Submitted:
21 February 2025
Posted:
25 February 2025
You are already at the latest version
Abstract
Keywords:
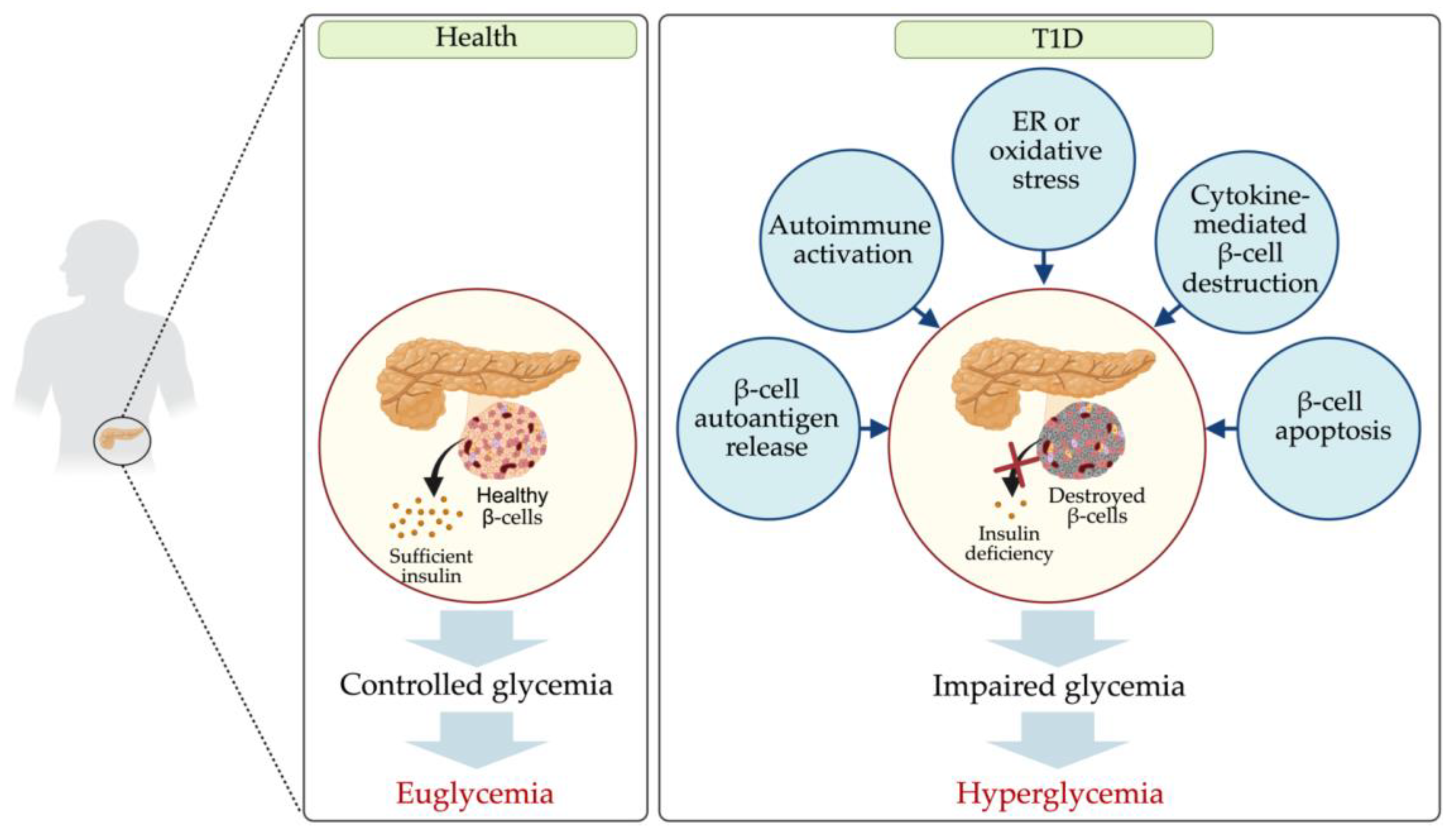
1. Introduction
2. Dysregulated miRNAs in T1D Patients
Dysregulated miRNAs in Various Samples from T1D Patients
Dysregulated miRNAs in Serum and Plasma
Dysregulated miRNAs in Blood Cells
Dysregulated miRNAs in T-Cells
Dysregulated miRNAs in Exosomes Derived from Plasma and Urine
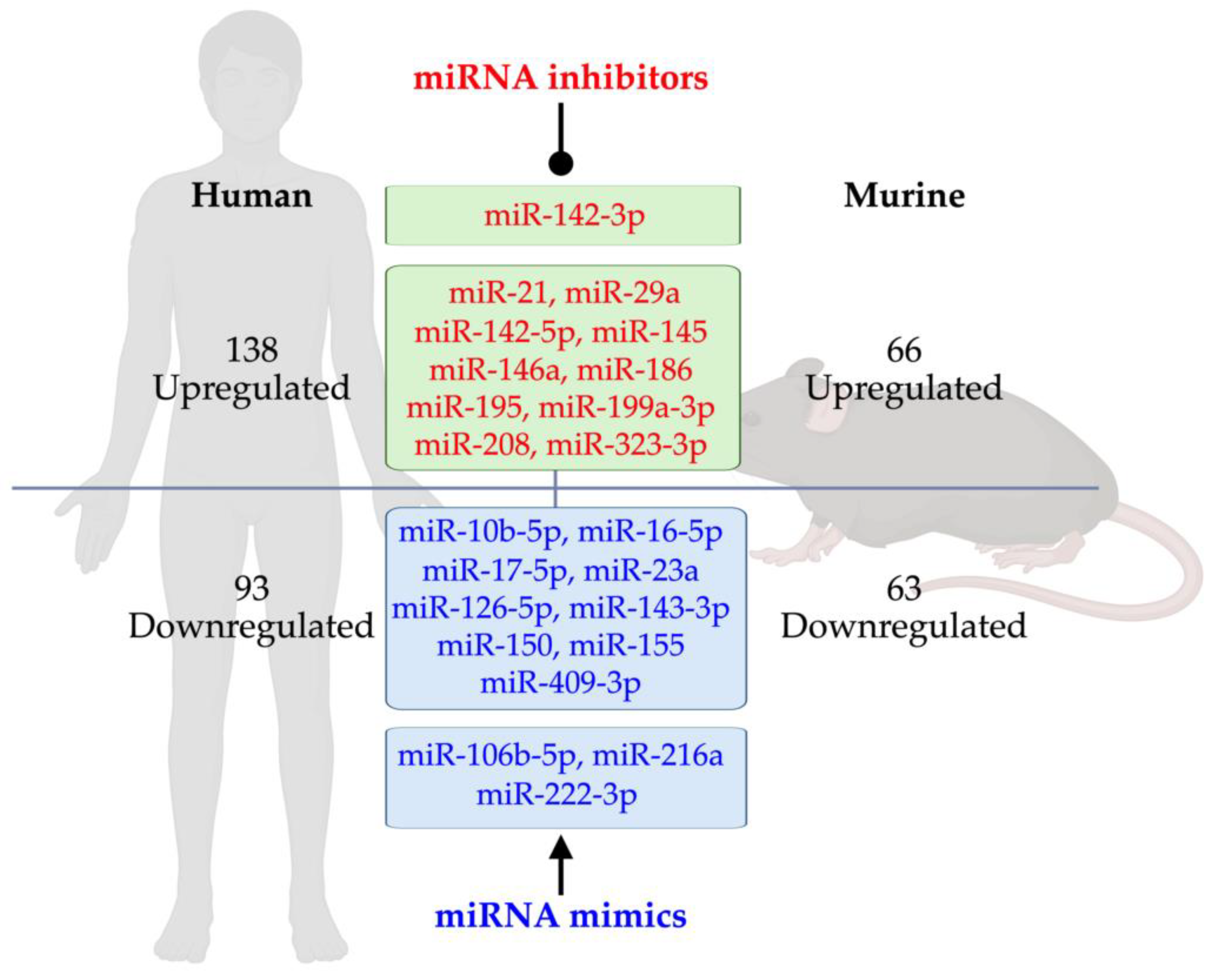
3. Dysregulated miRNAs in T1D Rodents
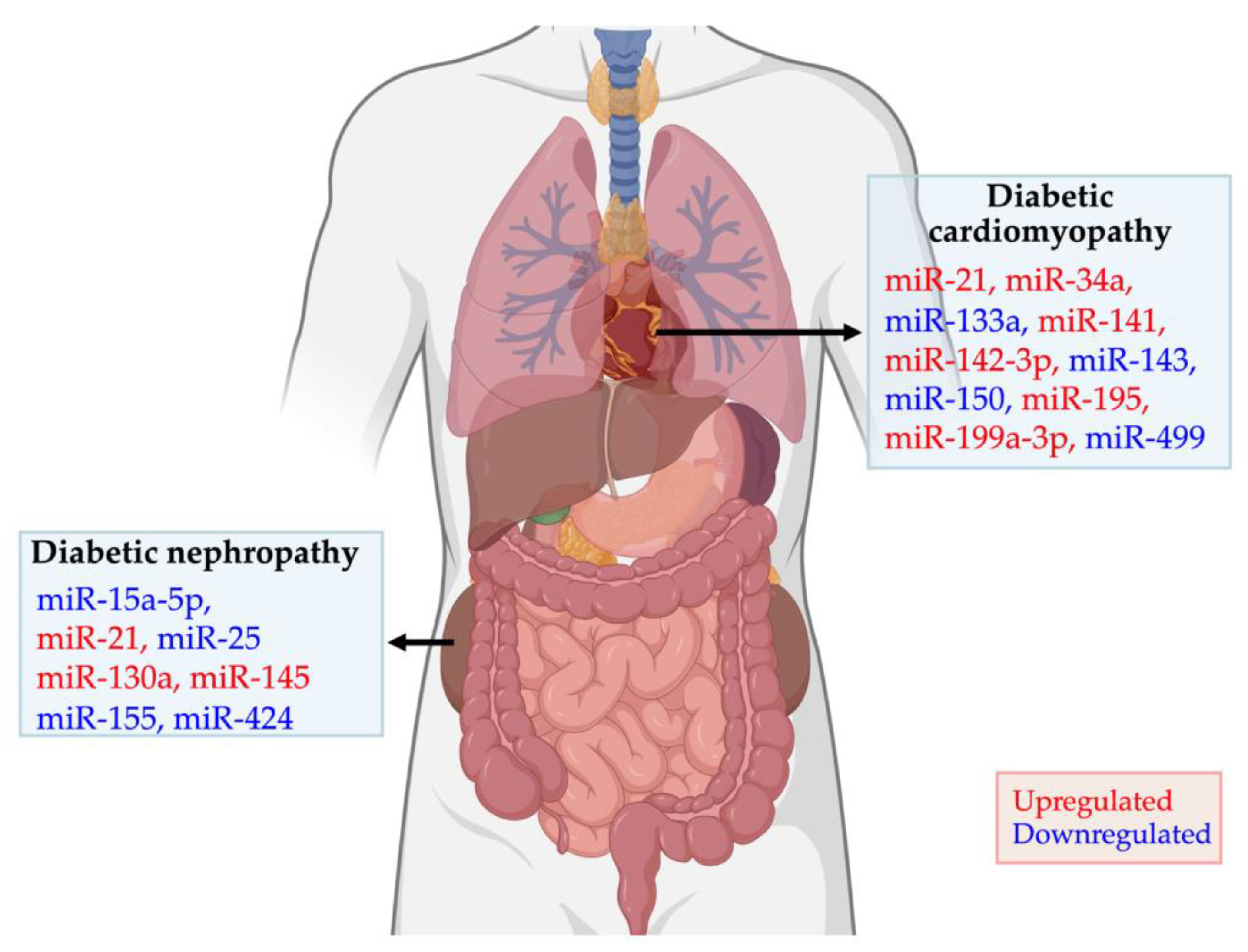
4. Dysregulated miRNAs in Diabetic Cardiomyopathy and Nephropathy
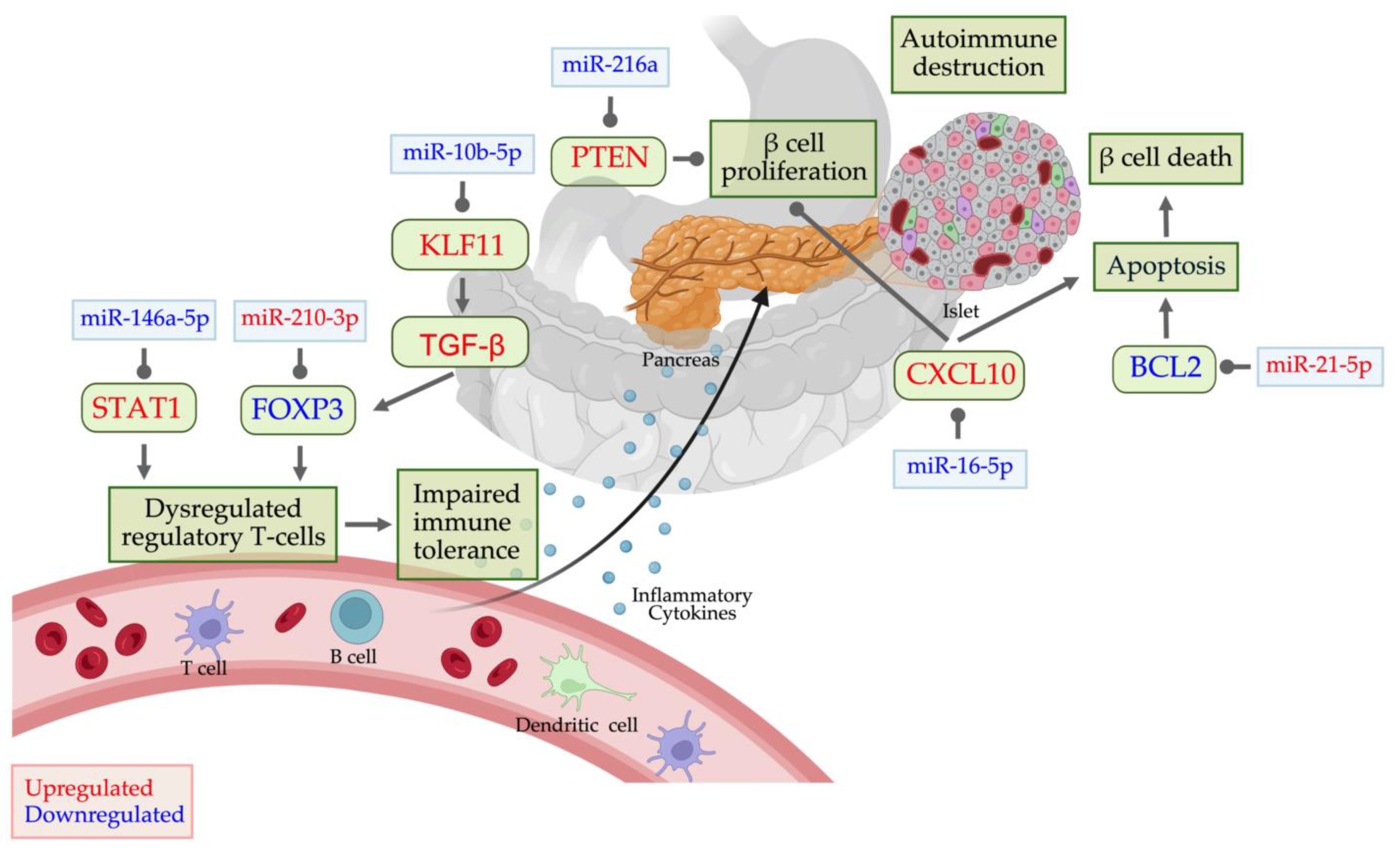
5. Dysregulated miRNAs in T1D and Their Potential Targets
β-Cell Autoantigen Release
Autoimmune Activation
Endoplasmic Reticulum and Oxidative Stress
Apoptosis
Insulin signaling
7. Conclusion and Future Study
Author Contributions
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Abbreviations
References
- Banday, M.Z.; Sameer, A.S.; Nissar, S. , Pathophysiology of diabetes: An overview. Avicenna J Med 2020, 10, 174–188. [Google Scholar] [CrossRef] [PubMed]
- Tuomi, T.; Santoro, N.; Caprio, S.; Cai, M.; Weng, J.; Groop, L. , The many faces of diabetes: a disease with increasing heterogeneity. Lancet 2014, 383, 1084–94. [Google Scholar] [CrossRef] [PubMed]
- Seyhan, A.A.; Nunez Lopez, Y.O.; Xie, H.; Yi, F.; Mathews, C.; Pasarica, M.; Pratley, R.E. , Pancreas-enriched miRNAs are altered in the circulation of subjects with diabetes: a pilot cross-sectional study. Sci Rep 2016, 6, 31479. [Google Scholar] [CrossRef]
- Gregory, G.A.; Robinson, T.I.G.; Linklater, S.E.; Wang, F.; Colagiuri, S.; de Beaufort, C.; Donaghue, K.C.; International Diabetes Federation Diabetes Atlas Type 1 Diabetes in Adults Special Interest, G. ; Magliano, D.J.; Maniam, J.; Orchard, T.J.; Rai, P.; Ogle, G.D., Global incidence, prevalence, and mortality of type 1 diabetes in 2021 with projection to 2040: a modelling study. Lancet Diabetes Endocrinol 2022, 10, 741–760. [Google Scholar] [CrossRef]
- Katsarou, A.; Gudbjornsdottir, S.; Rawshani, A.; Dabelea, D.; Bonifacio, E.; Anderson, B.J.; Jacobsen, L.M.; Schatz, D.A.; Lernmark, A. , Type 1 diabetes mellitus. Nat Rev Dis Primers 2017, 3, 17016. [Google Scholar] [CrossRef] [PubMed]
- Daneman, D. , Type 1 diabetes. Lancet 2006, 367, 847–58. [Google Scholar] [CrossRef]
- Margaritis, K.; Margioula-Siarkou, G.; Giza, S.; Kotanidou, E.P.; Tsinopoulou, V.R.; Christoforidis, A.; Galli-Tsinopoulou, A. , Micro-RNA Implications in Type-1 Diabetes Mellitus: A Review of Literature. Int J Mol Sci, 2021; 22. [Google Scholar]
- Assmann, T.S.; Recamonde-Mendoza, M.; De Souza, B.M.; Crispim, D. , MicroRNA expression profiles and type 1 diabetes mellitus: systematic review and bioinformatic analysis. Endocr Connect 2017, 6, 773–790. [Google Scholar] [CrossRef]
- Pirot, P.; Cardozo, A.K.; Eizirik, D.L. , Mediators and mechanisms of pancreatic beta-cell death in type 1 diabetes. Arq Bras Endocrinol Metabol 2008, 52, 156–65. [Google Scholar] [CrossRef]
- Ounissi-Benkalha, H.; Polychronakos, C. , The molecular genetics of type 1 diabetes: new genes and emerging mechanisms. Trends Mol Med 2008, 14, 268–75. [Google Scholar] [CrossRef]
- Ziegler, A.G.; Rewers, M.; Simell, O.; Simell, T.; Lempainen, J.; Steck, A.; Winkler, C.; Ilonen, J.; Veijola, R.; Knip, M.; Bonifacio, E.; Eisenbarth, G.S. , Seroconversion to multiple islet autoantibodies and risk of progression to diabetes in children. JAMA 2013, 309, 2473–9. [Google Scholar] [CrossRef]
- Guay, C.; Regazzi, R. , Circulating microRNAs as novel biomarkers for diabetes mellitus. Nat Rev Endocrinol 2013, 9, 513–21. [Google Scholar] [CrossRef] [PubMed]
- von Scholten, B.J.; Kreiner, F.F.; Gough, S.C.L.; von Herrath, M. , Current and future therapies for type 1 diabetes. Diabetologia 2021, 64, 1037–1048. [Google Scholar] [CrossRef] [PubMed]
- Natarajan, R.; Putta, S.; Kato, M. , MicroRNAs and diabetic complications. J Cardiovasc Transl Res 2012, 5, 413–22. [Google Scholar] [CrossRef]
- He, Z.; King, G.L. , Microvascular complications of diabetes. Endocrinol Metab Clin North Am 2004, 33, 215–38. [Google Scholar] [CrossRef]
- Beckman, J.A.; Creager, M.A.; Libby, P. , Diabetes and atherosclerosis: epidemiology, pathophysiology, and management. JAMA 2002, 287, 2570–81. [Google Scholar] [CrossRef]
- Esteller, M. , Non-coding RNAs in human disease. Nat Rev Genet 2011, 12, 861–74. [Google Scholar] [CrossRef] [PubMed]
- Satake, E.; Pezzolesi, M.G.; Md Dom, Z.I.; Smiles, A.M.; Niewczas, M.A.; Krolewski, A.S. , Circulating miRNA Profiles Associated With Hyperglycemia in Patients With Type 1 Diabetes. Diabetes 2018, 67, 1013–1023. [Google Scholar] [CrossRef]
- Erener, S.; Marwaha, A.; Tan, R.; Panagiotopoulos, C.; Kieffer, T.J. , Profiling of circulating microRNAs in children with recent onset of type 1 diabetes. JCI Insight 2017, 2, e89656. [Google Scholar] [CrossRef]
- Takahashi, P.; Xavier, D.J.; Evangelista, A.F.; Manoel-Caetano, F.S.; Macedo, C.; Collares, C.V.; Foss-Freitas, M.C.; Foss, M.C.; Rassi, D.M.; Donadi, E.A.; Passos, G.A.; Sakamoto-Hojo, E.T. , MicroRNA expression profiling and functional annotation analysis of their targets in patients with type 1 diabetes mellitus. Gene 2014, 539, 213–23. [Google Scholar] [CrossRef]
- Rupaimoole, R.; Slack, F.J. , MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov 2017, 16, 203–222. [Google Scholar] [CrossRef]
- Lu, Q.; Wu, R.; Zhao, M.; Garcia-Gomez, A.; Ballestar, E. , miRNAs as Therapeutic Targets in Inflammatory Disease. Trends Pharmacol Sci 2019, 40, 853–865. [Google Scholar] [CrossRef]
- Singh, R.; Ha, S.E.; Park, H.S.; Debnath, S.; Cho, H.; Baek, G.; Yu, T.Y.; Ro, S. , Sustained Effectiveness and Safety of Therapeutic miR-10a/b in Alleviating Diabetes and Gastrointestinal Dysmotility without Inducing Cancer or Inflammation in Murine Liver and Colon. Int J Mol Sci, 2024; 25. [Google Scholar]
- Zogg, H.; Singh, R.; Ro, S. , Current Advances in RNA Therapeutics for Human Diseases. Int J Mol Sci, 2022; 23. [Google Scholar]
- Niehrs, C.; Pollet, N. , Synexpression groups in eukaryotes. Nature 1999, 402, 483–7. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Maki, M.; Ding, R.; Yang, Y.; Zhang, B.; Xiong, L. , Genome-wide survey of tissue-specific microRNA and transcription factor regulatory networks in 12 tissues. Sci Rep 2014, 4, 5150. [Google Scholar] [CrossRef]
- Ambros, V. , The functions of animal microRNAs. Nature 2004, 431, 350–5. [Google Scholar] [CrossRef] [PubMed]
- Landgraf, P.; Rusu, M.; Sheridan, R.; Sewer, A.; Iovino, N.; Aravin, A.; Pfeffer, S.; Rice, A.; Kamphorst, A.O.; Landthaler, M.; Lin, C.; Socci, N.D.; Hermida, L.; Fulci, V.; Chiaretti, S.; Foa, R.; Schliwka, J.; Fuchs, U.; Novosel, A.; Muller, R.U.; Schermer, B.; Bissels, U.; Inman, J.; Phan, Q.; Chien, M.; Weir, D.B.; Choksi, R.; De Vita, G.; Frezzetti, D.; Trompeter, H.I.; Hornung, V.; Teng, G.; Hartmann, G.; Palkovits, M.; Di Lauro, R.; Wernet, P.; Macino, G.; Rogler, C.E.; Nagle, J.W.; Ju, J.; Papavasiliou, F.N.; Benzing, T.; Lichter, P.; Tam, W.; Brownstein, M.J.; Bosio, A.; Borkhardt, A.; Russo, J.J.; Sander, C.; Zavolan, M.; Tuschl, T. , A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 2007, 129, 1401–14. [Google Scholar] [CrossRef] [PubMed]
- Sayed, A.S.; Xia, K.; Salma, U.; Yang, T.; Peng, J. , Diagnosis, prognosis and therapeutic role of circulating miRNAs in cardiovascular diseases. Heart Lung Circ 2014, 23, 503–10. [Google Scholar] [CrossRef]
