C
8Mab-1, C
8Mab-2, and C
8Mab-3) [
29] using the CBIS method. Furthermore, we established specific mAbs against mouse CCR2 (mCCR2; clone C
2Mab-6) [
30], mCCR3 (clones C
3Mab-6 and C
3Mab-7) [
31], mouse CCR4 (mCCR4; clone C
4Mab-1) [
32], mouse CCR6 (mCCR6; clone C
6Mab-13) [
33], mouse CCR9 (mCCR9; clone C
9Mab-24) [
34], mouse CXCR5 (mCXCR5; clone Cx
5Mab-3) [
35], and mouse CXCR6 (mCXCR6; clone Cx
6Mab-1) [
36] using the N-terminal peptide immunization. In contrast, there are few reports for establishing anti-chemokine receptor mAbs by immunization of
ECL peptides.
In this study, we report a novel anti-mCCR7 mAb successfully developed by the ECL3 peptide immunization.
2. Materials and Methods
2.1. Cell Lines
Mouse myeloma P3X63Ag8.U1 (P3U1) and Chinese hamster ovary (CHO)-K1 were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). The mCCR7-overexpressed CHO-K1 (CHO/mCCR7) was previously established [
28]. Stable transfectants of the following chemokine receptors were previously established [
35]: CHO/mCCR1, CHO/mCCR2, CHO/mCCR3, CHO/PA-mCCR4, CHO/mCCR5, CHO/PA-mCCR6, CHO/mCCR8, CHO/mCCR9, CHO/PA-mCCR10, CHO/mCXCR1, CHO/mCXCR2, CHO/mCXCR3, CHO/mCXCR4, CHO/mCXCR5, CHO/mCXCR6, CHO/mCX3CR1, CHO/mXCR1. P3U1, CHO-K1, and chemokine receptors-expressed CHO-K1 were maintained in Roswell Park Memorial Institute (RPMI)-1640 medium (Nacalai Tesque Inc., Kyoto, Japan) with the same antibiotics described above and 10% heat-inactivated fetal bovine serum (FBS; Thermo Fisher Scientific, Inc., Waltham, MA, USA). All cells were cultured in a humidified incubator at 37°C with 5% CO
2.
2.2. Peptides
Eurofins Genomics KK (Tokyo, Japan) synthesized a partial sequence of the ECL of mCCR7 as follows.
mCCR7-1: SEAKSWIFGVYLC
mCCR7-2: ELLYSGLQKNSGEDTLRC
mCCR7-3: CETSKQLNIAYDVTYS
Subsequently, the keyhole limpet hemocyanin (KLH) was conjugated at the N-terminus of mCCR7-3 or the C-terminus of mCCR7-1 and mCCR7-2.
2.3. Development of Hybridomas
Three five-week-old female Sprague–Dawley (SD) rats were purchased from CLEA Japan (Tokyo, Japan). The Animal experiments were approved by the Animal Care and Use Committee of Tohoku University (Permit number: 2022MdA-001) and were carried out in accordance with the NIH (National Research Council) Guide for the Care and Use of Laboratory Animals. To develop mAbs against mCCR7, we intraperitoneally immunized three rats with 100 µg of the KLH-conjugated mCCR7 peptides plus Alhydrogel adjuvant 2% (InvivoGen, San Diego, CA, USA). The hybridomas were generated ad described previously [
36]. The hybridoma supernatants were subsequently screened using enzyme-linked immunosorbent assay (ELISA) with the mCCR7 peptides, followed by flow cytometry, using CHO/mCCR7 and CHO-K1. To produce purified mAbs, hybridomas were cultured in Hybridoma-SFM (Thermo Fisher Scientific, Inc.), and the purified mAbs were separated using Ab-Capcher (ProteNova Inc., Kagawa, Japan).
2.4. ELISA
The synthesized mCCR7 peptides were immobilized on Nunc Maxisorp 96 well immunoplates (Thermo Fisher Scientific Inc.) at 1 µg/mL concentration for 30 min at 37℃. After blocking with 1% bovine serum albumin (BSA)-in phosphate-buffered saline (PBS) containing 0.05% Tween20 (PBST; Nacalai Tesque, Inc.), the plates were incubated with supernatants of hybridomas for 30 min at 37℃. The enzymatic reactions were conducted and measured as described previously [
36].
2.5. Flow Cytometry
Cells were detached using 1 mM ethylenediaminetetraacetic acid (EDTA; Nacalai Tesque, Inc.) to prevent enzymatic degradation of mCCR7. The cells were washed with 0.1% BSA in PBS (blocking buffer) and incubated with C7Mab-2 at 4°C for 30 min. For peptide inhibition assay, C7Mab-2 (2 μg/mL) was pre-incubated with 1 μg/mL of mCCR7-3 peptide or dimethyl sulfoxide (DMSO) for 15 min, and incubated with the cells for 30 min at 4°C. After washing, the cells were incubated with Alexa Fluor 488-conjugated anti-rat IgG (1:1,000 dilution; Cell Signaling Technology, Inc., Danvers, MA, USA) at 4°C for 30 min. Data were collected and analyzed using the SA3800 Cell Analyzer and FlowJo software (BD Biosciences, Franklin Lakes, NJ, USA), respectively.
2.6. Determination of Dissociation Constant Using Flow Cytometry
CHO/mCCR7 cells were treated with serial dilutions of C7Mab-2 (0.006 to 100 μg/mL). The cells were incubated with Alexa Fluor 488-conjugated anti-rat IgG (1:200 dilution) at 4°C for 30 min. Data were collected and analyzed using the SA3800 Cell Analyzer and FlowJo software. By fitting one-site binding models in GraphPad Prism 6 software (GraphPad Software, Inc., La Jolla, CA, USA), the KD values of C7Mab-2 for CHO/mCCR7 were determined.
2.7. Immunohistochemistry
Cell blocks were prepared as described previously [
28]. The sections (4 µm thickness) were autoclaved in citrate buffer (pH 6.0; Nichirei Biosciences, Inc., Tokyo, Japan) for 20 min. After blocking with SuperBlock T20 Blocking Buffer (Thermo Fisher Scientific Inc.), the sections were incubated with C
7Mab-2 (20 μg/mL) for 1 h at room temperature. For peptide inhibition assay, C
7Mab-2 (20 μg/mL) was pre-incubated with 2 μg/mL of mCCR7-3 peptide or dimethyl sulfoxide (DMSO) for 15 min, and incubated with the cell blocks for 1 h. Color development was achieved as described previously [
28].
