Submitted:
05 March 2025
Posted:
06 March 2025
You are already at the latest version
Abstract
Keywords:
Brief Report
Furin Basics
Furin and SARS-CoV-2
Furin Arginine Pair Codon Usage Bias in 2025 SARS-CoV-2 Isolates
Methods
- To retrieve the genome region covering the S gene sequence (positions 20000-26000).
- Based on these coordinates, to retrieve the S gene region from the downloaded GISAID genome.
- To translate forward three frames of the retrieved S gene region (coding region).
- To identify the correct translation reading frame (no ambiguous characters, no stop signals).
- Based on the proper reading frame, to adjust the S gene coding sequence.
- To identify the furin site arginine pair. For each S protein sequence a 2-position RR-window was run. Using the RRAR pattern it was confirmed that the identified arginine pair corresponded to the arginine pair of the furin site.
- To identify the codon usage of the furin site argini pair. Knowing the positions of the arginine pair in the protein, multiplying by three the respective codons were extracted from the S gene sequences. The cases in which the arginine pair code was not CGG-CGG were recorded.
Competing interests
Acknowledgement
References
- Kristian G Andersen, Andrew Rambaut, W Ian Lipkin, Edward C Holmes, Robert F Garry. The proximal origin of SARS-CoV-2. Nat. Med. 26:450-452, 2020. [CrossRef] [PubMed]
- Murat Seyran, Damiano Pizzol, Parise Adadi, Tarek M A El-Aziz, Sk Sarif Hassan, Antonio Soares, Ramesh Kandimalla, Kenneth Lundstrom, Murtaza Tambuwala, Alaa A A Aljabali, Amos Lal, Gajendra K Azad, Pabitra P Choudhury, Vladimir N Uversky, Samendra P Sherchan, Bruce D Uhal, Nima Rezaei, Adam M Brufsky. Questions concerning the proximal origin of SARS-CoV-2. J. Med. Virol. 93(3):1204-1206, 2021. [CrossRef] [PubMed]
- Steven Quay, Richard Muller. The science suggests a Wuhan lab leak. The Wall Street Journal. Monday, June 7, 2021.
- Antonio, R. Romeu, Enric Ollé. SARS-CoV-2 and the Secret of the Furin Site. Preprints 2021, 2021020264. [CrossRef]
- Antonio, R. Romeu, Enric Ollé. The SARS-CoV-2 arginine dimers, 02 August 2021, PREPRINT (Version 1) available at Research Square. [CrossRef]
- Antonio R Romeu. Probable human origin of the SARS-CoV-2 polybasic furin cleavage motif. BMC Genom Data 24:71, 2023. [CrossRef] [PubMed]
- Antonio, R. Romeu. Synonymous base substitution in SARS-CoV-2 EE.2 lineage furin arginine CGG–CGG codons, 05 July 2023, PREPRINT (Version 1) available at Research Square. [CrossRef]
- Antonio, R. Romeu. SARS-CoV-2 CQ.1 and CQ.1.1 lineage isolates: 100% synonymous base substitution at furin arginine CGG–CGG codons, 13 July 2023, PREPRINT (Version 1) available at Research Square. [CrossRef]
- Antonio, R. Romeu. SARS-CoV-2 XBB.1.16.20 lineage study sample: 21% shows a synonymous base substitution in the S gene arginine codons CGG–CGG encoding the S protein furin cleavage site, 18 September 2023, PREPRINT (Version 1) available at Research Square. [CrossRef]
- FURIN furin, paired basic amino acid cleaving enzyme [ Homo sapiens (human) ]. NCBI. Available online: https://www.ncbi.nlm.nih.gov/gene/5045 (accessed on 1 March 2025).