- Zampetaki, A.; Willeit, P.; Drozdov, I.; Kiechl, S.; Mayr, M. , Profiling of circulating microRNAs: from single biomarkers to re-wired networks. Cardiovasc Res 2012, 93, 555–62. [Google Scholar] [CrossRef]
- Nielsen, L.B.; Wang, C.; Sorensen, K.; Bang-Berthelsen, C.H.; Hansen, L.; Andersen, M.L.; Hougaard, P.; Juul, A.; Zhang, C.Y.; Pociot, F.; Mortensen, H.B. , Circulating levels of microRNA from children with newly diagnosed type 1 diabetes and healthy controls: evidence that miR-25 associates to residual beta-cell function and glycaemic control during disease progression. Exp Diabetes Res 2012, 2012, 896362. [Google Scholar]
- Santos, A.S.; Ferreira, L.R.P.; da Silva, A.C.; Alves, L.I.; Damasceno, J.G.; Kulikowski, L.; Cunha-Neto, E.; da Silva, M.E.R. , Progression of Type 1 Diabetes: Circulating MicroRNA Expression Profiles Changes from Preclinical to Overt Disease. J Immunol Res 2022, 2022, 2734490. [Google Scholar] [CrossRef]
- Grieco, G.E.; Cataldo, D.; Ceccarelli, E.; Nigi, L.; Catalano, G.; Brusco, N.; Mancarella, F.; Ventriglia, G.; Fondelli, C.; Guarino, E.; Crisci, I.; Sebastiani, G.; Dotta, F. , Serum Levels of miR-148a and miR-21-5p Are Increased in Type 1 Diabetic Patients and Correlated with Markers of Bone Strength and Metabolism. Noncoding RNA, 2018; 4. [Google Scholar]
- Assmann, T.S.; Recamonde-Mendoza, M.; Punales, M.; Tschiedel, B.; Canani, L.H.; Crispim, D. , MicroRNA expression profile in plasma from type 1 diabetic patients: Case-control study and bioinformatic analysis. Diabetes Res Clin Pract 2018, 141, 35–46. [Google Scholar] [CrossRef]
- Osipova, J.; Fischer, D.C.; Dangwal, S.; Volkmann, I.; Widera, C.; Schwarz, K.; Lorenzen, J.M.; Schreiver, C.; Jacoby, U.; Heimhalt, M.; Thum, T.; Haffner, D. , Diabetes-associated microRNAs in pediatric patients with type 1 diabetes mellitus: a cross-sectional cohort study. J Clin Endocrinol Metab 2014, 99, E1661–E1665. [Google Scholar] [CrossRef] [PubMed]
- Ventriglia, G.; Mancarella, F.; Sebastiani, G.; Cook, D.P.; Mallone, R.; Mathieu, C.; Gysemans, C.; Dotta, F. , miR-409-3p is reduced in plasma and islet immune infiltrates of NOD diabetic mice and is differentially expressed in people with type 1 diabetes. Diabetologia 2020, 63, 124–136. [Google Scholar] [CrossRef]
- Ferraz, R.S.; Santos, L.C.B.; da-Silva-Cruz, R.L.; Braga-da-Silva, C.H.; Magalhaes, L.; Ribeiro-Dos-Santos, A.; Vidal, A.; Vinasco-Sandoval, T.; Reis-das-Merces, L.; Sena-Dos-Santos, C.; Pereira, A.L.; Silva, L.S.D.; de Melo, F.T.C.; de Souza, A.; Leal, V.S.G.; de Figueiredo, P.B.B.; Neto, J.F.A.; de Moraes, L.V.; de Lemos, G.N.; de Queiroz, N.N.M.; Felicio, K.M.; Cavalcante, G.C.; Ribeiro-Dos-Santos, A.; Felicio, J.S. , Global miRNA expression reveals novel nuclear and mitochondrial interactions in Type 1 diabetes mellitus. Front Endocrinol (Lausanne) 2022, 13, 1033809. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Ye, L.; Wang, B.; Gao, J.; Liu, R.; Hong, J.; Wang, W.; Gu, W.; Ning, G. , Decreased miR-146 expression in peripheral blood mononuclear cells is correlated with ongoing islet autoimmunity in type 1 diabetes patients 1miR-146. J Diabetes 2015, 7, 158–65. [Google Scholar] [CrossRef]
- Estrella, S.; Garcia-Diaz, D.F.; Codner, E.; Camacho-Guillen, P.; Perez-Bravo, F. , Expression of miR-22 and miR-150 in type 1 diabetes mellitus: Possible relationship with autoimmunity and clinical characteristics. Med Clin (Barc) 2016, 147, 245–7. [Google Scholar] [CrossRef]
- Wang, G.; Gu, Y.; Xu, N.; Zhang, M.; Yang, T. , Decreased expression of miR-150, miR146a and miR424 in type 1 diabetic patients: Association with ongoing islet autoimmunity. Biochem Biophys Res Commun 2018, 498, 382–387. [Google Scholar] [CrossRef] [PubMed]
- Massaro, J.D.; Polli, C.D.; Costa, E.S.M.; Alves, C.C.; Passos, G.A.; Sakamoto-Hojo, E.T.; Rodrigues de Holanda Miranda, W.; Bispo Cezar, N.J.; Rassi, D.M.; Crispim, F.; Dib, S.A.; Foss-Freitas, M.C.; Pinheiro, D.G.; Donadi, E.A. , Post-transcriptional markers associated with clinical complications in Type 1 and Type 2 diabetes mellitus. Mol Cell Endocrinol 2019, 490, 1–14. [Google Scholar] [CrossRef]
- Hezova, R.; Slaby, O.; Faltejskova, P.; Mikulkova, Z.; Buresova, I.; Raja, K.R.; Hodek, J.; Ovesna, J.; Michalek, J. , microRNA-342, microRNA-191 and microRNA-510 are differentially expressed in T regulatory cells of type 1 diabetic patients. Cell Immunol 2010, 260, 70–4. [Google Scholar] [CrossRef]
- Garcia-Contreras, M.; Shah, S.H.; Tamayo, A.; Robbins, P.D.; Golberg, R.B.; Mendez, A.J.; Ricordi, C. , Plasma-derived exosome characterization reveals a distinct microRNA signature in long duration Type 1 diabetes. Sci Rep 2017, 7, 5998. [Google Scholar] [CrossRef]
- Barutta, F.; Tricarico, M.; Corbelli, A.; Annaratone, L.; Pinach, S.; Grimaldi, S.; Bruno, G.; Cimino, D.; Taverna, D.; Deregibus, M.C.; Rastaldi, M.P.; Perin, P.C.; Gruden, G. , Urinary exosomal microRNAs in incipient diabetic nephropathy. PLoS One 2013, 8, e73798. [Google Scholar] [CrossRef]
- Wang, S.; Liu, Z.; Wang, J.; Ji, X.; Yao, Z.; Wang, X. , miR-21 promotes osteoclastogenesis through activation of PI3K/Akt signaling by targeting Pten in RAW264.7 cells. Mol Med Rep 2020, 21, 1125–1132. [Google Scholar] [CrossRef] [PubMed]
- Aghaei-Zarch, S.M. , Crosstalk between MiRNAs/lncRNAs and PI3K/AKT signaling pathway in diabetes mellitus: Mechanistic and therapeutic perspectives. Noncoding RNA Res 2024, 9, 486–507. [Google Scholar] [CrossRef]
- Melkman-Zehavi, T.; Oren, R.; Kredo-Russo, S.; Shapira, T.; Mandelbaum, A.D.; Rivkin, N.; Nir, T.; Lennox, K.A.; Behlke, M.A.; Dor, Y.; Hornstein, E. , miRNAs control insulin content in pancreatic beta-cells via downregulation of transcriptional repressors. EMBO J 2011, 30, 835–45. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Thielen, L.A.; Chen, J.; Grayson, T.B.; Grimes, T.; Bridges, S.L., Jr.; Tse, H.M.; Smith, B.; Patel, R.; Li, P.; Evans-Molina, C.; Ovalle, F.; Shalev, A. , Serum miR-204 is an early biomarker of type 1 diabetes-associated pancreatic beta-cell loss. Am J Physiol Endocrinol Metab 2019, 317, E723–E730. [Google Scholar] [CrossRef] [PubMed]
- Angelescu, M.A.; Andronic, O.; Dima, S.O.; Popescu, I.; Meivar-Levy, I.; Ferber, S.; Lixandru, D. , miRNAs as Biomarkers in Diabetes: Moving towards Precision Medicine. Int J Mol Sci, 2022; 23. [Google Scholar]
- Zi, Y.; Zhang, Y.; Wu, Y.; Zhang, L.; Yang, R.; Huang, Y. , Downregulation of microRNA-25-3p inhibits the proliferation and promotes the apoptosis of multiple myeloma cells via targeting the PTEN/PI3K/AKT signaling pathway. Int J Mol Med, 2021; 47. [Google Scholar]
- Kong, R.; Gao, J.; Ji, L.; Zhao, D. , MicroRNA-126 promotes proliferation, migration, invasion and endothelial differentiation while inhibits apoptosis and osteogenic differentiation of bone marrow-derived mesenchymal stem cells. Cell Cycle 2020, 19, 2119–2138. [Google Scholar] [CrossRef]
- Fang, S.; Ma, X.; Guo, S.; Lu, J. , MicroRNA-126 inhibits cell viability and invasion in a diabetic retinopathy model via targeting IRS-1. Oncol Lett 2017, 14, 4311–4318. [Google Scholar] [CrossRef]