3. Results
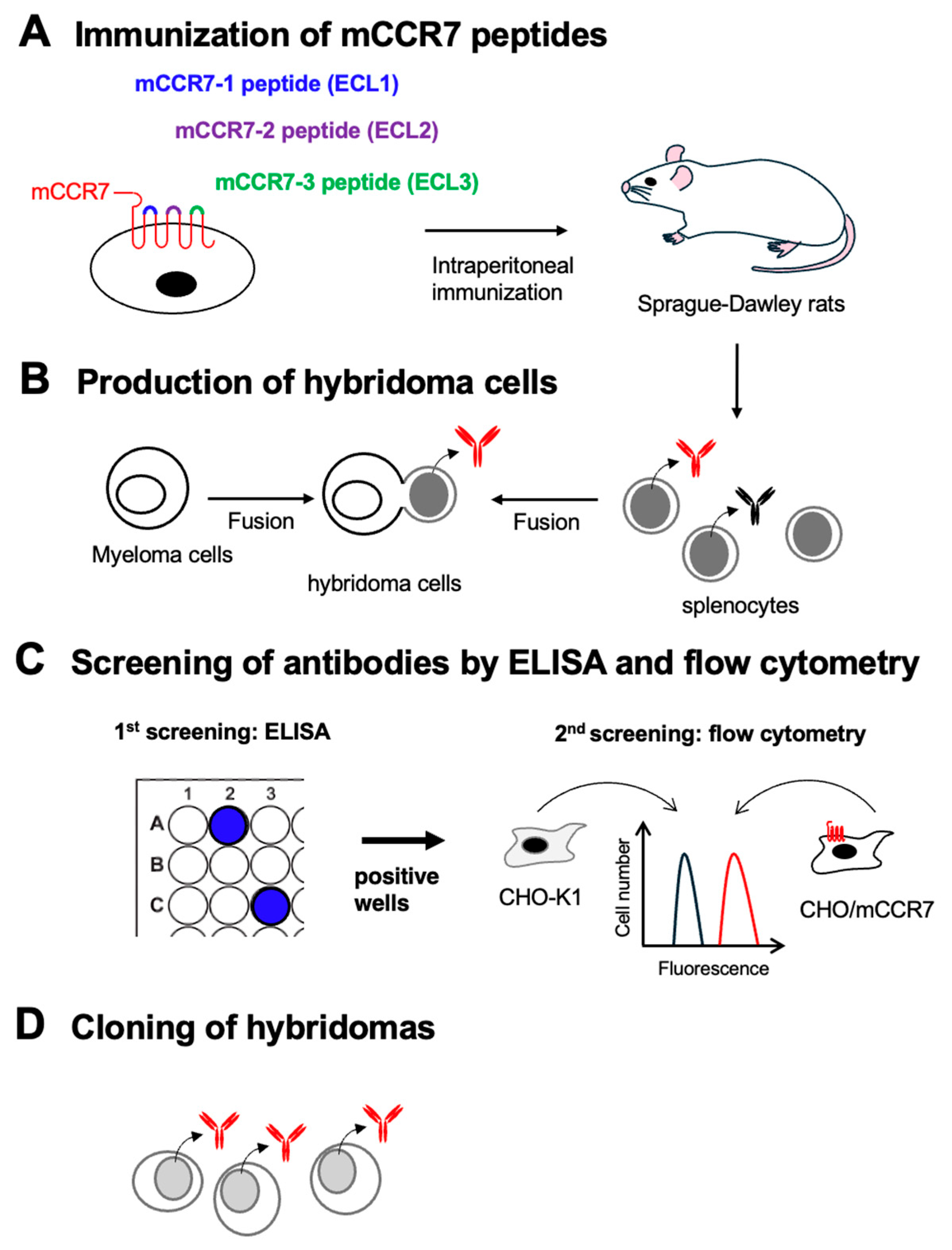
3.1. Development of Anti-Mouse CCR7 mAbs by Immunization of Three Extracellular Loop Peptides
Three SD rats were immunized with the KLH-conjugated mCCR7 peptides, respectively (
Figure 1A). The spleens were harvested from the immunized rats, and hybridomas were produced by fusion with P3U1 cells (
Figure 1B). Then, positive wells for each naked mCCR7 peptide were selected using ELISA (
Figure 1C). The ELISA screening identified 11 out of 1534 wells (to mCCR7-1, 0.7%), 78 out of 1534 wells (to mCCR7-2, 5.1%), and 93 out of 1438 wells (to mCCR7-3, 6.5%), which strongly reacted with the each mCCR7 peptide. Then 2
nd screenings were performed using flow cytometry (
Figure 1C). Among 93 ELISA-positive wells to mCCR7-3, 11 wells showed the reactivity to CHO/mCCR7, but not to CHO-K1 cells. We could not obtain the flow cytometry-positive wells in the hybridomas from mCCR7-1-KLH and mCCR7-2-KLH-immunized rats. The anti-mCCR7 mAb-producing hybridomas from KLH-mCCR7-3-immunized rats were further cloned by limiting dilution, and C