- Sun Tian, Qingsheng Huang, Ying Fang, Jianhua Wu. FurinDB: A database of 20-residue furin cleavage site motifs, substrates and their associated drugs. J. Mol. Sci. 8;12(2):1060-5, 2011. [CrossRef] [PubMed]
- Cody, B. Jackson, Michael Farzan, Bing Chen, Hyeryun Choe. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell. Biol. 23(1):3-20, 2022. [CrossRef] [PubMed]
- Sergey A Shiryaev, Andrei V Chernov, Vladislav S Golubkov, Elliot R Thomsen, Eugene Chudin, Mark S Chee, Igor A Kozlov, Alex Y Strongin, Piotr Cieplak. High-resolution analysis and functional mapping of cleavage sites and substrate proteins of furin in the human proteome. PLoS One 8(1):e54290, 2013. [CrossRef] [PubMed]
- Yuelong Shu, John McCauley. GISAID: Global initiative on sharing all influenza data – from vision to reality. Euro. Surveill. 2017;22:30494, 2017. [CrossRef] [PubMed]
- Global Initiative on Sharing Avian Influenza Data (GISAID) database. Available online: https://www.gisaid.org/ (accessed on 1 March 2025).
- Andrew Rambaut, Edward C Holmes, Áine O’Toole, Verity Hill, John T McCrone, Christopher Ruis, Louis du Plessis, Oliver G Pybus. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol. 2020;5:1403-7. [CrossRef] [PubMed]
- Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. (1-2):203-14, 2000. [CrossRef] [PubMed]
- 18 NCBI BLASTn https://blast.ncbi.nlm.nih.gov/BlastAlign.cgi. Accessed March 1, 2025.
- Fábio Madeira, Young Mi Park, Joon Lee, Nicola Buso, Tamer Gur, Nandana Madhusoodanan, Prasad Basutkar, Adrian R N Tivey, Simon C Potter, Robert D Finn, Rodrigo Lopez. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47(W1):W636-W641, 2019. doi10.1093/nar/gkz268. [PubMed]
- EMBL-EBI. Clustal Omega. https://www.ebi.ac.uk/jdispatcher/msa/clustalo. Accessed March 1, 2025.
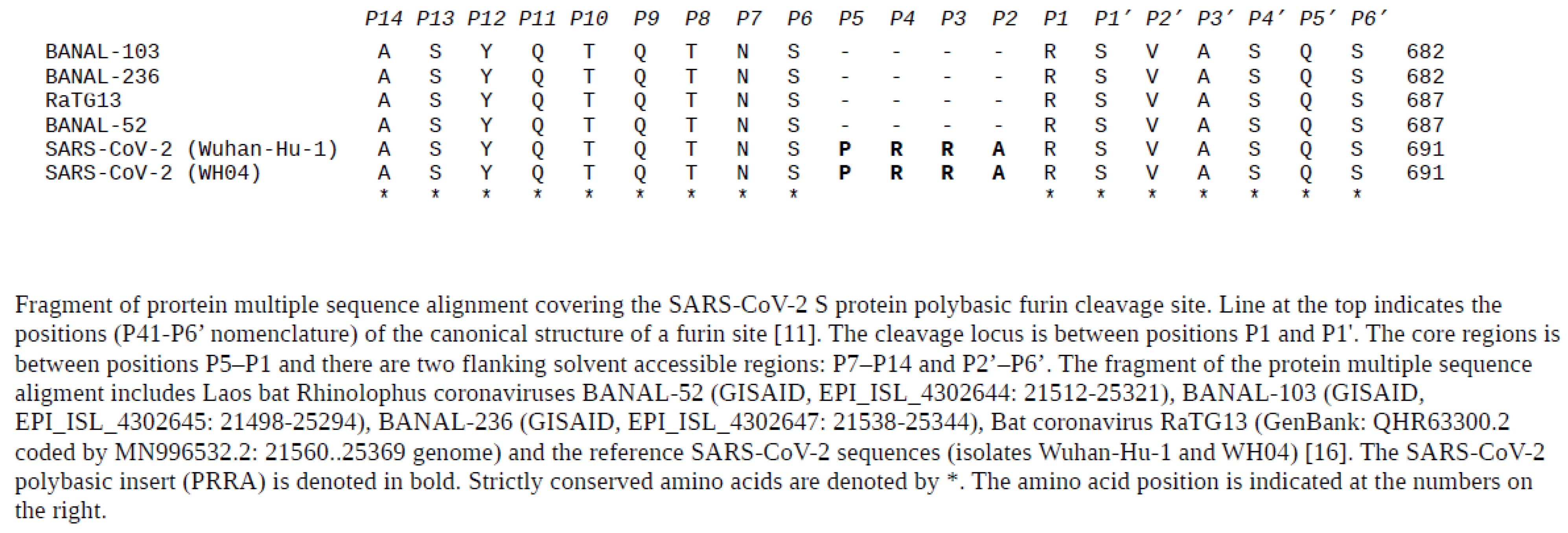
| |||||||||
| GISAID epi_isl id | Region | Country | Division | Length* | Lineage | Collect. Date | RR** | RR_Coding*** | Furin_Cleavage_Site** |
| NI_045512.2 (GenBank) | Asia | China | Hubei (Wuhan) | 29903 | B | 2019-12-26 | RR-683 | CGGCGG-23659 | 672-ASYQTQTNSPRRARSVASQS-691 |
| EPI_ISL_406801 | Asia | China | Hubei (Wuhan) | 29872 | A | 2020-01-30 | RR-682 | CGGCGG-23594 | 671-ASYQTQTNSPRRARSVASQS-690 |