- Rosell, R.; Wei, J.; Taron, M. , Circulating MicroRNA Signatures of Tumor-Derived Exosomes for Early Diagnosis of Non-Small-Cell Lung Cancer. Clin Lung Cancer 2009, 10, 8–9. [Google Scholar] [CrossRef]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O'Briant, K.C.; Allen, A.; Lin, D.W.; Urban, N.; Drescher, C.W.; Knudsen, B.S.; Stirewalt, D.L.; Gentleman, R.; Vessella, R.L.; Nelson, P.S.; Martin, D.B.; Tewari, M. , Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A 2008, 105, 10513–8. [Google Scholar] [CrossRef]
- Cortez, M.A.; Calin, G.A. , MicroRNA identification in plasma and serum: a new tool to diagnose and monitor diseases. Expert Opin Biol Ther 2009, 9, 703–711. [Google Scholar] [CrossRef]
- Gilad, S.; Meiri, E.; Yogev, Y.; Benjamin, S.; Lebanony, D.; Yerushalmi, N.; Benjamin, H.; Kushnir, M.; Cholakh, H.; Melamed, N.; Bentwich, Z.; Hod, M.; Goren, Y.; Chajut, A. , Serum microRNAs are promising novel biomarkers. PLoS One 2008, 3, e3148. [Google Scholar] [CrossRef]
- Wang, C.; Hu, J.; Lu, M.; Gu, H.; Zhou, X.; Chen, X.; Zen, K.; Zhang, C.Y.; Zhang, T.; Ge, J.; Wang, J.; Zhang, C. , A panel of five serum miRNAs as a potential diagnostic tool for early-stage renal cell carcinoma. Sci Rep 2015, 5, 7610. [Google Scholar] [CrossRef] [PubMed]
- Gui, J.; Tian, Y.; Wen, X.; Zhang, W.; Zhang, P.; Gao, J.; Run, W.; Tian, L.; Jia, X.; Gao, Y. , Serum microRNA characterization identifies miR-885-5p as a potential marker for detecting liver pathologies. Clin Sci (Lond) 2011, 120, 183–93. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Chen, H.; Si, H.; Li, X.; Ding, X.; Sheng, Q.; Chen, P.; Zhang, H. , Serum miR-23a, a potential biomarker for diagnosis of pre-diabetes and type 2 diabetes. Acta Diabetol 2014, 51, 823–31. [Google Scholar] [CrossRef]
- Zhou, S.S.; Jin, J.P.; Wang, J.Q.; Zhang, Z.G.; Freedman, J.H.; Zheng, Y.; Cai, L. , miRNAS in cardiovascular diseases: potential biomarkers, therapeutic targets and challenges. Acta Pharmacol Sin 2018, 39, 1073–1084. [Google Scholar] [CrossRef]
- Olivieri, F.; Rippo, M.R.; Procopio, A.D.; Fazioli, F. , Circulating inflamma-miRs in aging and age-related diseases. Front Genet 2013, 4, 121. [Google Scholar] [CrossRef]
- Du, M.; Liu, S.; Gu, D.; Wang, Q.; Zhu, L.; Kang, M.; Shi, D.; Chu, H.; Tong, N.; Chen, J.; Adams, T.S.; Zhang, Z.; Wang, M. , Clinical potential role of circulating microRNAs in early diagnosis of colorectal cancer patients. Carcinogenesis 2014, 35, 2723–30. [Google Scholar] [CrossRef]
- Wang, W.; Sun, G.; Zhang, L.; Shi, L.; Zeng, Y. , Circulating microRNAs as novel potential biomarkers for early diagnosis of acute stroke in humans. J Stroke Cerebrovasc Dis 2014, 23, 2607–2613. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.K.; Zhu, J.Q.; Zhang, J.T.; Li, Q.; Li, Y.; He, J.; Qin, Y.W.; Jing, Q. , Circulating microRNA: a novel potential biomarker for early diagnosis of acute myocardial infarction in humans. Eur Heart J 2010, 31, 659–66. [Google Scholar] [CrossRef]
- Sohel, M.M.H. , Circulating microRNAs as biomarkers in cancer diagnosis. Life Sci 2020, 248, 117473. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, Y.; Zhao, Y.; Wang, Y.; Ding, H.; Xue, S.; Li, P. , Circulating miRNAs as biomarkers for early diagnosis of coronary artery disease. Expert Opin Ther Pat 2018, 28, 591–601. [Google Scholar] [CrossRef]
- de Gonzalo-Calvo, D.; Vea, A.; Bar, C.; Fiedler, J.; Couch, L.S.; Brotons, C.; Llorente-Cortes, V.; Thum, T. , Circulating non-coding RNAs in biomarker-guided cardiovascular therapy: a novel tool for personalized medicine? Eur Heart J 2019, 40, 1643–1650. [Google Scholar] [CrossRef]
- Gao, W.; Liu, L.; Lu, X.; Shu, Y. , Circulating microRNAs: possible prediction biomarkers for personalized therapy of non-small-cell lung carcinoma. Clin Lung Cancer 2011, 12, 14–7. [Google Scholar] [CrossRef]
- Lu, J.; Xie, F.; Geng, L.; Shen, W.; Sui, C.; Yang, J. , Potential Role of MicroRNA-210 as Biomarker in Human Cancers Detection: A Meta-Analysis. Biomed Res Int 2015, 2015, 303987. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Luo, X.; Cui, H.; Ni, X.; Yuan, M.; Guo, Y.; Huang, X.; Zhou, H.; de Vries, N.; Tak, P.P.; Chen, S.; Shen, N. , MicroRNA-146A contributes to abnormal activation of the type I interferon pathway in human lupus by targeting the key signaling proteins. Arthritis Rheum 2009, 60, 1065–75. [Google Scholar] [CrossRef]
- Zhu, H.; Leung, S.W. , Identification of microRNA biomarkers in type 2 diabetes: a meta-analysis of controlled profiling studies. Diabetologia 2015, 58, 900–11. [Google Scholar] [CrossRef]
- Trajkovski, M.; Hausser, J.; Soutschek, J.; Bhat, B.; Akin, A.; Zavolan, M.; Heim, M.H.; Stoffel, M. , MicroRNAs 103 and 107 regulate insulin sensitivity. Nature 2011, 474, 649–53. [Google Scholar] [CrossRef] [PubMed]
- Liew, C.C.; Ma, J.; Tang, H.C.; Zheng, R.; Dempsey, A.A. , The peripheral blood transcriptome dynamically reflects system wide biology: a potential diagnostic tool. J Lab Clin Med 2006, 147, 126–32. [Google Scholar] [CrossRef]
- Luty, W.H.; Rodeberg, D.; Parness, J.; Vyas, Y.M. , Antiparallel segregation of notch components in the immunological synapse directs reciprocal signaling in allogeneic Th:DC conjugates. J Immunol 2007, 179, 819–29. [Google Scholar] [CrossRef] [PubMed]
- Pattu, V.; Qu, B.; Schwarz, E.C.; Strauss, B.; Weins, L.; Bhat, S.S.; Halimani, M.; Marshall, M.; Rettig, J.; Hoth, M. , SNARE protein expression and localization in human cytotoxic T lymphocytes. Eur J Immunol 2012, 42, 470–5. [Google Scholar] [CrossRef]
- Pelayo, R.; Hirose, J.; Huang, J.; Garrett, K.P.; Delogu, A.; Busslinger, M.; Kincade, P.W. , Derivation of 2 categories of plasmacytoid dendritic cells in murine bone marrow. Blood 2005, 105, 4407–15. [Google Scholar] [CrossRef]
- Murray, A.R.; Chen, Q.; Takahashi, Y.; Zhou, K.K.; Park, K.; Ma, J.X. , MicroRNA-200b downregulates oxidation resistance 1 (Oxr1) expression in the retina of type 1 diabetes model. Invest Ophthalmol Vis Sci 2013, 54, 1689–97. [Google Scholar] [CrossRef] [PubMed]
- Baseler, W.A.; Thapa, D.; Jagannathan, R.; Dabkowski, E.R.; Croston, T.L.; Hollander, J.M. , miR-141 as a regulator of the mitochondrial phosphate carrier (Slc25a3) in the type 1 diabetic heart. Am J Physiol Cell Physiol 2012, 303, C1244–C1251. [Google Scholar] [CrossRef]
- Silva, V.A.; Polesskaya, A.; Sousa, T.A.; Correa, V.M.; Andre, N.D.; Reis, R.I.; Kettelhut, I.C.; Harel-Bellan, A.; De Lucca, F.L. , Expression and cellular localization of microRNA-29b and RAX, an activator of the RNA-dependent protein kinase (PKR), in the retina of streptozotocin-induced diabetic rats. Mol Vis 2011, 17, 2228–40. [Google Scholar] [PubMed]
- Xiong, F.; Du, X.; Hu, J.; Li, T.; Du, S.; Wu, Q. , Altered retinal microRNA expression profiles in early diabetic retinopathy: an in silico analysis. Curr Eye Res 2014, 39, 720–9. [Google Scholar] [CrossRef] [PubMed]
- Zheng, D.; Ma, J.; Yu, Y.; Li, M.; Ni, R.; Wang, G.; Chen, R.; Li, J.; Fan, G.C.; Lacefield, J.C.; Peng, T. , Silencing of miR-195 reduces diabetic cardiomyopathy in C57BL/6 mice. Diabetologia 2015, 58, 1949–58. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Yao, D.; Yan, H.; Chen, X.; Wang, L.; Zhan, H. , The Role of MicroRNAs in the Pathogenesis of Diabetic Nephropathy. Int J Endocrinol 2019, 2019, 8719060. [Google Scholar] [CrossRef]
- Deshpande, S.; Abdollahi, M.; Wang, M.; Lanting, L.; Kato, M.; Natarajan, R. , Reduced Autophagy by a microRNA-mediated Signaling Cascade in Diabetes-induced Renal Glomerular Hypertrophy. Sci Rep 2018, 8, 6954. [Google Scholar] [CrossRef]