7Mab-2 (rat IgG
2b, kappa) was finally established (
Figure 1D).
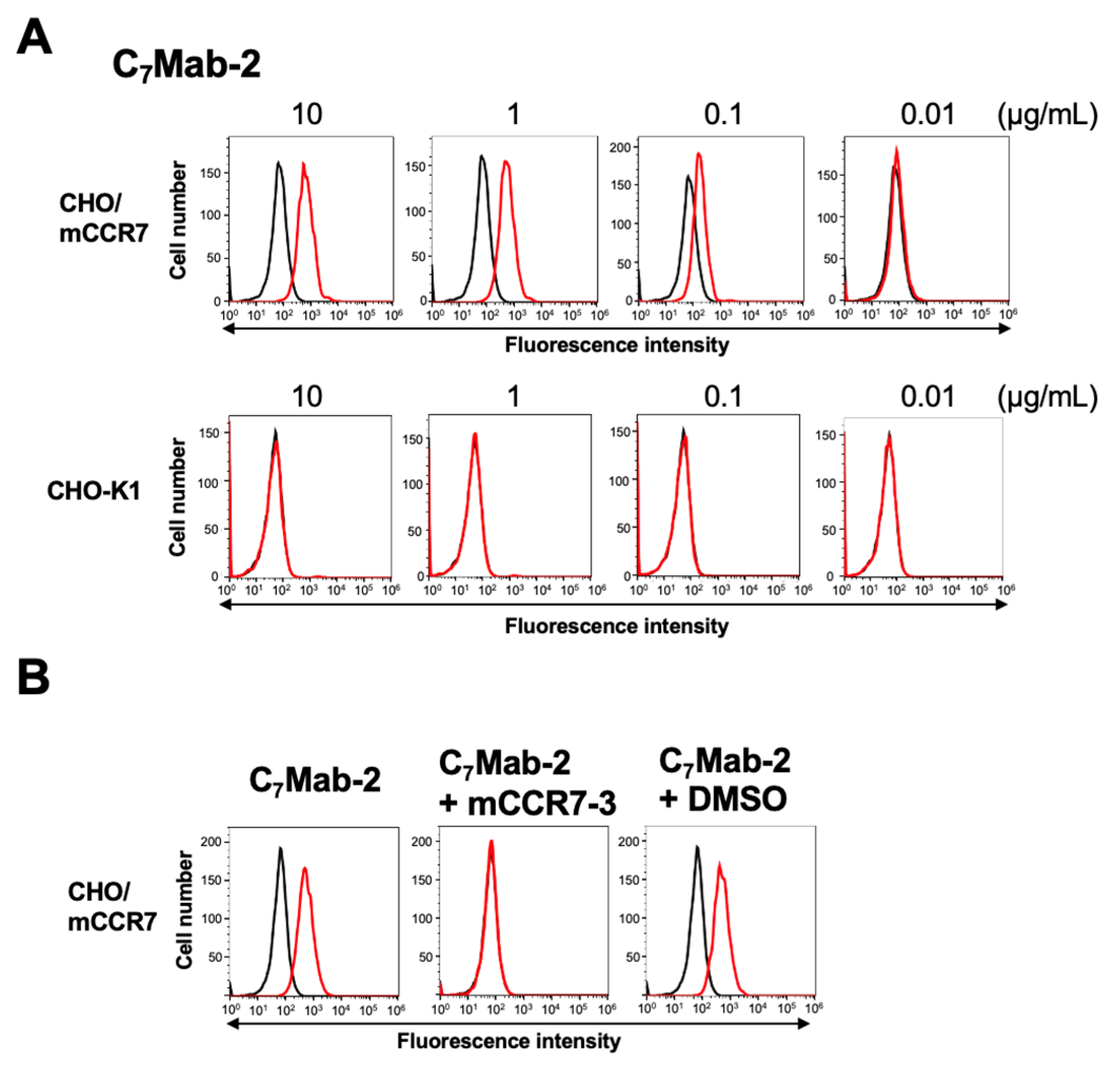
3.2. Flow Cytometry Using C7Mab-2
We conducted flow cytometry using C
7Mab-2 against CHO/mCCR7 and CHO-K1 cells. C
7Mab-2 recognized CHO/mCCR7 cells dose-dependently at 10, 1, 0.1, and 0.01 μg/mL (
Figure 2A). Parental CHO-K1 cells were not recognized by C
7Mab-2 even at 10 μg/mL (
Figure 2A). We next performed a peptide-blocking assay. As shown in
Figure 2B, C
7Mab-2 reacted with the CHO/mCCR7. The mCCR7-3 peptide completely neutralized the reactions.
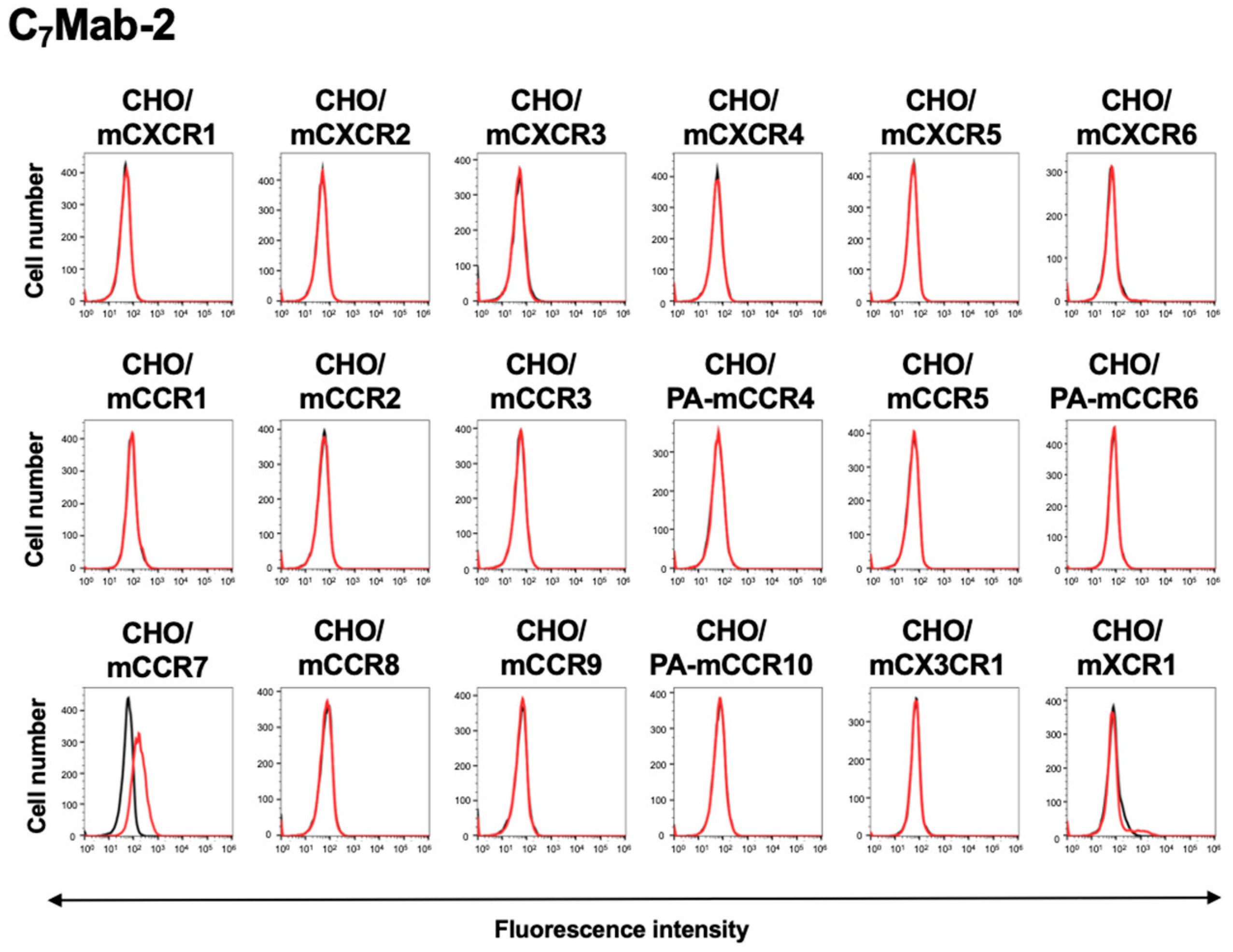
3.3. Reactivity of C7Mab-2 to CC, CXC, CX3C, and XC Chemokine Receptor-Expressed CHO-K1 Cells
We have established anti-mouse CC, CXC, CX3C, and XC chemokine receptor mAbs and evaluated them using these receptors-expressed CHO-K1 cells, as described previously [
35]. Among eighteen mouse CC, CXC, CX3C, and XC chemokine receptor-expressed CHO-K1 cells, C
7Mab-2 recognized only CHO/mCCR7, but not others (
Figure 3).