| EPI_ISL_19679698 | North America | Canada | British Columbia | 29553 | KP.3.1.1 | 2025-01-01 | RR-672 | CGGCGT-23531 | 661-ASYQTQTKSRRRARSVASQS-680 |
| EPI_ISL_19700434 | Asia | Israel | 29401 | LF.7.1.3 | 2025-01-02 | RR-673 | CGGCGA-23252 | 662-ASYQTQTKSRRRARSVASQS-681 | |
| EPI_ISL_19684930 | Asia | Japan | Hyogo | 29749 | KP.3.1.1 | 2025-01-02 | RR-672 | CGGCGT-23531 | 661-ASYQTQTKSRRRARSVASQS-680 |
| EPI_ISL_19682951 | North America | USA | Arizona | 29721 | XEC | 2025-01-02 | RR-673 | CGGCGT-23538 | 662-ASYQTQTKSRRRARSVASQS-681 |
| EPI_ISL_19666094 | North America | USA | Illinois | 29690 | XEC | 2025-01-02 | RR-673 | AGGCGG-23516 | 662-ASYQTQTKSRRRARSVASQS-681 |
| EPI_ISL_19663311 | North America | Canada | Ontario | 29722 | KP.3.1.1 | 2025-01-04 | RR-674 | CGGCGA-23513 | 663-XXXXXQTKSRRRARSVASQS-682 |
| EPI_ISL_19682995 | North America | USA | New York | 29698 | XEC | 2025-01-04 | RR-673 | CGTCGG-23538 | 662-ASYQTQTKSRRRARSVASQS-681 |
| EPI_ISL_19681312 | Europe | Germany | North Rhine-Westphalia | 29706 | XEC | 2025-01-06 | RR-673 | CGGCGT-23510 | 662-XXXXXQTKSRRRARSVASQS-681 |
| EPI_ISL_19666768 | Asia | Singapore | 29569 | LF.7.3.1 | 2025-01-06 | RR-673 | CGACGG-23310 | 662-ASYQTXTKSRRRARSVASQS-681 | |
| EPI_ISL_19706176 | Asia | South Korea | 29795 | KP.3.1.1 | 2025-01-06 | RR-668 | CGGCGT-23569 | 657-ASYQTQTKSRRRARSVASQS-676 | |
| EPI_ISL_19675573 | North America | Canada | Ontario | 29716 | KP.3.1.1 | 2025-01-07 | RR-672 | CGGCGT-23507 | 661-XXXXXQTKSRRRARSVASQS-680 |
| EPI_ISL_19683050 | North America | USA | New York | 29698 | XEC | 2025-01-07 | RR-673 | CGTCGG-23538 | 662-ASYQTQTKSRRRARSVASQS-681 |
| EPI_ISL_19676763 | Oceania | Australia | Queensland | 29721 | XEC | 2025-01-08 | RR-673 | CGTCGG-23538 | 662-ASYQTQTKSRRRARSVASQS-681 |
| EPI_ISL_19684954 | Asia | Japan | Hokkaido | 29723 | KP.3.1.1 | 2025-01-08 | RR-672 | CGGCGT-23531 | 661-ASYQTQTKSRRRARSVASQS-680 |
| EPI_ISL_19701204 | Europe | United Kingdom | England | 29738 | JN.1.8 | 2025-01-08 | RR-672 | CGGCGT-23521 | 661-XSYQTQTKSRRRARSVASQS-680 |
| EPI_ISL_19684105 | North America | USA | Colorado | 29703 | KP.3.1 | 2025-01-08 | RR-672 | CGTCGG-23507 | 661-ASYQTQTKSRRRARSVASQS-680 |