- Guo, R.; Nair, S. , Role of microRNA in diabetic cardiomyopathy: From mechanism to intervention. Biochim Biophys Acta Mol Basis Dis 2017, 1863, 2070–2077. [Google Scholar] [CrossRef]
- Thum, T.; Gross, C.; Fiedler, J.; Fischer, T.; Kissler, S.; Bussen, M.; Galuppo, P.; Just, S.; Rottbauer, W.; Frantz, S.; Castoldi, M.; Soutschek, J.; Koteliansky, V.; Rosenwald, A.; Basson, M.A.; Licht, J.D.; Pena, J.T.; Rouhanifard, S.H.; Muckenthaler, M.U.; Tuschl, T.; Martin, G.R.; Bauersachs, J.; Engelhardt, S. , MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature 2008, 456, 980–4. [Google Scholar] [CrossRef]
- Fomison-Nurse, I.; Saw, E.E.L.; Gandhi, S.; Munasinghe, P.E.; Van Hout, I.; Williams, M.J.A.; Galvin, I.; Bunton, R.; Davis, P.; Cameron, V.; Katare, R. , Diabetes induces the activation of pro-ageing miR-34a in the heart, but has differential effects on cardiomyocytes and cardiac progenitor cells. Cell Death Differ 2018, 25, 1336–1349. [Google Scholar] [CrossRef]
- Shen, E.; Diao, X.; Wang, X.; Chen, R.; Hu, B. , MicroRNAs involved in the mitogen-activated protein kinase cascades pathway during glucose-induced cardiomyocyte hypertrophy. The American journal of pathology 2011, 179, 639–50. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Zhou, B.; Su, H.; Liu, Y.; Du, C. , miR-150 regulates high glucose-induced cardiomyocyte hypertrophy by targeting the transcriptional co-activator p300. Experimental cell research 2013, 319, 173–84. [Google Scholar] [CrossRef] [PubMed]
- Dhas, Y.; Arshad, N.; Biswas, N.; Jones, L.D.; Ashili, S. , MicroRNA-21 Silencing in Diabetic Nephropathy: Insights on Therapeutic Strategies. Biomedicines, 2023; 11. [Google Scholar]
- Dieter, C.; Assmann, T.S.; Costa, A.R.; Canani, L.H.; de Souza, B.M.; Bauer, A.C.; Crispim, D. , MiR-30e-5p and MiR-15a-5p Expressions in Plasma and Urine of Type 1 Diabetic Patients With Diabetic Kidney Disease. Front Genet 2019, 10, 563. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, H.; Liu, J.; Han, P.; Li, X.; Bai, H.; Zhang, C.; Sun, X.; Teng, Y.; Zhang, Y.; Yuan, X.; Chu, Y.; Zhao, B. , Variations in MicroRNA-25 Expression Influence the Severity of Diabetic Kidney Disease. J Am Soc Nephrol 2017, 28, 3627–3638. [Google Scholar] [CrossRef]
- Yildirim, S.S.; Akman, D.; Catalucci, D.; Turan, B. , Relationship between downregulation of miRNAs and increase of oxidative stress in the development of diabetic cardiac dysfunction: junctin as a target protein of miR-1. Cell biochemistry and biophysics 2013, 67, 1397–408. [Google Scholar] [CrossRef]
- Tsai, S.; Shameli, A.; Santamaria, P. , CD8+ T cells in type 1 diabetes. Adv Immunol 2008, 100, 79–124. [Google Scholar]
- ElEssawy, B.; Li, X.C. , Type 1 diabetes and T regulatory cells. Pharmacol Res 2015, 98, 22–30. [Google Scholar] [CrossRef]
- Garzon, R.; Volinia, S.; Liu, C.G.; Fernandez-Cymering, C.; Palumbo, T.; Pichiorri, F.; Fabbri, M.; Coombes, K.; Alder, H.; Nakamura, T.; Flomenberg, N.; Marcucci, G.; Calin, G.A.; Kornblau, S.M.; Kantarjian, H.; Bloomfield, C.D.; Andreeff, M.; Croce, C.M. , MicroRNA signatures associated with cytogenetics and prognosis in acute myeloid leukemia. Blood 2008, 111, 3183–9. [Google Scholar] [CrossRef]
- Hui, A.B.; Shi, W.; Boutros, P.C.; Miller, N.; Pintilie, M.; Fyles, T.; McCready, D.; Wong, D.; Gerster, K.; Waldron, L.; Jurisica, I.; Penn, L.Z.; Liu, F.F. , Robust global micro-RNA profiling with formalin-fixed paraffin-embedded breast cancer tissues. Lab Invest 2009, 89, 597–606. [Google Scholar] [CrossRef]
- Gao, S.; Zhao, Z.Y.; Wu, R.; Zhang, Y.; Zhang, Z.Y. , Prognostic value of microRNAs in colorectal cancer: a meta-analysis. Cancer Manag Res 2018, 10, 907–929. [Google Scholar] [CrossRef]
- Schmidt, W.M.; Spiel, A.O.; Jilma, B.; Wolzt, M.; Muller, M. , In vivo profile of the human leukocyte microRNA response to endotoxemia. Biochem Biophys Res Commun 2009, 380, 437–41. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Sharples, R.A.; Scicluna, B.J.; Hill, A.F. , Exosomes provide a protective and enriched source of miRNA for biomarker profiling compared to intracellular and cell-free blood. J Extracell Vesicles 2014, 3. [Google Scholar] [CrossRef]
- Yang, J.; Wei, F.; Schafer, C.; Wong, D.T. , Detection of tumor cell-specific mRNA and protein in exosome-like microvesicles from blood and saliva. PLoS One 2014, 9, e110641. [Google Scholar] [CrossRef] [PubMed]
- Guay, C.; Menoud, V.; Rome, S.; Regazzi, R. , Horizontal transfer of exosomal microRNAs transduce apoptotic signals between pancreatic beta-cells. Cell Commun Signal 2015, 13, 17. [Google Scholar] [CrossRef]
- Setyowati Karolina, D.; Sepramaniam, S.; Tan, H.Z.; Armugam, A.; Jeyaseelan, K. , miR-25 and miR-92a regulate insulin I biosynthesis in rats. RNA Biol 2013, 10, 1365–78. [Google Scholar] [CrossRef] [PubMed]
- Knezevic, I.; Patel, A.; Sundaresan, N.R.; Gupta, M.P.; Solaro, R.J.; Nagalingam, R.S.; Gupta, M. , A novel cardiomyocyte-enriched microRNA, miR-378, targets insulin-like growth factor 1 receptor: implications in postnatal cardiac remodeling and cell survival. J Biol Chem 2012, 287, 12913–26. [Google Scholar] [CrossRef]
- Ludwig, N.; Leidinger, P.; Becker, K.; Backes, C.; Fehlmann, T.; Pallasch, C.; Rheinheimer, S.; Meder, B.; Stahler, C.; Meese, E.; Keller, A. , Distribution of miRNA expression across human tissues. Nucleic Acids Res 2016, 44, 3865–77. [Google Scholar] [CrossRef]
- Cabiati, M.; Federico, G.; Del Ry, S. , Importance of Studying Non-Coding RNA in Children and Adolescents with Type 1 Diabetes. Biomedicines, 2024; 12. [Google Scholar]
- Roggli, E.; Gattesco, S.; Caille, D.; Briet, C.; Boitard, C.; Meda, P.; Regazzi, R. , Changes in microRNA expression contribute to pancreatic beta-cell dysfunction in prediabetic NOD mice. Diabetes 2012, 61, 1742–51. [Google Scholar] [CrossRef]
- Guay, C.; Kruit, J.K.; Rome, S.; Menoud, V.; Mulder, N.L.; Jurdzinski, A.; Mancarella, F.; Sebastiani, G.; Donda, A.; Gonzalez, B.J.; Jandus, C.; Bouzakri, K.; Pinget, M.; Boitard, C.; Romero, P.; Dotta, F.; Regazzi, R. , Lymphocyte-Derived Exosomal MicroRNAs Promote Pancreatic beta Cell Death and May Contribute to Type 1 Diabetes Development. Cell Metab 2019, 29, 348–361. [Google Scholar] [CrossRef]
- Sims, E.K.; Lakhter, A.J.; Anderson-Baucum, E.; Kono, T.; Tong, X.; Evans-Molina, C. , MicroRNA 21 targets BCL2 mRNA to increase apoptosis in rat and human beta cells. Diabetologia 2017, 60, 1057–1065. [Google Scholar] [CrossRef]
- Tian, C.; Ouyang, X.; Lv, Q.; Zhang, Y.; Xie, W. , Cross-talks between microRNAs and mRNAs in pancreatic tissues of streptozotocin-induced type 1 diabetic mice. Biomed Rep 2015, 3, 333–342. [Google Scholar] [CrossRef] [PubMed]
- Ban, E.; Jeong, S.; Park, M.; Kwon, H.; Park, J.; Song, E.J.; Kim, A. , Accelerated wound healing in diabetic mice by miRNA-497 and its anti-inflammatory activity. Biomed Pharmacother 2020, 121, 109613. [Google Scholar] [CrossRef] [PubMed]
- Diao, X.; Shen, E.; Wang, X.; Hu, B. , Differentially expressed microRNAs and their target genes in the hearts of streptozotocin-induced diabetic mice. Mol Med Rep 2011, 4, 633–40. [Google Scholar]
- Alipour, M.R.; Khamaneh, A.M.; Yousefzadeh, N.; Mohammad-nejad, D.; Soufi, F.G. , Upregulation of microRNA-146a was not accompanied by downregulation of pro-inflammatory markers in diabetic kidney. Mol Biol Rep 2013, 40, 6477–83. [Google Scholar] [CrossRef] [PubMed]