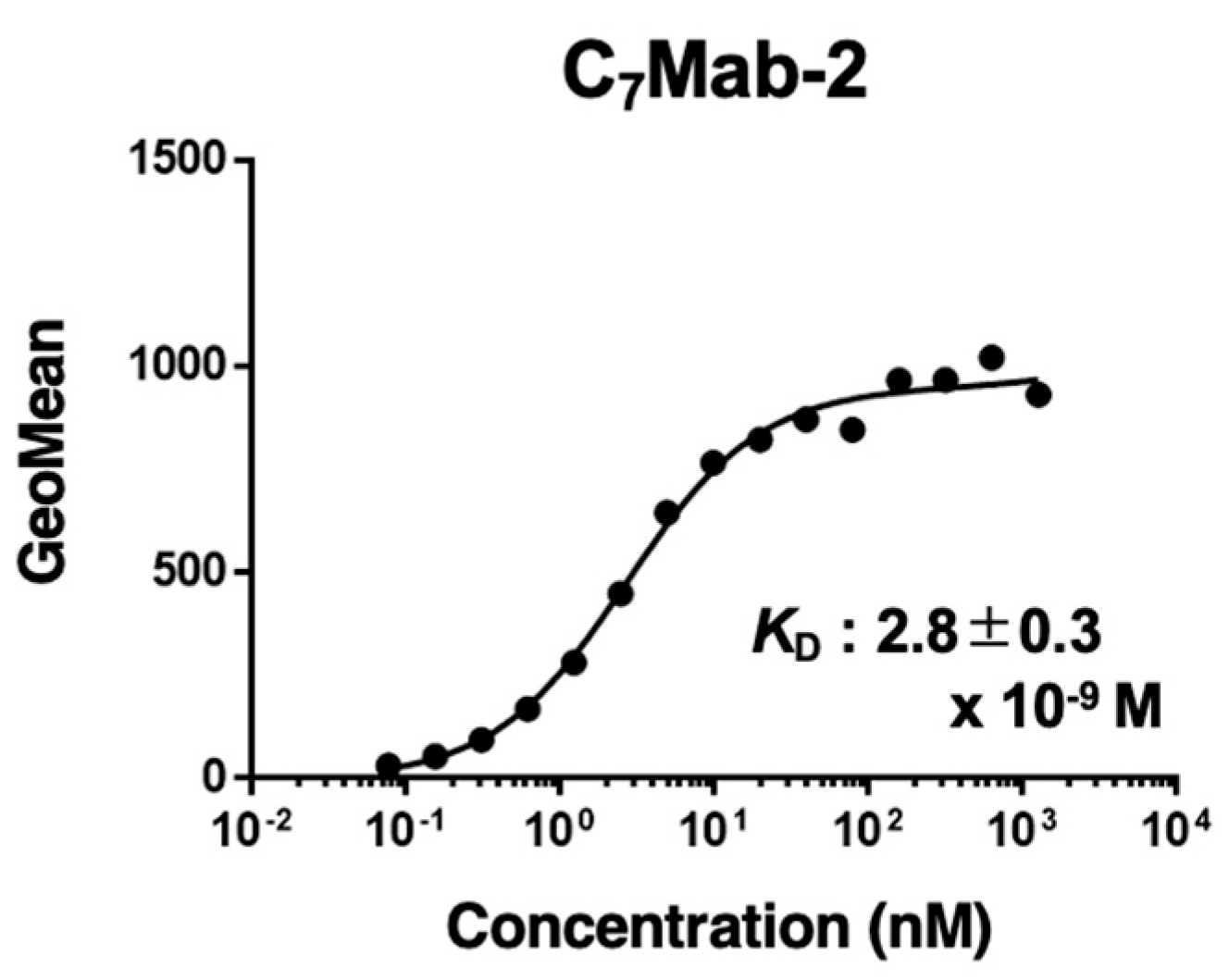
3.4. Determination of KD Value of C7Mab-2 by Flow Cytometry
The binding affinity of C
7Mab-2 was evaluated using flow cytometry. The average
KD value of C
7Mab-2 for CHO/mCCR7 cells from three independent measurements (supplementary Fig. S1) was 2.8 ± 0.3 × 10⁻⁹ M (
Figure 4).
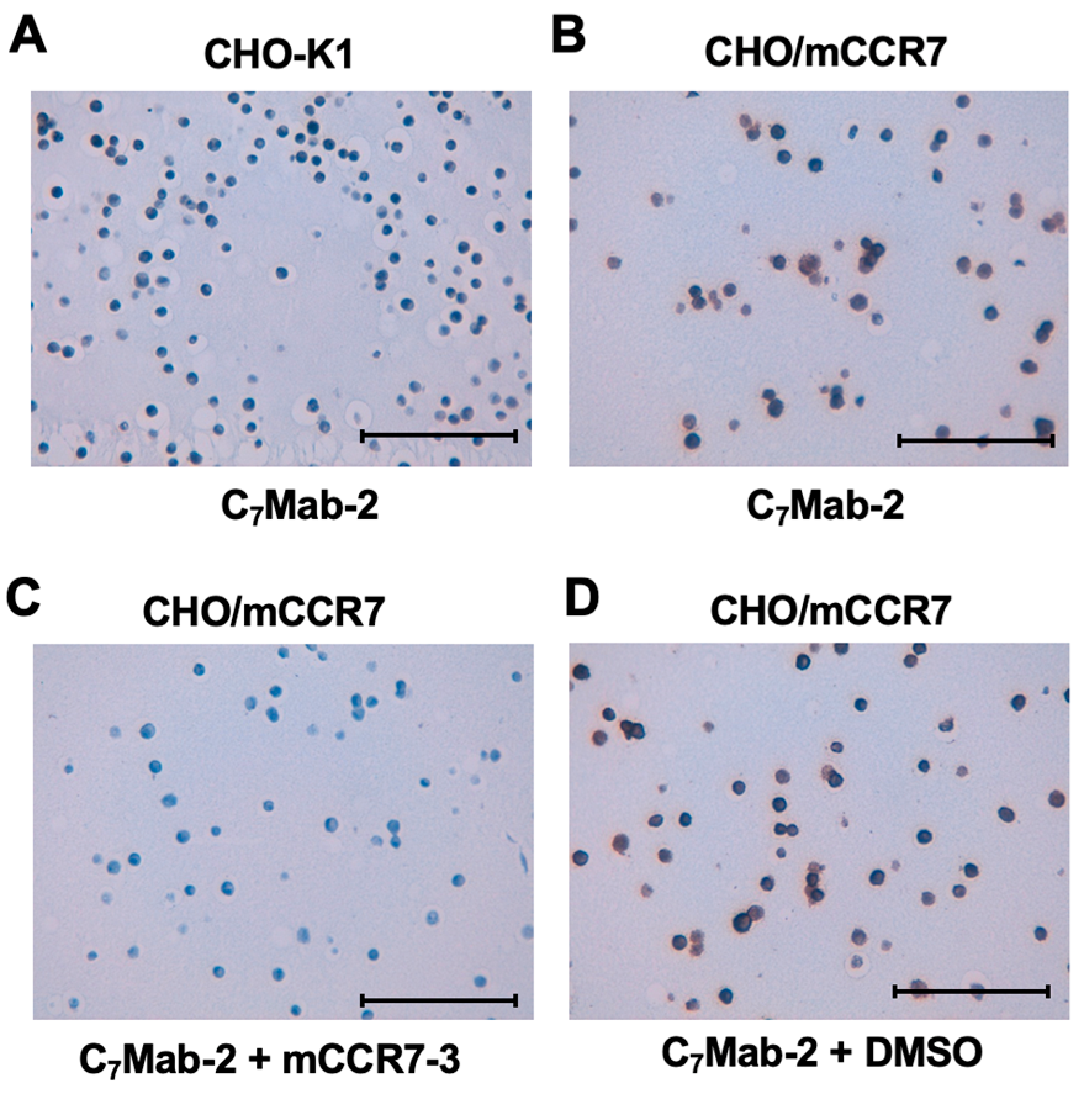
3.5. Immunohistochemistry Using C7Mab-2 in Mouse CCR7-Overexpressed CHO-K1 Cells
To evaluate the suitability of C
7Mab-2 for immunohistochemistry in formalin-fixed paraffin-embedded (FFPE) samples, the sections of CHO/mCCR7 and CHO-K1 cells were stained with C
7Mab-2. The cytoplasmic and membranous staining of mCCR7 was observed in CHO/mCCR7 cells (
Figure 5A). In contrast, no staining was detected in CHO-K1 cells (
Figure 5B). Furthermore, the reaction was completely neutralized by the mCCR7-3 peptide (
Figure 5C and D).