| EPI_ISL_19691313 | North America | Canada | Ontario | 29716 | KP.3.1.1 | 2025-01-10 | RR-672 | CGGCGT-23507 | 661-XXXXXQTKSRRRARSVASQS-680 |
| EPI_ISL_19696722 | Asia | Japan | Miyagi | 29723 | LP.8.1 | 2025-01-10 | RR-672 | CGGCGA-23531 | 661-ASYQTQTKSRRRARSVASQS-680 |
| EPI_ISL_19707783 | North America | USA | Hawaii | 29703 | KP.3.1.1 | 2025-01-10 | RR-672 | CGGCGT-23507 | 661-ASYQTQTKSRRRARSVASQS-680 |
| EPI_ISL_19729037 | Asia | Japan | Toyama | 29723 | KP.3.1.1 | 2025-01-11 | RR-672 | CGGCGT-23531 | 661-ASYQTQTKSRRRARSVASQS-680 |
| EPI_ISL_19693631 | North America | Canada | Quebec | 29716 | KP.3.1.1 | 2025-01-12 | RR-672 | CGGCGT-23507 | 661-ASYQTQTKSRRRARSVASQS-680 |
| EPI_ISL_19684922 | Asia | Japan | Hyogo | 29723 | KP.3.1.1 | 2025-01-12 | RR-672 | CGGCGT-23531 | 661-ASYQTQTKSRRRARSVASQS-680 |
| EPI_ISL_19684921 | Asia | Japan | Hyogo | 29723 | KP.3.1.1 | 2025-01-12 | RR-672 | CGGCGT-23531 | 661-ASYQTQTKSRRRARSVASQS-680 |
| EPI_ISL_19691627 | North America | Canada | Ontario | 29716 | KP.3.1.1 | 2025-01-14 | RR-672 | CGGCGT-23507 | 661-XXXXXQTKSRRRARSVASQS-680 |
| EPI_ISL_19731362 | South America | Ecuador | Cotopaxi | 29830 | JN.1.11 | 2025-01-14 | RR-669 | AGGCGG-23572 | 658-ASYQTQTKSRRRARSVASQS-677 |
| EPI_ISL_19700994 | Asia | Japan | Gifu | 29723 | KP.3.1.1 | 2025-01-14 | RR-672 | CGGCGT-23531 | 661-ASYQTQTKSRRRARSVASQS-680 |
| EPI_ISL_19707721 | Asia | Japan | Saitama | 29723 | KP.3.1.1 | 2025-01-14 | RR-672 | CGGCGT-23531 | 661-ASYQTQTKSRRRARSVASQS-680 |
| EPI_ISL_19697568 | Asia | Japan | Tokushima | 29723 | KP.3.1.1 | 2025-01-14 | RR-672 | CGGCGT-23531 | 661-ASYQTQTKSRRRARSVASQS-680 |
| EPI_ISL_19712229 | Oceania | Australia | Queensland | 29721 | KP.3.3 | 2025-01-15 | RR-673 | CGGCGA-23538 | 662-ASYQTQTKSRRRARSVASQS-681 |
| EPI_ISL_19696525 | Oceania | Australia | South Australia | 29673 | KP.3.3 | 2025-01-15 | RR-672 | AGGCGG-23512 | 661-ASYQTQTKSRRRARSVASQS-680 |
| EPI_ISL_19695415 | Europe | Germany | Baden-Wurttemberg | 29703 | KP.3.1.1 | 2025-01-15 | RR-672 | AGGCGG-23507 | 661-ASYQTQTKSRRRARSVASQS-680 |
| EPI_ISL_19681098 | Asia | Japan | Kanagawa | 29723 | KP.3.1.1 | 2025-01-15 | RR-672 | CGGCGT-23531 | 661-ASYQTQTKSRRRARSVASQS-680 |
| EPI_ISL_19691828 | North America | Canada | Ontario | 29716 | KP.3.1.1 | 2025-01-16 | RR-672 | CGGCGT-23507 | 661-XXXXXQTKSRRRARSVASQS-680 |