- Tsukita, S.; Yamada, T.; Takahashi, K.; Munakata, Y.; Hosaka, S.; Takahashi, H.; Gao, J.; Shirai, Y.; Kodama, S.; Asai, Y.; Sugisawa, T.; Chiba, Y.; Kaneko, K.; Uno, K.; Sawada, S.; Imai, J.; Katagiri, H. , MicroRNAs 106b and 222 Improve Hyperglycemia in a Mouse Model of Insulin-Deficient Diabetes via Pancreatic beta-Cell Proliferation. EBioMedicine 2017, 15, 163–172. [Google Scholar] [CrossRef]
- Bushati, N.; Cohen, S.M. , microRNA functions. Annu Rev Cell Dev Biol 2007, 23, 175–205. [Google Scholar] [CrossRef] [PubMed]
- Rosengren, A.; Dikaiou, P. , Cardiovascular outcomes in type 1 and type 2 diabetes. Diabetologia 2023, 66, 425–437. [Google Scholar] [CrossRef]
- Lee, Y.B.; Han, K.; Kim, B.; Lee, S.E.; Jun, J.E.; Ahn, J.; Kim, G.; Jin, S.M.; Kim, J.H. , Risk of early mortality and cardiovascular disease in type 1 diabetes: a comparison with type 2 diabetes, a nationwide study. Cardiovasc Diabetol 2019, 18, 157. [Google Scholar] [CrossRef]
- Rawshani, A.; Sattar, N.; Franzen, S.; Rawshani, A.; Hattersley, A.T.; Svensson, A.M.; Eliasson, B.; Gudbjornsdottir, S. , Excess mortality and cardiovascular disease in young adults with type 1 diabetes in relation to age at onset: a nationwide, register-based cohort study. Lancet 2018, 392, 477–486. [Google Scholar] [CrossRef]
- Lakhter, A.J.; Pratt, R.E.; Moore, R.E.; Doucette, K.K.; Maier, B.F.; DiMeglio, L.A.; Sims, E.K. , Beta cell extracellular vesicle miR-21-5p cargo is increased in response to inflammatory cytokines and serves as a biomarker of type 1 diabetes. Diabetologia 2018, 61, 1124–1134. [Google Scholar] [CrossRef]
- Zhu, Y.; Sun, Y.; Zhou, Y.; Zhang, Y.; Zhang, T.; Li, Y.; You, W.; Chang, X.; Yuan, L.; Han, X. , MicroRNA-24 promotes pancreatic beta cells toward dedifferentiation to avoid endoplasmic reticulum stress-induced apoptosis. J Mol Cell Biol 2019, 11, 747–760. [Google Scholar] [CrossRef] [PubMed]
- Taheri, M.; Eghtedarian, R.; Dinger, M.E.; Ghafouri-Fard, S. , Emerging roles of non-coding RNAs in the pathogenesis of type 1 diabetes mellitus. Biomed Pharmacother 2020, 129, 110509. [Google Scholar] [CrossRef]
- Jankauskas, S.S.; Gambardella, J.; Sardu, C.; Lombardi, A.; Santulli, G. , Functional Role of miR-155 in the Pathogenesis of Diabetes Mellitus and Its Complications. Noncoding RNA, 2021; 7. [Google Scholar]
- Xue, L.; Xiong, C.; Li, J.; Ren, Y.; Zhang, L.; Jiao, K.; Chen, C.; Ding, P. , miR-200-3p suppresses cell proliferation and reduces apoptosis in diabetic retinopathy via blocking the TGF-beta2/Smad pathway. Biosci Rep, 2020; 40. [Google Scholar]
- Erener, S.; Mojibian, M.; Fox, J.K.; Denroche, H.C.; Kieffer, T.J. , Circulating miR-375 as a biomarker of beta-cell death and diabetes in mice. Endocrinology 2013, 154, 603–8. [Google Scholar] [CrossRef]
- Coulson, D.J.; Bakhashab, S.; Latief, J.S.; Weaver, J.U. , MiR-126, IL-7, CXCR1/2 receptors, inflammation and circulating endothelial progenitor cells: The study on targets for treatment pathways in a model of subclinical cardiovascular disease (type 1 diabetes mellitus). J Transl Med 2021, 19, 140. [Google Scholar] [CrossRef] [PubMed]
- Nizam, R.; Malik, M.Z.; Jacob, S.; Alsmadi, O.; Koistinen, H.A.; Tuomilehto, J.; Alkandari, H.; Al-Mulla, F.; Thanaraj, T.A. , Circulating hsa-miR-320a and its regulatory network in type 1 diabetes mellitus. Front Immunol 2024, 15, 1376416. [Google Scholar] [CrossRef]
- Rasmi, Y.; Mohamed, Y.A.; Alipour, S.; Ahmed, S.; Abdelmajed, S.S. , The role of miR-143/miR-145 in the development, diagnosis, and treatment of diabetes. J Diabetes Metab Disord 2024, 23, 39–47. [Google Scholar] [CrossRef]
- Swolin-Eide, D.; Forsander, G.; Pundziute Lycka, A.; Novak, D.; Grillari, J.; Diendorfer, A.B.; Hackl, M.; Magnusson, P. , Circulating microRNAs in young individuals with long-duration type 1 diabetes in comparison with healthy controls. Sci Rep 2023, 13, 11634. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.B.; Li, Y.; Liu, G.Y.; Li, T.H.; Li, F.; Guan, J.; Wang, H.J. , Long non-coding RNA PVT1, a molecular sponge of miR-26b, is involved in the progression of hyperglycemia-induced collagen degradation in human chondrocytes by targeting CTGF/TGF-beta signal ways. Innate Immun 2020, 26, 204–214. [Google Scholar] [CrossRef]
- Singh, R.; Ha, S.E.; Wei, L.; Jin, B.; Zogg, H.; Poudrier, S.M.; Jorgensen, B.G.; Park, C.; Ronkon, C.F.; Bartlett, A.; Cho, S.; Morales, A.; Chung, Y.H.; Lee, M.Y.; Park, J.K.; Gottfried-Blackmore, A.; Nguyen, L.; Sanders, K.M.; Ro, S. , miR-10b-5p Rescues Diabetes and Gastrointestinal Dysmotility. Gastroenterology 2021, 160, 1662–1678. [Google Scholar] [CrossRef]
- Douvris, A.; Vinas, J.; Burns, K.D. , miRNA-486-5p: signaling targets and role in non-malignant disease. Cell Mol Life Sci 2022, 79, 376. [Google Scholar] [CrossRef]
- Garavelli, S.; Bruzzaniti, S.; Tagliabue, E.; Di Silvestre, D.; Prattichizzo, F.; Mozzillo, E.; Fattorusso, V.; La Sala, L.; Ceriello, A.; Puca, A.A.; Mauri, P.; Strollo, R.; Marigliano, M.; Maffeis, C.; Petrelli, A.; Bosi, E.; Franzese, A.; Galgani, M.; Matarese, G.; de Candia, P. , Plasma circulating miR-23~27~24 clusters correlate with the immunometabolic derangement and predict C-peptide loss in children with type 1 diabetes. Diabetologia 2020, 63, 2699–2712. [Google Scholar] [CrossRef]
- Otmani, K.; Rouas, R.; Lagneaux, L.; Krayem, M.; Duvillier, H.; Berehab, M.; Lewalle, P. , Acute myeloid leukemia-derived exosomes deliver miR-24-3p to hinder the T-cell immune response through DENN/MADD targeting in the NF-kappaB signaling pathways. Cell Commun Signal 2023, 21, 253. [Google Scholar] [CrossRef] [PubMed]
- Ray, S.L.; Coulson, D.J.; Yeoh, M.L.Y.; Tamara, A.; Latief, J.S.; Bakhashab, S.; Weaver, J.U. , The Role of miR-342 in Vascular Health. Study in Subclinical Cardiovascular Disease in Mononuclear Cells, Plasma, Inflammatory Cytokines and PANX2. Int J Mol Sci, 2020; 21. [Google Scholar]
- Daamouch, S.; Bluher, M.; Vazquez, D.C.; Hackl, M.; Hofbauer, L.C.; Rauner, M. , MiR-144-5p and miR-21-5p do not drive bone disease in a mouse model of type 1 diabetes mellitus. JBMR Plus 2024, 8, ziae036. [Google Scholar] [CrossRef] [PubMed]
- Ventriglia, G.; Nigi, L.; Sebastiani, G.; Dotta, F. , MicroRNAs: Novel Players in the Dialogue between Pancreatic Islets and Immune System in Autoimmune Diabetes. Biomed Res Int 2015, 2015, 749734. [Google Scholar] [CrossRef]
- Dooley, J.; Garcia-Perez, J.E.; Sreenivasan, J.; Schlenner, S.M.; Vangoitsenhoven, R.; Papadopoulou, A.S.; Tian, L.; Schonefeldt, S.; Serneels, L.; Deroose, C.; Staats, K.A.; Van der Schueren, B.; De Strooper, B.; McGuinness, O.P.; Mathieu, C.; Liston, A. , The microRNA-29 Family Dictates the Balance Between Homeostatic and Pathological Glucose Handling in Diabetes and Obesity. Diabetes 2016, 65, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.Y.; Yang, C.Y.; Rui, Z.L. , MicroRNA-125b-5p improves pancreatic beta-cell function through inhibiting JNK signaling pathway by targeting DACT1 in mice with type 2 diabetes mellitus. Life Sci 2019, 224, 67–75. [Google Scholar] [CrossRef]
- Liu, L.; Yan, J.; Xu, H.; Zhu, Y.; Liang, H.; Pan, W.; Yao, B.; Han, X.; Ye, J.; Weng, J. , Two Novel MicroRNA Biomarkers Related to beta-Cell Damage and Their Potential Values for Early Diagnosis of Type 1 Diabetes. J Clin Endocrinol Metab 2018, 103, 1320–1329. [Google Scholar] [CrossRef]
- Bagge, A.; Dahmcke, C.M.; Dalgaard, L.T. , Syntaxin-1a is a direct target of miR-29a in insulin-producing beta-cells. Horm Metab Res 2013, 45, 463–6. [Google Scholar]