4. Discussion
This study established an anti-mCCR7 mAb, C
7Mab-2 by the ECL3 peptide immunization. C
7Mab-2 can be applied to flow cytometry (
Figure 2) and immunohistochemistry (
Figure 5) to detect mCCR7-positive cells. The reactivity (
Figure 2) and affinity (
Figure 4) of C
7Mab-2 are similar to another anti-mCCR7 mAb, C
7Mab-7, which was established by the CBIS method [
28]. Furthermore, we also confirmed that C
7Mab-2 recognizes mCCR7, but not other CC, CXC, CX3C, and XC chemokine receptors (
Figure 3). We immunized the ECL1–3 peptides and obtained the ELISA-positive wells in hybridomas from each peptide-immunized rat. However, we could not obtain flow cytometry-positive wells in hybridomas derived from ECL1 and ECL2 peptide-immunized rats. In hybridomas from ECL3 peptide-immunized rat, only 10% of ELISA-positive supernatants recognized CHO/mCCR7 in flow cytometry, indicating that conformational changes and modification including glycosylation [
37] or
disulfide bonds [
4] would restrict the recognition of mAbs. We will determine the critical epitope of C
7Mab-2, which helps the understanding of the recognition of mCCR7. We previously determined the Cx
6Mab-1 epitope using 1 × and 2 × alanine scanning methods [
38].
Structural information of chemokine receptors is required to develop drugs that fulfill the requirements. Much effort has been made to determine the structures in complex with either synthetic ligands [
39,
40,
41] or native chemokines [
42,
43] by X-ray crystallography. The cryo-electron microscopy (cryo-EM) has been solved several chemokine receptor-ligand complexes [
44,
45,
46,
47]. Although the CCR7-ligand complex has not been solved, the crystal structure of CCR7 with Cmp2105, an intracellular allosteric CCR7 receptor antagonist was previously determined [
48]. Recently, chemokine receptor-mAb complexes have been solved by cryo-EM
, which provides a detailed structural and mechanistic framework of chemokine receptor activation and inhibition [
49]. Since C
7Mab-2 is known to recognize ECL3 of mCCR7, it could help the structural analysis of mCCR7 in the future studies.
Several
in vitro and
in vivo preclinical tumor models have demonstrated that the increased CCR7 expression promotes tumor growth and metastasis, whereas reduced CCR7 expression suppresses these processes [
50]. For instance, in an orthotopic model, mCCR7-overexpressed mouse mammary tumor cells (PyVmT) demonstrated enhanced metastasis to the lymph nodes. In contrast, the control cells did not migrate to the lymph nodes but metastasized to the lungs. Additionally, mCCR7 overexpression significantly increased tumor growth in PyVmT both
in vitro and
in vivo compared to the control [
51]. Furthermore, in mouse melanoma model, mCCR7-overexpressed B16 melanoma cells exhibited a significantly higher rate of lymph node metastasis than control cells, although the primary tumor size remained unchanged [
52]. To target the mCCR7-positive tumors
in vivo, C
7Mab-2 (rat IgG
2b) should be converted to mouse IgG
2a mAb. We have already determined the V
H and V
L sequence of C
7Mab-2. Therefore, there is an advantage in generating a large amount of recombinant mAbs for therapeutic uses in preclinical models.
In a syngeneic mouse model of oral cancers, the growth of tumors was significantly decreased in mCCR7-knockout (KO) mice [
53]. Single-cell RNA sequence analysis showed that the M2 macrophage proportion in the KO group was low compared to control [
53]. mCCR7 stimulates the polarization of M2 macrophage, which promotes the migration, invasion and proliferation of tumor cells [
53]. Therefore, the depletion of mCCR7-expressing cells by anti-mCCR7 mAbs such as class-switched and defucosylated mouse IgG
2a-type C
7Mab-2 could be helpful to investigate the effect of depletion of mCCR7-expressing cells on the tumor growth.
In conclusion, C7Mab-2 is expected to obtain proof-of-concept in preclinical models to develop antibody therapies.
Author Contributions
Haruto Yamamoto: Investigation. Hiroyuki Suzuki: Investigation, Funding acquisition, Writing – original draft. Tomohiro Tanaka: Investigation, Funding acquisition. Hiroyuki Satofuka: Investigation, Funding acquisition. Mika K. Kaneko: Conceptualization, Funding acquisition. Yukinari Kato: Conceptualization, Funding acquisition, Project administration, Writing – review and editing. All authors have read and agreed to the published version of the manuscript
Funding
This research was supported in part by Japan Agency for Medical Research and Development (AMED) under Grant Numbers: JP24am0521010 (to Y.K.), JP24ama121008 (to Y.K.), JP24ama221339 (to Y.K.), JP24bm1123027 (to Y.K.), and JP24ck0106730 (to Y.K.), and by the Japan Society for the Promotion of Science (JSPS) Grants-in-Aid for Scientific Research (KAKENHI) grant nos. 22K06995 (to H.Suzuki), 24K18268 (to T.T), 24K11652 (to H.Satofuka), and 22K07224 (to Y.K.).
Institutional Review Board Statement
The animal study protocol was approved by the Animal Care and Use Committee of Tohoku University (Permit number: 2022MdA-001) for studies involving animals.
Informed Consent Statement
Not applicable.
Data Availability Statement
All related data and methods are presented in this paper. Additional inquiries should be addressed to the corresponding authors.