| EPI_ISL_19691829 | North America | Canada | Ontario | 29716 | KP.3.1.1 | 2025-01-16 | RR-672 | CGGCGT-23507 | 661-XXXXXQTKSRRRARSVASQS-680 |
| EPI_ISL_19700973 | Asia | Japan | Gifu | 29723 | KP.3.1.1 | 2025-01-16 | RR-672 | CGGCGT-23531 | 661-ASYQTQTKSRRRARSVASQS-680 |
| EPI_ISL_19692038 | North America | Canada | Ontario | 29716 | KP.3.1.1 | 2025-01-17 | RR-672 | CGGCGT-23507 | 661-XXXXXQTKSRRRARSVVSQS-680 |
| EPI_ISL_19692035 | North America | Canada | Ontario | 29716 | KP.3.1.1 | 2025-01-17 | RR-672 | CGGCGT-23507 | 661-XXXXXQTKSRRRARSVVSQS-680 |
| EPI_ISL_19692037 | North America | Canada | Ontario | 29716 | KP.3.1.1 | 2025-01-17 | RR-672 | CGGCGT-23507 | 661-XXXXXQTKSRRRARSVVSQS-680 |
| EPI_ISL_19702183 | North America | Canada | Ontario | 29722 | KP.3.1.1 | 2025-01-18 | RR-674 | CGGCGT-23513 | 663-XXXXXQTKSRRRARSVVSQS-682 |
| EPI_ISL_19693229 | Asia | Hong Kong | 29706 | XDV.1 | 2025-01-18 | RR-673 | AGGCGG-23510 | 662-ASYQTQTKSRRRARSVASQS-681 | |
| EPI_ISL_19714873 | Oceania | Australia | South Australia | 29673 | KP.3.3 | 2025-01-19 | RR-672 | AGGCGG-23512 | 661-ASYQTQTKSRRRARSVASQS-680 |
| EPI_ISL_19714872 | Oceania | Australia | South Australia | 29673 | KP.3.3 | 2025-01-19 | RR-672 | AGGCGG-23512 | 661-ASYQTQTKSRRRARSVASQS-680 |
| EPI_ISL_19704613 | Asia | Japan | Hyogo | 29723 | KP.3.1.1 | 2025-01-19 | RR-672 | CGGCGT-23531 | 661-ASYQTQTKSRRRARSVASQS-680 |
| EPI_ISL_19700970 | Asia | Japan | Gifu | 29723 | KP.3.1.1 | 2025-01-20 | RR-672 | CGGCGT-23531 | 661-ASYQTQTKSRRRARSVASQS-680 |
| EPI_ISL_19696435 | Asia | Japan | Hokkaido | 29723 | KP.3.1.1 | 2025-01-20 | RR-672 | CGGCGT-23531 | 661-ASYQTQTKSRRRARSVASQS-680 |
| EPI_ISL_19697628 | Asia | Japan | Hokkaido | 29723 | KP.3.1.1 | 2025-01-20 | RR-672 | CGGCGT-23531 | 661-ASYQTQTKSRRRARSVASQS-680 |
| EPI_ISL_19729193 | Asia | Japan | Miyagi | 29723 | KP.3.1.1 | 2025-01-20 | RR-672 | CGGCGT-23531 | 661-ASYQTQTKSRRRARSVASQS-680 |
| EPI_ISL_19729288 | Asia | Japan | Saitama | 29723 | KP.3.1.1 | 2025-01-20 | RR-672 | CGTCGG-23531 | 661-ASYQTQTKSRRRARSVASQS-680 |
| EPI_ISL_19712131 | Asia | Japan | Ehime | 29723 | KP.3.1.1 | 2025-01-22 | RR-672 | CGGCGT-23531 | 661-ASYQTQTKSRRRARSVASQS-680 |