- Ma, X.; Becker Buscaglia, L.E.; Barker, J.R.; Li, Y. , MicroRNAs in NF-kappaB signaling. J Mol Cell Biol 2011, 3, 159–66. [Google Scholar] [CrossRef]
- Wang, P.; Hou, J.; Lin, L.; Wang, C.; Liu, X.; Li, D.; Ma, F.; Wang, Z.; Cao, X. , Inducible microRNA-155 feedback promotes type I IFN signaling in antiviral innate immunity by targeting suppressor of cytokine signaling 1. J Immunol 2010, 185, 6226–33. [Google Scholar] [CrossRef]
- Faraoni, I.; Antonetti, F.R.; Cardone, J.; Bonmassar, E. , miR-155 gene: a typical multifunctional microRNA. Biochim Biophys Acta 2009, 1792, 497–505. [Google Scholar] [CrossRef] [PubMed]
- Belgardt, B.F.; Ahmed, K.; Spranger, M.; Latreille, M.; Denzler, R.; Kondratiuk, N.; von Meyenn, F.; Villena, F.N.; Herrmanns, K.; Bosco, D.; Kerr-Conte, J.; Pattou, F.; Rulicke, T.; Stoffel, M. , The microRNA-200 family regulates pancreatic beta cell survival in type 2 diabetes. Nat Med 2015, 21, 619–27. [Google Scholar] [CrossRef] [PubMed]
- Fayyad-Kazan, H.; Rouas, R.; Fayyad-Kazan, M.; Badran, R.; El Zein, N.; Lewalle, P.; Najar, M.; Hamade, E.; Jebbawi, F.; Merimi, M.; Romero, P.; Burny, A.; Badran, B.; Martiat, P. , MicroRNA profile of circulating CD4-positive regulatory T cells in human adults and impact of differentially expressed microRNAs on expression of two genes essential to their function. J Biol Chem 2012, 287, 9910–9922. [Google Scholar] [CrossRef]
- Devlin, C.; Greco, S.; Martelli, F.; Ivan, M. , miR-210: More than a silent player in hypoxia. IUBMB Life 2011, 63, 94–100. [Google Scholar] [CrossRef]
- Molitch, M.E.; DeFronzo, R.A.; Franz, M.J.; Keane, W.F.; Mogensen, C.E.; Parving, H.H.; Steffes, M.W.; American Diabetes, A. , Nephropathy in diabetes. Diabetes Care 2004, 27 Suppl 1, S79–83. [Google Scholar]
- Argyropoulos, C.; Wang, K.; McClarty, S.; Huang, D.; Bernardo, J.; Ellis, D.; Orchard, T.; Galas, D.; Johnson, J. , Urinary microRNA profiling in the nephropathy of type 1 diabetes. PLoS One 2013, 8, e54662. [Google Scholar] [CrossRef]
- Wang, G.; Kwan, B.C.; Lai, F.M.; Chow, K.M.; Li, P.K.; Szeto, C.C. , Urinary sediment miRNA levels in adult nephrotic syndrome. Clin Chim Acta 2013, 418, 5–11. [Google Scholar] [CrossRef]
- Wang, S.; Aurora, A.B.; Johnson, B.A.; Qi, X.; McAnally, J.; Hill, J.A.; Richardson, J.A.; Bassel-Duby, R.; Olson, E.N. , The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell 2008, 15, 261–71. [Google Scholar] [CrossRef]
- Long, J.; Wang, Y.; Wang, W.; Chang, B.H.; Danesh, F.R. , Identification of microRNA-93 as a novel regulator of vascular endothelial growth factor in hyperglycemic conditions. J Biol Chem 2010, 285, 23457–65. [Google Scholar] [CrossRef]
- Samandari, N.; Mirza, A.H.; Kaur, S.; Hougaard, P.; Nielsen, L.B.; Fredheim, S.; Mortensen, H.B.; Pociot, F. , Influence of Disease Duration on Circulating Levels of miRNAs in Children and Adolescents with New Onset Type 1 Diabetes. Noncoding RNA, 2018; 4. [Google Scholar]
- Wallberg, M.; Recino, A.; Phillips, J.; Howie, D.; Vienne, M.; Paluch, C.; Azuma, M.; Wong, F.S.; Waldmann, H.; Cooke, A. , Anti-CD3 treatment up-regulates programmed cell death protein-1 expression on activated effector T cells and severely impairs their inflammatory capacity. Immunology 2017, 151, 248–260. [Google Scholar] [CrossRef]
- Benson, R.A.; Garcon, F.; Recino, A.; Ferdinand, J.R.; Clatworthy, M.R.; Waldmann, H.; Brewer, J.M.; Okkenhaug, K.; Cooke, A.; Garside, P.; Wallberg, M. , Non-Invasive Multiphoton Imaging of Islets Transplanted Into the Pinna of the NOD Mouse Ear Reveals the Immediate Effect of Anti-CD3 Treatment in Autoimmune Diabetes. Front Immunol 2018, 9, 1006. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.H.; Wu, Z.; Hu, C.; Zhang, M.; Wang, W.Z.; Hu, X.B. , Endoplasmic reticulum stress and destruction of pancreatic beta cells in type 1 diabetes. Chin Med J (Engl) 2020, 133, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Padgett, L.E.; Broniowska, K.A.; Hansen, P.A.; Corbett, J.A.; Tse, H.M. , The role of reactive oxygen species and proinflammatory cytokines in type 1 diabetes pathogenesis. Ann N Y Acad Sci 2013, 1281, 16–35. [Google Scholar] [CrossRef] [PubMed]
- Leenders, F.; Groen, N.; de Graaf, N.; Engelse, M.A.; Rabelink, T.J.; de Koning, E.J.P.; Carlotti, F. , Oxidative Stress Leads to beta-Cell Dysfunction Through Loss of beta-Cell Identity. Front Immunol 2021, 12, 690379. [Google Scholar] [CrossRef]
- Wei, J.; Zhang, Y.; Luo, Y.; Wang, Z.; Bi, S.; Song, D.; Dai, Y.; Wang, T.; Qiu, L.; Wen, L.; Yuan, L.; Yang, J.Y. , Aldose reductase regulates miR-200a-3p/141-3p to coordinate Keap1-Nrf2, Tgfbeta1/2, and Zeb1/2 signaling in renal mesangial cells and the renal cortex of diabetic mice. Free Radic Biol Med 2014, 67, 91–102. [Google Scholar] [CrossRef]
- Ghaffari, M.; Razi, S.; Zalpoor, H.; Nabi-Afjadi, M.; Mohebichamkhorami, F.; Zali, H. , Association of MicroRNA-146a with Type 1 and 2 Diabetes and their Related Complications. J Diabetes Res 2023, 2023, 2587104. [Google Scholar] [CrossRef]
- Velosa, A.P.; Teodoro, W.R.; dos Anjos, D.M.; Konno, R.; Oliveira, C.C.; Katayama, M.L.; Parra, E.R.; Capelozzi, V.L.; Yoshinari, N.H. , Collagen V-induced nasal tolerance downregulates pulmonary collagen mRNA gene and TGF-beta expression in experimental systemic sclerosis. Respir Res 2010, 11, 1. [Google Scholar] [CrossRef] [PubMed]
- Salas-Perez, F.; Codner, E.; Valencia, E.; Pizarro, C.; Carrasco, E.; Perez-Bravo, F. , MicroRNAs miR-21a and miR-93 are down regulated in peripheral blood mononuclear cells (PBMCs) from patients with type 1 diabetes. Immunobiology 2013, 218, 733–7. [Google Scholar] [CrossRef]
- Qadir, M.M.F.; Klein, D.; Alvarez-Cubela, S.; Dominguez-Bendala, J.; Pastori, R.L. , The Role of MicroRNAs in Diabetes-Related Oxidative Stress. Int J Mol Sci, 2019; 20. [Google Scholar]
- Gao, X.; Zhao, S. , miRNA-16-5p inhibits the apoptosis of high glucose-induced pancreatic beta cells via targeting of CXCL10: potential biomarkers in type 1 diabetes mellitus. Endokrynol Pol 2020, 71, 404–410. [Google Scholar] [CrossRef]
- Roggli, E.; Britan, A.; Gattesco, S.; Lin-Marq, N.; Abderrahmani, A.; Meda, P.; Regazzi, R. , Involvement of microRNAs in the cytotoxic effects exerted by proinflammatory cytokines on pancreatic beta-cells. Diabetes 2010, 59, 978–86. [Google Scholar] [CrossRef]
- Lin, L.; Mahner, S.; Jeschke, U.; Hester, A. , The Distinct Roles of Transcriptional Factor KLF11 in Normal Cell Growth Regulation and Cancer as a Mediator of TGF-beta Signaling Pathway. Int J Mol Sci, 2020; 21. [Google Scholar]
- Pezzolesi, M.G.; Satake, E.; McDonnell, K.P.; Major, M.; Smiles, A.M.; Krolewski, A.S. , Circulating TGF-beta1-Regulated miRNAs and the Risk of Rapid Progression to ESRD in Type 1 Diabetes. Diabetes 2015, 64, 3285–93. [Google Scholar] [CrossRef] [PubMed]
- Pooja Rathan, V.; Bhuvaneshwari, K.; Nideesh Adit, G.; Kavyashree, S.; Thulasi, N.; Geetha, A.V.S.; Milan, K.L.; Ramkumar, K.M. , Therapeutic potential of SMAD7 targeting miRNA in the pathogenesis of diabetic nephropathy. Arch Biochem Biophys 2024, 764, 110265. [Google Scholar] [CrossRef] [PubMed]
- Qin, W.; Shi, Y.; Zhao, B.; Yao, C.; Jin, L.; Ma, J.; Jin, Y. , miR-24 regulates apoptosis by targeting the open reading frame (ORF) region of FAF1 in cancer cells. PLoS One 2010, 5, e9429. [Google Scholar] [CrossRef] [PubMed]