Conflicts of Interest
The authors declare no conflict of interest involving this article.
References
- Griffith, J.W.; Sokol, C.L.; Luster, A.D. Chemokines and chemokine receptors: positioning cells for host defense and immunity. Annu Rev Immunol 2014, 32, 659–702. [Google Scholar] [PubMed]
- Schulz, O.; Hammerschmidt, S.I.; Moschovakis, G.L.; Förster, R. Chemokines and Chemokine Receptors in Lymphoid Tissue Dynamics. Annu Rev Immunol 2016, 34, 203–242. [Google Scholar] [PubMed]
- Allen, S.J.; Crown, S.E.; Handel, T.M. Chemokine: receptor structure, interactions, and antagonism. Annu Rev Immunol 2007, 25, 787–820. [Google Scholar] [PubMed]
- Arimont, M.; Sun, S.L.; Leurs, R.; et al. Structural Analysis of Chemokine Receptor-Ligand Interactions. J Med Chem 2017, 60, 4735–4779. [Google Scholar]
- Kohli, K.; Pillarisetty, V.G.; Kim, T.S. Key chemokines direct migration of immune cells in solid tumors. Cancer Gene Ther 2022, 29, 10–21. [Google Scholar]
- Hughes, C.E.; Nibbs, R.J.B. A guide to chemokines and their receptors. Febs j 2018, 285, 2944–2971. [Google Scholar]
- Han, L.; Zhang, L. CCL21/CCR7 axis as a therapeutic target for autoimmune diseases. Int Immunopharmacol 2023, 121, 110431. [Google Scholar]
- Forster, R.; Davalos-Misslitz, A.C.; Rot, A. CCR7 and its ligands: balancing immunity and tolerance. Nat Rev Immunol 2008, 8, 362–371. [Google Scholar]
- Cuesta-Mateos, C.; Terron, F.; Herling, M. CCR7 in Blood Cancers - Review of Its Pathophysiological Roles and the Potential as a Therapeutic Target. Front Oncol 2021, 11, 736758. [Google Scholar]
- Brandum, E.P.; Jorgensen, A.S.; Rosenkilde, M.M.; Hjorto, G.M. Dendritic Cells and CCR7 Expression: An Important Factor for Autoimmune Diseases, Chronic Inflammation, and Cancer. Int J Mol Sci 2021, 22, 15. [Google Scholar]
- Bill, C.A.; Allen, C.M.; Vines, C.M. C-C Chemokine Receptor 7 in Cancer. Cells 2022, 11, 4. [Google Scholar] [CrossRef] [PubMed]
- Alrumaihi, F. The Multi-Functional Roles of CCR7 in Human Immunology and as a Promising Therapeutic Target for Cancer Therapeutics. Front Mol Biosci 2022, 9, 834149. [Google Scholar]
- Ji, H.; Hu, C.; Yang, X.; et al. Lymph node metastasis in cancer progression: molecular mechanisms, clinical significance and therapeutic interventions. Signal Transduct Target Ther 2023, 8, 367. [Google Scholar] [PubMed]
- Irino, T.; Takeuchi, H.; Matsuda, S.; et al. CC-Chemokine receptor CCR7: a key molecule for lymph node metastasis in esophageal squamous cell carcinoma. BMC Cancer 2014, 14, 291. [Google Scholar]
- Mashino, K.; Sadanaga, N.; Yamaguchi, H.; et al. Expression of chemokine receptor CCR7 is associated with lymph node metastasis of gastric carcinoma. Cancer Res 2002, 62, 2937–2941. [Google Scholar]
- Shi, W.; Zou, R.; Yang, M.; et al. Analysis of Genes Involved in Ulcerative Colitis Activity and Tumorigenesis Through Systematic Mining of Gene Co-expression Networks. Front Physiol 2019, 10, 662. [Google Scholar]
- Li, K.; Xu, B.; Xu, G.; Liu, R. CCR7 regulates Twist to induce the epithelial-mesenchymal transition in pancreatic ductal adenocarcinoma. Tumour Biol 2016, 37, 419–424. [Google Scholar]
- Wagner, P.L.; Moo, T.A.; Arora, N.; et al. The chemokine receptors CXCR4 and CCR7 are associated with tumor size and pathologic indicators of tumor aggressiveness in papillary thyroid carcinoma. Ann Surg Oncol 2008, 15, 2833–2841. [Google Scholar]
- Tsuzuki, H.; Takahashi, N.; Kojima, A.; et al. Oral and oropharyngeal squamous cell carcinomas expressing CCR7 have poor prognoses. Auris Nasus Larynx 2006, 33, 37–42. [Google Scholar]
- Basile, J.; Thiers, B.; Maize, J., Sr.; Lathers, D.M. Chemokine receptor expression in non-melanoma skin cancer. J Cutan Pathol 2008, 35, 623–629. [Google Scholar]
- Muller, A.; Homey, B.; Soto, H.; et al. Involvement of chemokine receptors in breast cancer metastasis. Nature 2001, 410, 50–56. [Google Scholar] [PubMed]
- Li, G.; Tanaka, T.; Suzuki, H.; Kaneko, M.K.; Kato, Y. Cx1Mab-1: A Novel Anti-mouse CXCR1 Monoclonal Antibody for Flow Cytometry. Monoclon Antib Immunodiagn Immunother 2024, in press.
- Ouchida, T.; Isoda, Y.; Tanaka, T.; et al. Cx(3)Mab-4: A Novel Anti-Mouse CXCR3 Monoclonal Antibody for Flow Cytometry. Monoclon Antib Immunodiagn Immunother 2024.
- Ouchida, T.; Suzuki, H.; Tanaka, T.; Kaneko, M.K.; Kato, Y. Cx(4)Mab-1: A Novel Anti-Mouse CXCR4 Monoclonal Antibody for Flow Cytometry. Monoclon Antib Immunodiagn Immunother 2023.
- Ouchida, T.; Isoda, Y.; Nakamura, T.; et al. Establishment of a Novel Anti-Mouse CCR1 Monoclonal Antibody C(1)Mab-6. Monoclon Antib Immunodiagn Immunother 2024.