| EPI_ISL_19702323 | North America | Canada | Ontario | 29716 | JN.1.8 | 2025-01-23 | RR-674 | CGGAGG-23507 | 663-ASYQTQTKSRRRARSVASQS-682 |
| EPI_ISL_19729202 | Asia | Japan | Miyagi | 29723 | KP.3.1.1 | 2025-01-23 | RR-672 | CGGCGT-23531 | 661-ASYQTQTKSRRRARSVASQS-680 |
| EPI_ISL_19716895 | North America | USA | New Jersey | 29681 | JN.1.8 | 2025-01-24 | RR-672 | CGTCGG-23507 | 661-ASYQTQTKSRRRARSVASQS-680 |
| EPI_ISL_19724894 | Asia | Japan | Tokyo | 29749 | KP.3.1.1 | 2025-01-26 | RR-672 | CGGCGT-23531 | 661-ASYQTQTKSRRRARSVASQS-680 |
| EPI_ISL_19731882 | Europe | United Kingdom | England | 29749 | XEC | 2025-01-30 | RR-672 | CGTCGG-23530 | 661-ASYQTQTKSRRRARSVVSQS-680 |
| EPI_ISL_19718119 | North America | Canada | Ontario | 29703 | KP.3.1.1 | 2025-02-03 | RR-672 | CGGCGT-23507 | 661-ASYQTQTKSRRRARSVVSQS-680 |
| EPI_ISL_19718127 | North America | Canada | Ontario | 29703 | KP.3.1.1 | 2025-02-03 | RR-672 | CGGCGT-23507 | 661-ASYQTQTKSRRRARSVVSQS-680 |
| EPI_ISL_19718123 | North America | Canada | Ontario | 29716 | KP.3.1.1 | 2025-02-03 | RR-672 | CGGCGT-23507 | 661-XXXXXQTKSRRRARSVVSQS-680 |
| EPI_ISL_19722193 | Asia | Japan | Hokkaido | 29723 | KP.3.1.1 | 2025-02-03 | RR-672 | CGGCGT-23531 | 661-ASYQTQTKSRRRARSVASQS-680 |
| EPI_ISL_19731403 | Asia | Japan | Saitama | 29725 | KP.3.1.1 | 2025-02-03 | RR-672 | CGGCGT-23533 | 661-ASYQTQTKSRRRARSVASQS-680 |
| EPI_ISL_19718129 | North America | Canada | Ontario | 29703 | KP.3.1.1 | 2025-02-04 | RR-672 | CGGCGT-23507 | 661-ASYQTQTKSRRRARSVVSQS-680 |
| EPI_ISL_19722192 | Asia | Japan | Hokkaido | 29723 | KP.3.1.1 | 2025-02-05 | RR-672 | CGGCGT-23531 | 661-ASYQTQTKSRRRARSVASQS-680 |
| EPI_ISL_19722186 | Asia | Japan | Hokkaido | 29723 | KP.3.1.1 | 2025-02-06 | RR-672 | CGGCGT-23531 | 661-ASYQTQTKSRRRARSVASQS-680 |
| ||
| Lineage | Percent | Number of Isolates |
| KP.3.1.1 | 1.74 | 42 |
| XEC | 0.09 | 7 |
| KP.3.3 | 2.07 | 4 |
| JN.1.8 | 0.64 | 3 |
| LF.7.1.3 | 6.67 | 1 |
| LF.7.3.1 | 6.25 | 1 |
| XDV.1 | 1.59 | 1 |
| KP.3.1 | 0.51 | 1 |
| JN.1.11 | 0.35 | 1 |
| LP.8.1 | 0.12 | 1 |
| ||
| Lineage | Percent | Number of Isolates |
| XEC | 44.5105 | 7792 |
| KP.3.1.1 | 13.7896 | 2414 |
| MC.1 | 5.2839 | 925 |
| LP.8.1 | 4.8783 | 854 |