- Chan, M.C.; Hilyard, A.C.; Wu, C.; Davis, B.N.; Hill, N.S.; Lal, A.; Lieberman, J.; Lagna, G.; Hata, A. , Molecular basis for antagonism between PDGF and the TGFbeta family of signalling pathways by control of miR-24 expression. EMBO J 2010, 29, 559–73. [Google Scholar] [CrossRef]
- Donath, M.Y.; Storling, J.; Maedler, K.; Mandrup-Poulsen, T. , Inflammatory mediators and islet beta-cell failure: a link between type 1 and type 2 diabetes. J Mol Med (Berl) 2003, 81, 455–70. [Google Scholar] [CrossRef]
- Chen, X.; Ba, Y.; Ma, L.; Cai, X.; Yin, Y.; Wang, K.; Guo, J.; Zhang, Y.; Chen, J.; Guo, X.; Li, Q.; Li, X.; Wang, W.; Zhang, Y.; Wang, J.; Jiang, X.; Xiang, Y.; Xu, C.; Zheng, P.; Zhang, J.; Li, R.; Zhang, H.; Shang, X.; Gong, T.; Ning, G.; Wang, J.; Zen, K.; Zhang, J.; Zhang, C.Y. , Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 2008, 18, 997–1006. [Google Scholar] [CrossRef]
- Hu, Z.; Dong, J.; Wang, L.E.; Ma, H.; Liu, J.; Zhao, Y.; Tang, J.; Chen, X.; Dai, J.; Wei, Q.; Zhang, C.; Shen, H. , Serum microRNA profiling and breast cancer risk: the use of miR-484/191 as endogenous controls. Carcinogenesis 2012, 33, 828–34. [Google Scholar] [CrossRef]
- Wu, Q.; Wang, C.; Lu, Z.; Guo, L.; Ge, Q. , Analysis of serum genome-wide microRNAs for breast cancer detection. Clin Chim Acta, 2012; 413, 1058–1065. [Google Scholar]
- Li, L.M.; Hu, Z.B.; Zhou, Z.X.; Chen, X.; Liu, F.Y.; Zhang, J.F.; Shen, H.B.; Zhang, C.Y.; Zen, K. , Serum microRNA profiles serve as novel biomarkers for HBV infection and diagnosis of HBV-positive hepatocarcinoma. Cancer research 2010, 70, 9798–807. [Google Scholar] [CrossRef]
- Razumilava, N.; Bronk, S.F.; Smoot, R.L.; Fingas, C.D.; Werneburg, N.W.; Roberts, L.R.; Mott, J.L. , miR-25 targets TNF-related apoptosis inducing ligand (TRAIL) death receptor-4 and promotes apoptosis resistance in cholangiocarcinoma. Hepatology 2012, 55, 465–75. [Google Scholar] [CrossRef]
- Zhang, H.; Zuo, Z.; Lu, X.; Wang, L.; Wang, H.; Zhu, Z. , MiR-25 regulates apoptosis by targeting Bim in human ovarian cancer. Oncol Rep 2012, 27, 594–8. [Google Scholar]
- Fu, Y.; Zhang, Y.; Wang, Z.; Wang, L.; Wei, X.; Zhang, B.; Wen, Z.; Fang, H.; Pang, Q.; Yi, F. , Regulation of NADPH oxidase activity is associated with miRNA-25-mediated NOX4 expression in experimental diabetic nephropathy. Am J Nephrol 2010, 32, 581–9. [Google Scholar] [CrossRef]
- Gonzalez-Martin, A.; Adams, B.D.; Lai, M.; Shepherd, J.; Salvador-Bernaldez, M.; Salvador, J.M.; Lu, J.; Nemazee, D.; Xiao, C. , The microRNA miR-148a functions as a critical regulator of B cell tolerance and autoimmunity. Nat Immunol 2016, 17, 433–40. [Google Scholar] [CrossRef] [PubMed]
- Ha, S.E.; Singh, R.; Jin, B.; Baek, G.; Jorgensen, B.G.; Zogg, H.; Debnath, S.; Park, H.S.; Cho, H.; Watkins, C.M.; Cho, S.; Kim, M.S.; Lee, M.Y.; Yu, T.Y.; Jeong, J.W.; Ro, S. , miR-10a/b-5p-NCOR2 Regulates Insulin-Resistant Diabetes in Female Mice. Int J Mol Sci, 2024; 25. [Google Scholar]
- Martello, G.; Rosato, A.; Ferrari, F.; Manfrin, A.; Cordenonsi, M.; Dupont, S.; Enzo, E.; Guzzardo, V.; Rondina, M.; Spruce, T.; Parenti, A.R.; Daidone, M.G.; Bicciato, S.; Piccolo, S. , A MicroRNA targeting dicer for metastasis control. Cell 2010, 141, 1195–207. [Google Scholar] [CrossRef] [PubMed]
- Kfir-Erenfeld, S.; Haggiag, N.; Biton, M.; Stepensky, P.; Assayag-Asherie, N.; Yefenof, E. , miR-103 inhibits proliferation and sensitizes hemopoietic tumor cells for glucocorticoid-induced apoptosis. Oncotarget 2017, 8, 472–489. [Google Scholar] [CrossRef]
- Barutta, F.; Corbetta, B.; Bellini, S.; Guarrera, S.; Matullo, G.; Scandella, M.; Schalkwijk, C.; Stehouwer, C.D.; Chaturvedi, N.; Soedamah-Muthu, S.S.; Durazzo, M.; Gruden, G. , MicroRNA 146a is associated with diabetic complications in type 1 diabetic patients from the EURODIAB PCS. J Transl Med 2021, 19, 475. [Google Scholar] [CrossRef]
- Lu, L.F.; Boldin, M.P.; Chaudhry, A.; Lin, L.L.; Taganov, K.D.; Hanada, T.; Yoshimura, A.; Baltimore, D.; Rudensky, A.Y. , Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell 2010, 142, 914–29. [Google Scholar] [CrossRef]
- Ellenrieder, V.; Buck, A.; Harth, A.; Jungert, K.; Buchholz, M.; Adler, G.; Urrutia, R.; Gress, T.M. , KLF11 mediates a critical mechanism in TGF-beta signaling that is inactivated by Erk-MAPK in pancreatic cancer cells. Gastroenterology 2004, 127, 607–20. [Google Scholar] [CrossRef]
- Guasparri, I.; Keller, S.A.; Cesarman, E. , KSHV vFLIP is essential for the survival of infected lymphoma cells. J Exp Med 2004, 199, 993–1003. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Jin, W.; Hardegen, N.; Lei, K.J.; Li, L.; Marinos, N.; McGrady, G.; Wahl, S.M. , Conversion of peripheral CD4+CD25- naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. J Exp Med 2003, 198, 1875–86. [Google Scholar] [CrossRef]
- Erener, S.; Ellis, C.E.; Ramzy, A.; Glavas, M.M.; O'Dwyer, S.; Pereira, S.; Wang, T.; Pang, J.; Bruin, J.E.; Riedel, M.J.; Baker, R.K.; Webber, T.D.; Lesina, M.; Bluher, M.; Algul, H.; Kopp, J.L.; Herzig, S.; Kieffer, T.J. , Deletion of pancreas-specific miR-216a reduces beta-cell mass and inhibits pancreatic cancer progression in mice. Cell Rep Med 2021, 2, 100434. [Google Scholar] [CrossRef]
- Wang, P.; Liu, Q.; Zhao, H.; Bishop, J.O.; Zhou, G.; Olson, L.K.; Moore, A. , miR-216a-targeting theranostic nanoparticles promote proliferation of insulin-secreting cells in type 1 diabetes animal model. Sci Rep 2020, 10, 5302. [Google Scholar] [CrossRef] [PubMed]
- Scherm, M.G.; Serr, I.; Zahm, A.M.; Schug, J.; Bellusci, S.; Manfredini, R.; Salb, V.K.; Gerlach, K.; Weigmann, B.; Ziegler, A.G.; Kaestner, K.H.; Daniel, C. , miRNA142-3p targets Tet2 and impairs Treg differentiation and stability in models of type 1 diabetes. Nature communications 2019, 10, 5697. [Google Scholar] [CrossRef] [PubMed]
| Source | Detection method | miRNA profile | Ref. | ||||
|---|---|---|---|---|---|---|---|
| Sample | T1D patient characteristic | Expression | miRNA | ||||
| Diagnosis | Age (years) | n | n | ||||
| Source | Detection method | miRNA profile | Ref. | ||||
| Serum | 21 to 42 days | 9.0 ± 1.8 | 10 | qPCR | Up | let-7e/g-5p miR-18a-5p miR-23b-3p miR-25-3p miR-30e-5p miR-93-5p miR-103a-2-5p miR-125a-3p miR-140-5p miR-144-5p miR-182-5p miR-183-5p miR-192-5p miR-214-5p miR-221-3p miR-222-3p miR-324-3p/5p miR-331-3p miR-345-5p miR-377-3p miR-454-3p miR-500a-5p miR-502-3p miR-1468 |
[19] |
| Down | miR-100-5p miR-154-3p miR-490-5p miR-630 miR-636 miR-639 miR-675-3p miR-720 |
||||||
| 12 months |
404 | Sequencing qPCR |
Up | miR-10a miR-21 miR-24 miR-25 miR-26a/b miR-27a/b miR-29a/b miR-30a-5p miR-103 miR-125b miR-148a miR-152 miR-181a miR-199a miR-200a/c miR-210 miR-222 miR-320a miR-340 |
[31] | ||
| <6 months or 2 to 5 yrs |
26 | qPCR | Up | miR-10a miR-21 miR-27a miR-92a miR-100 miR-148a miR-200a miR-208 miR-212 miR-323-3p miR-346 miR-451 miR-886-3p |
[32] | ||
| Down | miR-16-5p miR-125a-5p miR-126 miR-146a miR-155 miR-197 miR-342-3p miR-374 miR-454 miR-518d |
||||||
| 15.71 ± 11.33 yrs | 33.57 ± 8.17 | 15 | qPCR | Up | miR-21-5p miR-148a |
[33] | |
| Plasma | <5 yrs | 19.2 ± 6.4 | 16 | Microarray qPCR | Up | miR-15b-5p miR-21-3p/5p miR-25-3p miR-29a-3p miR-101-3p miR-103a-3p miR-133a-5p miR-148a-3p miR-148b-3p miR-155-5p miR-200a/c-3p miR-210-3p miR-222-3p miR-320 miR-342 miR-1275 |
[34] |
| Down | miR-29b-3p miR-146a-5p miR-181a-5p miR-338-3p |
||||||
| ≥5 yrs | 19.9 ± 4.6 | 17 | Microarray qPCR | Up | miR-26b-5p miR-146a-5p miR-148b-3p miR-338-3p miR-340-5p miR-1275 |