- Asano, T.; Suzuki, H.; Tanaka, T.; et al. C(3)Mab-3: A Monoclonal Antibody for Mouse CC Chemokine Receptor 3 for Flow Cytometry. Monoclon Antib Immunodiagn Immunother 2022, 41, 74–79. [Google Scholar]
- Suzuki, H.; Tanaka, T.; Li, G.; et al. Development of a Sensitive Anti-Mouse CCR5 Monoclonal Antibody for Flow Cytometry. Monoclon Antib Immunodiagn Immunother 2024, 43, 96–100. [Google Scholar]
- Satofuka, H.; Suzuki, H.; Tanaka, T.; et al. A novel anti-mouse CCR7 monoclonal antibody, C7Mab-7, demonstrates high sensitivity in flow cytometry, western blot, and immunohistochemistry. Biochemistry and Biophysics Reports 2025, 41, 101948. [Google Scholar]
- Tanaka, T.; Nanamiya, R.; Takei, J.; et al. Development of Anti-Mouse CC Chemokine Receptor 8 Monoclonal Antibodies for Flow Cytometry. Monoclon Antib Immunodiagn Immunother 2021, 40, 65–70. [Google Scholar]
- Tanaka, T.; Li, G.; Asano, T.; et al. Development of a Novel Anti-Mouse CCR2 Monoclonal Antibody (C(2)Mab-6) by N-Terminal Peptide Immunization. Monoclon Antib Immunodiagn Immunother 2022, 41, 80–86. [Google Scholar]
- Asano, T.; Suzuki, H.; Goto, N.; et al. Establishment of Novel Anti-Mouse CCR3 Monoclonal Antibodies (C(3)Mab-6 and C(3)Mab-7) by N-terminal Peptide Immunization. Monoclon Antib Immunodiagn Immunother 2022, 41, 94–100. [Google Scholar] [PubMed]
- Takei, J.; Suzuki, H.; Asano, T.; et al. Development of a Novel Anti-Mouse CCR4 Monoclonal Antibody (C(4)Mab-1) by N-Terminal Peptide Immunization. Monoclon Antib Immunodiagn Immunother 2022, 41, 87–93. [Google Scholar]
- Asano, T.; Tanaka, T.; Suzuki, H.; et al. Development of a Novel Anti-Mouse CCR6 Monoclonal Antibody (C(6)Mab-13) by N-Terminal Peptide Immunization. Monoclon Antib Immunodiagn Immunother 2022, 41, 343–349. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, H.; Asano, T.; Suzuki, H.; et al. Establishment of a Sensitive Monoclonal Antibody Against Mouse CCR9 (C(9)Mab-24) for Flow Cytometry. Monoclon Antib Immunodiagn Immunother 2023, 42, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, K.; Suzuki, H.; Tanaka, T.; Kaneko, M.K.; Kato, Y. Establishment of a high-affinity anti-mouse CXCR5 monoclonal antibody for flow cytometry. MI 2024, 2, 1. [Google Scholar] [CrossRef]
- Kitamura, K.; Suzuki, H.; Kaneko, M.K.; Kato, Y. Cx(6)Mab-1: A Novel Anti-Mouse CXCR6 Monoclonal Antibody Established by N-Terminal Peptide Immunization. Monoclon Antib Immunodiagn Immunother 2022, 41, 133–141. [Google Scholar]
- Hauser, M.A.; Kindinger, I.; Laufer, J.M.; et al. Distinct CCR7 glycosylation pattern shapes receptor signaling and endocytosis to modulate chemotactic responses. J Leukoc Biol 2016, 99, 993–1007. [Google Scholar]
- Isoda, Y.; Tanaka, T.; Suzuki, H.; et al. Epitope Mapping of an Anti-Mouse CXCR6 Monoclonal Antibody (Cx(6)Mab-1) Using the 2 × Alanine Scanning Method. Monoclon Antib Immunodiagn Immunother 2022, 41, 275–278. [Google Scholar] [CrossRef]
- Zheng, Y.; Qin, L.; Zacarías, N.V.; et al. Structure of CC chemokine receptor 2 with orthosteric and allosteric antagonists. Nature 2016, 540, 458–461. [Google Scholar]
- Oswald, C.; Rappas, M.; Kean, J.; et al. Intracellular allosteric antagonism of the CCR9 receptor. Nature 2016, 540, 462–465. [Google Scholar]
- Tan, Q.; Zhu, Y.; Li, J.; et al. Structure of the CCR5 chemokine receptor-HIV entry inhibitor maraviroc complex. Science 2013, 341, 1387–1390. [Google Scholar] [PubMed]
- Qin, L.; Kufareva, I.; Holden, L.G.; et al. Structural biology. Crystal structure of the chemokine receptor CXCR4 in complex with a viral chemokine. Science 2015, 347, 1117–1122. [Google Scholar] [PubMed]
- Burg, J.S.; Ingram, J.R.; Venkatakrishnan, A.J.; et al. Structural biology. Structural basis for chemokine recognition and activation of a viral G protein-coupled receptor. Science 2015, 347, 1113–1117. [Google Scholar]
- Saha, S.; Sano, F.K.; Sharma, S.; et al. Molecular basis of promiscuous chemokine binding and structural mimicry at the C-X-C chemokine receptor, CXCR2. Mol Cell 2025.
- Peng, Q.; Jiang, H.; Cheng, X.; et al. Cryo-EM Structure and Biochemical Analysis of the Human Chemokine Receptor CCR8. Biochemistry 2024, 63, 1892–1900. [Google Scholar]
- Wang, J.; Chen, G.; Liao, Q.; et al. Cryo-EM structure of the human chemerin receptor 1-Gi protein complex bound to the C-terminal nonapeptide of chemerin. Proc Natl Acad Sci U S A 2023, 120, e2214324120. [Google Scholar]
- Ishimoto, N.; Park, J.H.; Kawakami, K.; et al. Structural basis of CXC chemokine receptor 1 ligand binding and activation. Nat Commun 2023, 14, 4107. [Google Scholar]
- Jaeger, K.; Bruenle, S.; Weinert, T.; et al. Structural Basis for Allosteric Ligand Recognition in the Human CC Chemokine Receptor 7. Cell 2019, 178, 1222–1230.e1210. [Google Scholar]
- Sun, D.; Sun, Y.; Janezic, E.; et al. Structural basis of antibody inhibition and chemokine activation of the human CC chemokine receptor 8. Nat Commun 2023, 14, 7940. [Google Scholar]
- Salem, A.; Alotaibi, M.; Mroueh, R.; Basheer, H.A.; Afarinkia, K. CCR7 as a therapeutic target in Cancer. Biochim Biophys Acta Rev Cancer 2021, 1875, 188499. [Google Scholar]
- Cunningham, H.D.; Shannon, L.A.; Calloway, P.A.; et al. Expression of the C-C chemokine receptor 7 mediates metastasis of breast cancer to the lymph nodes in mice. Transl Oncol 2010, 3, 354–361. [Google Scholar] [PubMed]
- Takekoshi, T.; Fang, L.; Paragh, G.; Hwang, S.T. CCR7-expressing B16 melanoma cells downregulate interferon-γ-mediated inflammation and increase lymphangiogenesis in the tumor microenvironment. Oncogenesis 2012, 1, e9. [Google Scholar] [PubMed]
- Wang, Z.; Kirkwood, K.L.; Wang, Y.; et al. Analysis of the effect of CCR7 on the microenvironment of mouse oral squamous cell carcinoma by single-cell RNA sequencing technology. J Exp Clin Cancer Res 2024, 43, 94. [Google Scholar] [PubMed]
Figure 1.