| LP.8.1.1 | 4.4899 | 786 |
| JN.1.8 | 2.6962 | 472 |
| JN.1.16.1 | 2.4106 | 422 |
| JN.1.16 | 1.7365 | 304 |
| JN.1.11 | 1.6223 | 284 |
| KP.3.3.2 | 1.1882 | 208 |
| KP.3.1 | 1.1310 | 198 |
| KP.3.3 | 1.1025 | 193 |
| MC.10.1 | 0.9540 | 167 |
| JN.1.16.3 | 0.9482 | 166 |
| LF.7.2.1 | 0.7712 | 135 |
| MB.1.1 | 0.6626 | 116 |
| KP.3 | 0.5655 | 99 |
| LF.7.1 | 0.5370 | 94 |
| LP.8.1.2 | 0.5027 | 88 |
| KP.2 | 0.4913 | 86 |
| LF.7 | 0.4513 | 79 |
| XEC.1 | 0.4456 | 78 |
| MC.10.2.1 | 0.4341 | 76 |
| MC.1.2 | 0.3713 | 65 |
| XDV.1 | 0.3599 | 63 |
| XDY | 0.3427 | 60 |
| JN.1 | 0.2856 | 50 |
| KP.1.1.3 | 0.2799 | 49 |
| LF.7.3 | 0.2742 | 48 |
| MC.21.1 | 0.2399 | 42 |
| JN.1.40 | 0.2171 | 38 |
| JN.1.8.1 | 0.2114 | 37 |
| KS.1.1 | 0.2114 | 37 |
| LF.7.1.2 | 0.1942 | 34 |
| MC.13 | 0.1942 | 34 |
| KP.3.3.1 | 0.1828 | 32 |
| MC.2 | 0.1828 | 32 |
| MC.10.1.6 | 0.1599 | 28 |
| MC.1.3 | 0.1542 | 27 |
| MC.10.1.1 | 0.1485 | 26 |
| JN.11 | 0.1371 | 24 |
| MC.13.2 | 0.1257 | 22 |
| MC.1.5 | 0.1200 | 21 |
| MC.1.6 | 0.1028 | 18 |
| MC.24 | 0.1028 | 18 |
| KP.1.1 | 0.0971 | 17 |
| MC.8.1 | 0.0971 | 17 |
| LF.7.3.1 | 0.0914 | 16 |
| LP.7 | 0.0914 | 16 |
| KP.1 | 0.0857 | 15 |
| LF.7.1.3 | 0.0857 | 15 |
| LP.5 | 0.0857 | 15 |
| MC.9 | 0.0857 | 15 |
| KP.1.1.5 | 0.0800 | 14 |
| KP.3.2 | 0.0800 | 14 |
| MC.10.1.2 | 0.0800 | 14 |
| MC.10.2 | 0.0743 | 13 |
| JN.1.3 | 0.0685 | 12 |
| KP.3.1.4 | 0.0685 | 12 |
| MC.1.1 | 0.0685 | 12 |
| JN.1.18 | 0.0628 | 11 |
| KP.1.1.1 | 0.0628 | 11 |
| LF.7.2 | 0.0628 | 11 |
| MC.11 | 0.0628 | 11 |
| JN.1.13 | 0.0571 | 10 |
| BA.2.86.1 | 0.0514 | 9 |
| JN.1.37 | 0.0514 | 9 |
| LF.7.6.1 | 0.0514 | 9 |
| MC.1.4 | 0.0514 | 9 |
| MC.33.1 | 0.0514 | 9 |
| JN.1.11.1 | 0.0457 | 8 |
| JN.1.15 | 0.0457 | 8 |
| KP.2.2 | 0.0457 | 8 |
| KP.2.3.12 | 0.0457 | 8 |
| MC.10 | 0.0457 | 8 |
| MC.35 | 0.0457 | 8 |
| JN.1.18.6 | 0.0400 | 7 |
| KP.2.9 | 0.0400 | 7 |
| KP.3.3.3 | 0.0400 | 7 |
| MC.1.7 | 0.0400 | 7 |
| MC.10.1.4 | 0.0400 | 7 |
| MC.28 | 0.0400 | 7 |
| PA.1 | 0.0400 | 7 |
| XDQ | 0.0400 | 7 |
| JN.1.32 | 0.0343 | 6 |
| KP.3.1.3 | 0.0343 | 6 |
| KS.1 | 0.0343 | 6 |
| LU.2 | 0.0343 | 6 |
| MC.16 | 0.0343 | 6 |
| MC.19 | 0.0343 | 6 |
| MC.8 | 0.0343 | 6 |
| Unassigned | 0.0343 | 6 |
| BA.2.86 | 0.0286 | 5 |