[34] | |
| Down | miR-15b-5p miR-103a-3p miR-126-3p miR-148a-3p miR-155-5p miR-181a-5p miR-200a/c-3p miR-210-3p miR-222-3p |
||||||
| < 1 yr | 12.93 ± 3.34 | 16 | qPCR |
Up | miR-21 miR-210 |
[35] | |
| - | 25.9 ± 5.7 | 16 | qPCR | Up | miR-21 miR-24 miR-29a miR-30d miR-34a miR-126 miR-146a miR-148a miR-375 miR-376a |
[3] | |
| - | 37 | 95 | qPCR | Up | miR-125b-5p miR-365a-3p miR-770-5p |
[18] | |
| Down | miR-5190 | ||||||
| < 1 yr | 31.0±10.2 | 34 | qPCR | Down | miR-409-3p | [36] | |
| Peripheral blood | - | 26.93 ± 9.62 | 12 | Sequencing qPCR |
Up | let-7i-5p miR-26b-5p miR-99b-5p miR-342-3p miR-501-3p miR-652-3p |
[37] |
| Down | miR-15a-5p miR-15b-3p/5p miR-16-5p miR-16-2-3p miR-17-5p miR-21-5p miR-25-3p miR-26a/b-5p miR-27b-3p miR-30e-5p miR-93-5p miR-98-5p miR-100-5p miR-101-3p miR-103a-5p miR-107 miR-106b-3p/5p miR-126-3p/5p miR-130a/b-3p miR-143-3p miR-144-3p/5p miR-181a-5p miR-185-5p miR-221-3p miR-363-3p miR-532-5p |
||||||
| PBMCs | - | 23.5 ± 3.9 | 11 | Microarray | Up | let-7f/g miR-7 miR-10a miR-15b miR-16 miR-18b miR-19a miR-20a miR-20b miR-21 miR-26b miR-27b miR-30e miR-32 miR-33a miR-98 miR-101 miR-126 miR-148a/b miR-186 miR-195 miR-199a-3p miR-301a miR-335 miR-338-3p miR-340 miR-424 miR-450a miR-454 miR-542-3p miR-548c-3p |
[20] |
| Down | miR-140-3p miR-324-5p miR-342-3p/5p miR-423-5p miR-720 miR-766 miR-940 miR-1275 |
||||||
| Newly diagnosed T1D patients | 17.50 ± 3.68 | 60 | Microarray | Up | miR-320b miR-486-5p miR-652 miR-1275 miR-1301 |
[38] | |
| Down | miR-15b miR-19b miR-22 miR-23a miR-25 miR-28-5p miR-29a miR-30b/c miR-146a miR-146b-5p miR-200c miR-221 miR-342-5p |
||||||
| - | - | 20 | qPCR | Up | miR-22 | [39] | |
| Down | miR-150 | ||||||
| - | - | 78 | qPCR | Down | miR-146a miR-150 miR-424 |
[40] | |
| ≥10 yrs | 37.35 ± 12.82 | 31 | Sequencing qPCR |
Up | miR-133a-3p miR-142-5p miR-144-5p miR-145-3p miR-193a-5p miR-199a-5p miR-382-5p miR-409-5p miR-486-5p miR-543 miR-873-5p miR-1249-3p miR-1299 miR-3150b-3p miR-4531 |
[41] | |
| Down | miR-16-5p miR-144-3p miR-409-5p miR-501-3p miR-1271-5p miR-4485-3p |
||||||
| T-cells and Tregs isolated from peripheral blood | - | - | 5 | qPCR | Up | miR-510 | [42] |
| Down | miR-191 miR-342 |
||||||
| Urine | < 1 yrs | 12.93 ± 3.34 | 68 | qPCR | Up | miR-21 miR-210 |
[35] |
| Down | miR-126 | ||||||
| Plasma-derived exosome | 25.3 ± 15.9 yrs | 46.1 ± 14.4 | 12 | Microarray qPCR |
Up | miR-25-3p | [43] |
| Down | miR-16 miR-302d-3p miR-378e miR-570-3p miR-574-5p miR-579 |
||||||
| Urine-derived exosome | 30.7 ± 6.3 yrs | 57.9 ± 3.7 |
12 | qPCR | Up | miR-130a miR-145 |
[44] |
| Down | miR-155 miR-424 |
||||||
| Species | Diabetic Animal Models |
Source | Detection Methods | miRNA Expression Alteration | Ref. | |
|---|---|---|---|---|---|---|
| Expression | miRNAs | |||||
| Mice | Akita spontaneous mutation (Ins2Akita) mice | Retina |
Microarray qPCR |
Up | miR-7a miR-28 miR-124 miR-186 miR-199a-3p miR-200b miR-369-5p miR-410 miR-429 |
[77] |
| Down | miR-184 miR-296-5p miR-467b miR-539 miR-1196 miR-1224 |
|||||
| Pre-diabetic non-obese diabetic (NOD) mice | Pancreatic islet, cultured islet, infiltrating lymphocytes | qPCR | Up | miR-29a miR-29b miR-29c miR-142-5p miR-155 |
[106,107] | |
| Down | miR-142-3p miR-150 |
|||||
| Pancreatic β-cells | qPCR | Up | miR-142-5p | [107] |
||
| Down | miR-150 miR-155 |
|||||
| Diabetic NOD mice | Pancreatic islet/plasma | qPCR | Up | miR-21 | [108] | |
| Microarray qPCR | Down | miR-126a-3p miR-126a-5p miR-155 miR-188-3p miR-204 miR-218 miR-409-3p |
[36] | |||
| C57BL/6J mice induced with Streptozotocin (STZ) | Pancreatic tissue | Microarray | Up | miR-154-3p miR-296-3p miR-323-3p miR-491-5p miR-669m-3p miR-670-5p miR-697 miR-881-3p miR-3058-3p miR-5119 miR-5130 miR-5622-3p |
[109] | |
| Down | miR-7a-5p miR-7b-5p let-7a-5p let-7f-5p miR-10b-5p miR-16-5p miR-17-5p miR-26b-5p miR-28a-5p miR-28c miR-101a-3p miR-101b-3p miR-101c miR-126-3p miR-126-5p miR-143-3p miR-151-5p miR-184-3p miR-410-5p miR-451a miR-466 miR-467c-3p miR-467f miR-467g miR-467h miR-669 miR-1187 miR-3086-3p miR-5625-5p |
|||||
| Pancreatic islet | qPCR | Up | miR-21 | [108] | ||
| Full-thickness skin lesion | qPCR | Up | miR-29 | [110] | ||
| Down | miR-16 miR-21 miR-23a miR-24 miR-27b miR-31 miR-132 miR-195 miR-497 |
|||||
| Glomeruli | qPCR | Up | miR-145 | [44] | ||
| Heart | Microarray | Up | miR-21 miR-24 miR-142-3p miR-195 miR-199a-3p miR-208 miR-221 miR-499-3p miR-700 miR-705 |
[111] | ||
| Down | miR-1 miR-20a miR-29a miR-143 miR-220b miR-373 |
|||||
| FVB mice induced with STZ |
Heart | qPCR | Up | miR-107 miR-122 miR-125b-3p miR-134 miR-139-5p miR-141 miR-185 miR-193b miR-197 miR-200c miR-208-b miR-221 miR-222 miR-295 miR-298 miR-329 miR-346 miR-409-3p miR-431 miR-466g miR-467a miR-541 miR-542-5p miR-666-3p miR-702 miR-770-3p |
[78] | |
| Down | miR-302a miR-882 miR-883-5p |
|||||
| Rats | Sprague-Dawley or Wistar rats induced with STZ | Retina |
Microarray qPCR | Up | miR-29b miR-34 miR-203 miR-216 miR-410 |
[79] [80] |
| Down | miR-212 | |||||
| Kidney | qPCR | Up | miR-146a | [112] | ||
| Group | Pathway | Upregulated miRNAs |
Downregulated miRNAs |
|---|---|---|---|
| Apoptosis | β-cell apoptosis | miR-21-5p [33,118] miR-24 [119] miR-34a [120] miR-155 [34,121] miR-181a-5p [8] miR-195 [81] miR-200a-3p [34,122] miR-375 [123] miR-424 [20] |
miR-100-5p [42] miR-126 [124] miR-146a-5p [34,40] miR-150-5p [40] miR-320a-3p [125] miR-324-5p [20,109] miR-342-3p [42] miR-424 [40] |
| TP53 signaling | miR-145 [126] |
miR-324-5p [20] miR-342-3p [20] miR-423-5p [20] |
|
| Wnt signaling | miR-143-3p [127] miR-144-5p [19] miR-148a-3p [33,34] miR-365a-3p [18] |
miR-140-3p [20] miR-324-5p [20] miR-342-3p [20] miR-766 [20] miR-940 [20] |
|
| TGF-β signaling | miR-26b [34,128] miR-382-5p [41] |
miR-10b-5p [129] | |
| mTOR signaling | miR-323-3p [109] | ||
| ER or oxidative stress |
FOXO signaling | miR-21-5p [33] miR-148a-3p [34] miR-323-3p [109] miR-486-5p [32,130] |
|
| NF-κB signaling | miR-24-3p [131,132] miR-155-5p [34] |
miR-146a-5p [34] miR-342 [133] |
|
| NRF2 | miR-21-5p [134] miR-144-5p [134] |
||
| Notch signaling | miR-140-3p [20] miR-146a-5p [34] miR-324-5p [20] miR-423-5p [20] miR-1275 [20] |
||
| Endocytosis | miR-21-5p [118,135] miR-29a-3p [34,136] |
miR-324-5p [20] miR-342-3p [20,42] |
|
| Immune system activation |
Immune system-related | miR-103a-3p [34] miR-155-5p [34] miR-200a-3p [34] |
|
| T-cell regulation | miR-31 [42] miR-342 [42] |
||
| Chemokine signaling [20] | miR-18b miR-20b miR-33a miR-101 miR-186 miR-338-3p |
miR-940 | |
| β-cell autoantigen release |
Jak-STAT signaling | miR-21-5p [8] miR-24-3p [8,105] miR-125b-5p [18,137] miR-181-5p [8] miR-323-3p [109] miR-210-5p [8] |
|
| MAPK signaling | miR-199a [20] miR-342 [34] miR-450a [20] miR-548c-3p [20] |
miR-100-5p [8] miR-150-5p [8] |
|
| β-cell insulin release |
Insulin signaling | miR-21 [20] miR-26b [37] miR-32 [20] miR-103a-3p [34] miR-143-3p [37] miR-148a [20] miR-155-5p [34] miR-200a-3p [34] miR-210-3p [34] miR-320c [138] miR-424 [20] miR-1225-5p [138] |
miR-29a-3p [139] miR-146a-5p [34] miR-324-5p [20] miR-342-3p [20] miR-423-5p [20] |
| Axon guidance [20] | miR-21 miR-26b miR-32 miR-126 miR-424 |
miR-766 miR-940 |
|
| Focal adhesion [20] | miR-7 miR-19a miR-27b miR-98 miR-148b miR-195 miR-301a miR-335 miR-454 miR-542-3p |
miR-1275 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