Schematic representation of anti-mCCR7 mAbs production. (A) The KLH-conjugated mCCR7 ECL peptides (mCCR7-1, mCCR7-2, and mCCR7-3) were immunized into three Sprague–Dawley rats. (B) The spleen cells were fused with P3U1 cells. (C) To select anti-mCCR7 mAb-producing hybridomas, the supernatants were screened by ELISA and flow cytometry using CHO-K1 and CHO/mCCR7 cells. (D) The anti-mCCR7 mAb-producing hybridomas from KLH-mCCR7-3-immunized rats were further cloned by limiting dilution, and C7Mab-2 (rat IgG2b, kappa) was finally established.
Figure 1.
Schematic representation of anti-mCCR7 mAbs production. (A) The KLH-conjugated mCCR7 ECL peptides (mCCR7-1, mCCR7-2, and mCCR7-3) were immunized into three Sprague–Dawley rats. (B) The spleen cells were fused with P3U1 cells. (C) To select anti-mCCR7 mAb-producing hybridomas, the supernatants were screened by ELISA and flow cytometry using CHO-K1 and CHO/mCCR7 cells. (D) The anti-mCCR7 mAb-producing hybridomas from KLH-mCCR7-3-immunized rats were further cloned by limiting dilution, and C7Mab-2 (rat IgG2b, kappa) was finally established.
Figure 2.
Flow cytometry analysis of C7Mab-2 against CHO/mCCR7 and CHO-K1 cells. (A) CHO/mCCR7 and CHO-K1 cells were treated with 0.01, 0.1, 1 and 10 µg/mL of C7Mab-2 (red line). The mAb-treated cells were further incubated with anti-rat IgG conjugated with Alexa Fluor 488. The black line represents the negative control (blocking buffer). The dose-dependent reactivities of C7Mab-2 to CHO/mCCR7 were investigated at least three times. (B) A peptide-blocking assay using C7Mab-2 with mCCR7-3 peptide. C7Mab-2 (2 µg/mL) plus mCCR7-3 (1 μg/mL, blue line) or control (1% DMSO in blocking buffer, red line) were reacted with CHO/mCCR7 for 30 min at 4°C, followed by treatment with Alexa Fluor 488-conjugated anti-rat IgG. The black line represents the negative control (blocking buffer). DMSO, dimethyl sulfoxide.
Figure 2.
Flow cytometry analysis of C7Mab-2 against CHO/mCCR7 and CHO-K1 cells. (A) CHO/mCCR7 and CHO-K1 cells were treated with 0.01, 0.1, 1 and 10 µg/mL of C7Mab-2 (red line). The mAb-treated cells were further incubated with anti-rat IgG conjugated with Alexa Fluor 488. The black line represents the negative control (blocking buffer). The dose-dependent reactivities of C7Mab-2 to CHO/mCCR7 were investigated at least three times. (B) A peptide-blocking assay using C7Mab-2 with mCCR7-3 peptide. C7Mab-2 (2 µg/mL) plus mCCR7-3 (1 μg/mL, blue line) or control (1% DMSO in blocking buffer, red line) were reacted with CHO/mCCR7 for 30 min at 4°C, followed by treatment with Alexa Fluor 488-conjugated anti-rat IgG. The black line represents the negative control (blocking buffer). DMSO, dimethyl sulfoxide.
Figure 3.
Flow cytometry analysis of C7Mab-2 in CC, CXC, CX3C, and XC chemokine receptor-expressed CHO-K1 cells. Eighteen mouse CC, CXC, CX3C, and XC chemokine receptor-expressed CHO-K1 cells were treated with 1 µg/mL of C7Mab-2 (red line) or control blocking buffer (black line), followed by the treatment with anti-rat IgG conjugated with Alexa Fluor 488. Fluorescence data were collected using the SA3800 Cell Analyzer. Note that each receptor expression was previously confirmed by flow cytometry.
Figure 3.
Flow cytometry analysis of C7Mab-2 in CC, CXC, CX3C, and XC chemokine receptor-expressed CHO-K1 cells. Eighteen mouse CC, CXC, CX3C, and XC chemokine receptor-expressed CHO-K1 cells were treated with 1 µg/mL of C7Mab-2 (red line) or control blocking buffer (black line), followed by the treatment with anti-rat IgG conjugated with Alexa Fluor 488. Fluorescence data were collected using the SA3800 Cell Analyzer. Note that each receptor expression was previously confirmed by flow cytometry.
Figure 4.
The binding affinity of C7Mab-2. CHO/mCCR7 cells were suspended in serially diluted C7Mab-2. The cells were treated with anti-rat IgG conjugated with Alexa Fluor 488. Subsequently, the fluorescence data were collected using the SA3800 Cell Analyzer, followed by calculating the KD using GraphPad PRISM 6. The representative results were shown. The three independent experiments determined the KD values (mean ± SD [M]).
Figure 4.
The binding affinity of C7Mab-2. CHO/mCCR7 cells were suspended in serially diluted C7Mab-2. The cells were treated with anti-rat IgG conjugated with Alexa Fluor 488. Subsequently, the fluorescence data were collected using the SA3800 Cell Analyzer, followed by calculating the KD using GraphPad PRISM 6. The representative results were shown. The three independent experiments determined the KD values (mean ± SD [M]).
Figure 5.
Immunohistochemistry of paraffin-embedded cell sections of CHO/mCCR7 and CHO-K1 cells using C7Mab-2. (A, B) Sections of CHO-K1 (A) and CHO/mCCR7 (B) were treated with 20 μg/mL of C7Mab-2, followed by treatment with the Histofine Simple Stain Mouse MAX PO (Rat). (C, D) A peptide-blocking assay using C7Mab-2 with mCCR7-3 peptide. C7Mab-2 (20 µg/mL) plus mCCR7-3 (2 μg/mL, C) or control (1% DMSO in blocking buffer, D) were reacted with the sections of CHO/mCCR7, followed by treatment with the Histofine Simple Stain Mouse MAX PO (Rat). Color was developed using DAB, and counterstaining was performed using hematoxylin. Scale bar = 100 μm.
Figure 5.
Immunohistochemistry of paraffin-embedded cell sections of CHO/mCCR7 and CHO-K1 cells using C7Mab-2. (A, B) Sections of CHO-K1 (A) and CHO/mCCR7 (B) were treated with 20 μg/mL of C7Mab-2, followed by treatment with the Histofine Simple Stain Mouse MAX PO (Rat). (C, D) A peptide-blocking assay using C7Mab-2 with mCCR7-3 peptide. C7Mab-2 (20 µg/mL) plus mCCR7-3 (2 μg/mL, C) or control (1% DMSO in blocking buffer, D) were reacted with the sections of CHO/mCCR7, followed by treatment with the Histofine Simple Stain Mouse MAX PO (Rat). Color was developed using DAB, and counterstaining was performed using hematoxylin. Scale bar = 100 μm.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).