| JN.1.10 | 0.0286 | 5 |
| KP.2.3 | 0.0286 | 5 |
| LB.1.1 | 0.0286 | 5 |
| LP.9 | 0.0286 | 5 |
| MB.1 | 0.0286 | 5 |
| MC.10.1.5 | 0.0286 | 5 |
| MC.13.1 | 0.0286 | 5 |
| MC.31 | 0.0286 | 5 |
| MC.4 | 0.0286 | 5 |
| BA.3 | 0.0228 | 4 |
| KP.5 | 0.0228 | 4 |
| LW.1 | 0.0228 | 4 |
| MC.13.3 | 0.0228 | 4 |
| MC.13.4 | 0.0228 | 4 |
| MC.23 | 0.0228 | 4 |
| MC.26 | 0.0228 | 4 |
| MC.34 | 0.0228 | 4 |
| JN.1.20 | 0.0171 | 3 |
| JN.1.4 | 0.0171 | 3 |
| JN.1.59 | 0.0171 | 3 |
| JN.1.9 | 0.0171 | 3 |
| JN.10 | 0.0171 | 3 |
| KP.2.3.4 | 0.0171 | 3 |
| KP.3.2.3 | 0.0171 | 3 |
| LP.8.2 | 0.0171 | 3 |
| MC.1.3.1 | 0.0171 | 3 |
| MC.10.1.3 | 0.0171 | 3 |
| MC.13.2.1 | 0.0171 | 3 |
| MC.3 | 0.0171 | 3 |
| MC.30 | 0.0171 | 3 |
| MC.6 | 0.0171 | 3 |
| ND.1.1.2 | 0.0171 | 3 |
| JN.1.13.1 | 0.0114 | 2 |
| JN.1.4.4 | 0.0114 | 2 |
| JN.1.9.2 | 0.0114 | 2 |
| KP.2.3.6 | 0.0114 | 2 |
| KP.3.5 | 0.0114 | 2 |
| MC.17.1 | 0.0114 | 2 |
| MC.19.1 | 0.0114 | 2 |
| MC.2.1 | 0.0114 | 2 |
| MC.27 | 0.0114 | 2 |
| ND.1.1 | 0.0114 | 2 |
| NT.1 | 0.0114 | 2 |
| XDQ.1 | 0.0114 | 2 |
| BA.1.1 | 0.0057 | 1 |
| BA.2 | 0.0057 | 1 |
| BQ.1.1 | 0.0057 | 1 |
| BQ.1.1.18 | 0.0057 | 1 |
| BQ.1.18 | 0.0057 | 1 |
| FL.4.2 | 0.0057 | 1 |
| HF.1.1 | 0.0057 | 1 |
| HV.1 | 0.0057 | 1 |
| JG.3 | 0.0057 | 1 |
| JN.1.18.3 | 0.0057 | 1 |
| JN.1.22 | 0.0057 | 1 |
| JN.1.4.6 | 0.0057 | 1 |
| JN.1.49 | 0.0057 | 1 |
| JN.1.49.1 | 0.0057 | 1 |
| JN.1.51 | 0.0057 | 1 |
| JN.1.52 | 0.0057 | 1 |
| JN.1.53 | 0.0057 | 1 |
| JN.1.7.3 | 0.0057 | 1 |
| KP.2.14 | 0.0057 | 1 |
| KP.2.3.10 | 0.0057 | 1 |
| KP.3.1.2 | 0.0057 | 1 |
| KP.3.2.5 | 0.0057 | 1 |
| KP.4 | 0.0057 | 1 |
| KP.4.2 | 0.0057 | 1 |
| KS.1.2 | 0.0057 | 1 |
| LB.1.3.1 | 0.0057 | 1 |
| LF.3.1 | 0.0057 | 1 |
| LF.7.4 | 0.0057 | 1 |
| LF.7.6 | 0.0057 | 1 |
| LP.10.1.1 | 0.0057 | 1 |
| LV.1 | 0.0057 | 1 |
| MA.1 | 0.0057 | 1 |
| MC.14 | 0.0057 | 1 |
| MC.17 | 0.0057 | 1 |
| MC.21 | 0.0057 | 1 |
| MC.24.1 | 0.0057 | 1 |
| MC.26.1 | 0.0057 | 1 |
| MC.28.1 | 0.0057 | 1 |
| MC.28.1.1 | 0.0057 | 1 |
| MC.32.1 | 0.0057 | 1 |
| MC.33.2 | 0.0057 | 1 |
| MC.37 | 0.0057 | 1 |
| ML.1 | 0.0057 | 1 |
| ML.2 | 0.0057 | 1 |
| MM.1 | 0.0057 | 1 |
| XDK.3 | 0.0057 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




