Submitted:
21 February 2025
Posted:
24 February 2025
You are already at the latest version
Abstract
Wetlands are essential carbon sinks in the global ecosystem, absorbing CO2 in their biomass and soils and mitigating global warming. Accurate above-ground biomass (AGB) and organic carbon (Corg) estimation is crucial for wetland carbon sink research. Remote sensing (RS) data effectively estimates and maps AGB and Corg in wetlands using various techniques, but there is still room to improve the efficiency of Machine Learning (ML) based approaches. This study examined how different sample data treatments and plot sizes impact a Random Forest model’s performance based on RS for AGB and Corg prediction. The model was trained with samples of emergent vegetation collected in a palustrine wetland in southern Brazil and spectral variables (single bands and Vegetation Indices — VI) from medium and high-resolution optical images, Sentinel 2 and PlanetScope, respectively. The treatments involve the AGB and Corg values dimensioned for three different plot sizes (Group 1) and the same subjected to the Natural Logarithmic normalization — NL (Group 2). Therefore, six AGB and Corg models were created for each sensor. Models and sensor performances and spectral variable importance were compared. In our results, NL-normalized sample data RF models proved more accurate. Larger plots produced smaller prediction errors with S2 models, indicating the influence of plot size on the reliability of the estimate. S2 surpassed PS in AGB/Corg prediction, but PS was superior in mapping spatial variability. The VI CO2Flux and S2’s SWIR, Blue, Green, and RE bands 6 and 7, were more importance for AGB/Corg prediction. The innovation of this study is that, in addition to optimizing RF model parameters, optimizing the AGB and Corg dataset collected in the field, i.e., evaluating normalization and plot sizes, is crucial to obtain more accurate estimates with RS and ML-based models. This approach, integrated with Sentinel 2’s medium-resolution data and the combination of VIs and bands, enhances AGB/Corg stock estimation and monitoring in wetlands, and the highlighted predictors can act as spectral indicators of these ecological functions.
Keywords:
1. Introduction
2. Data and Methods
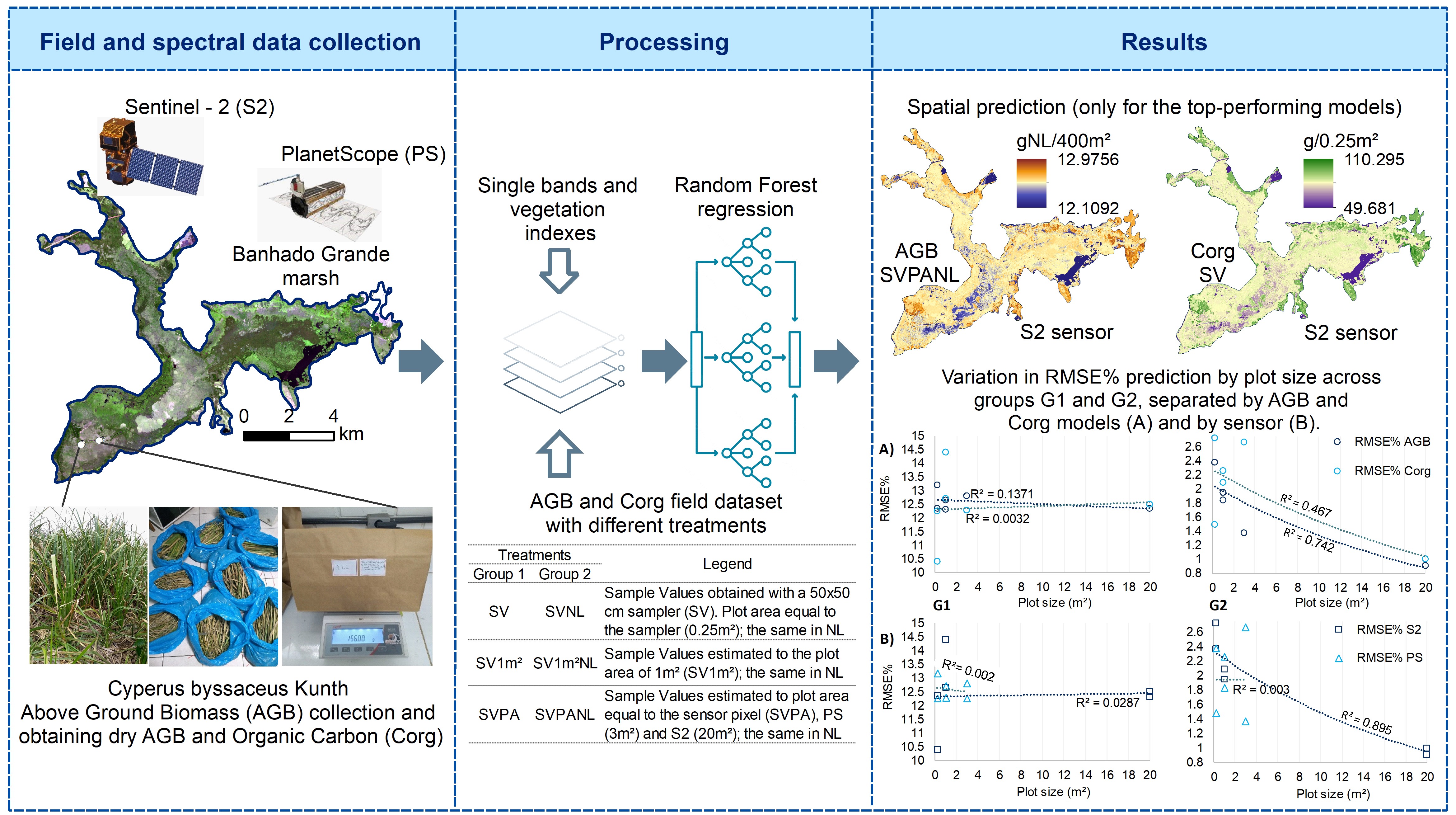
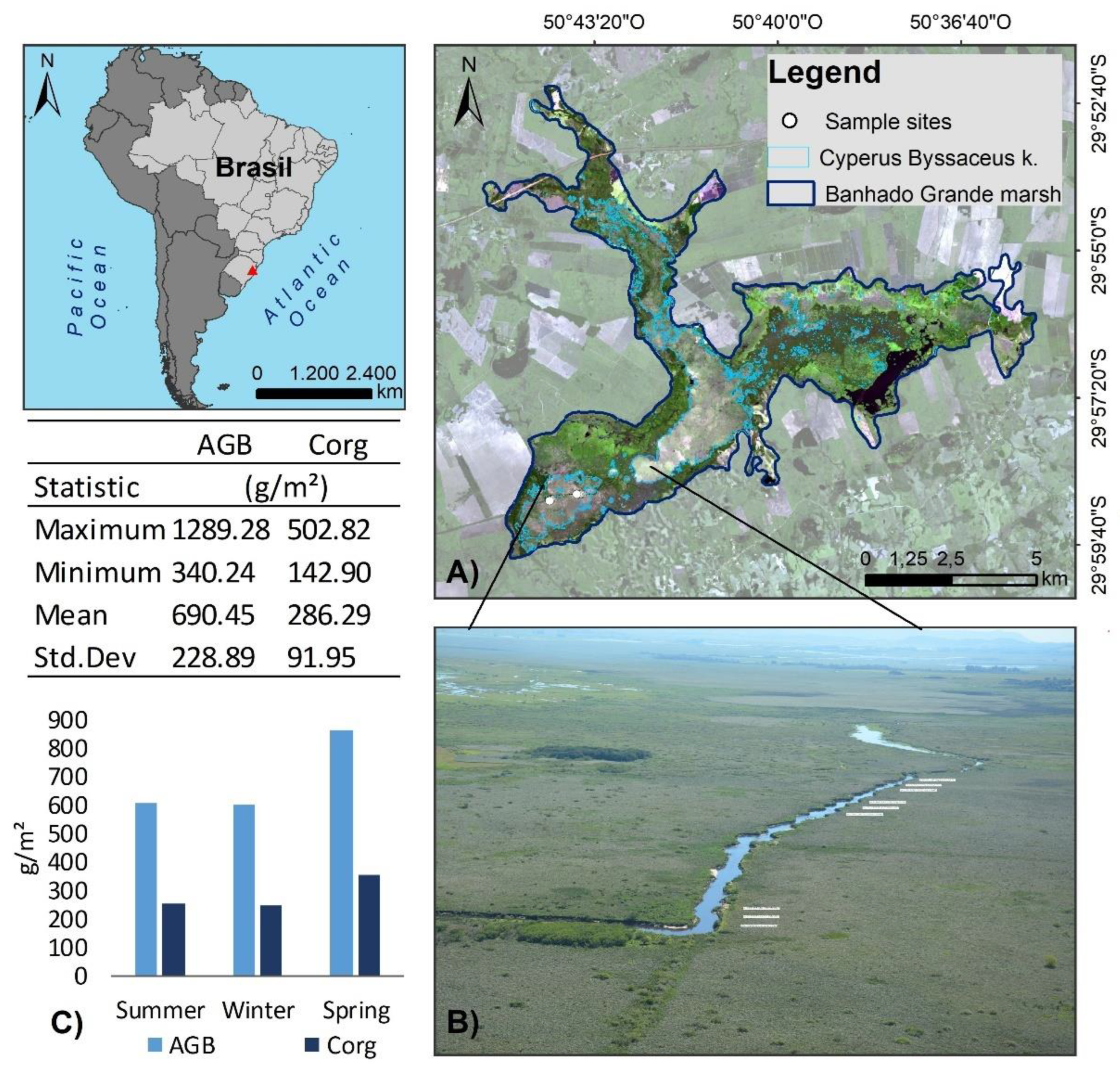
2.1. Study Area and Field Data Collection
2.2. Remote Sensing Datasets
2.3. Development of the Prediction Models
2.4. Evaluation of Models
3. Results
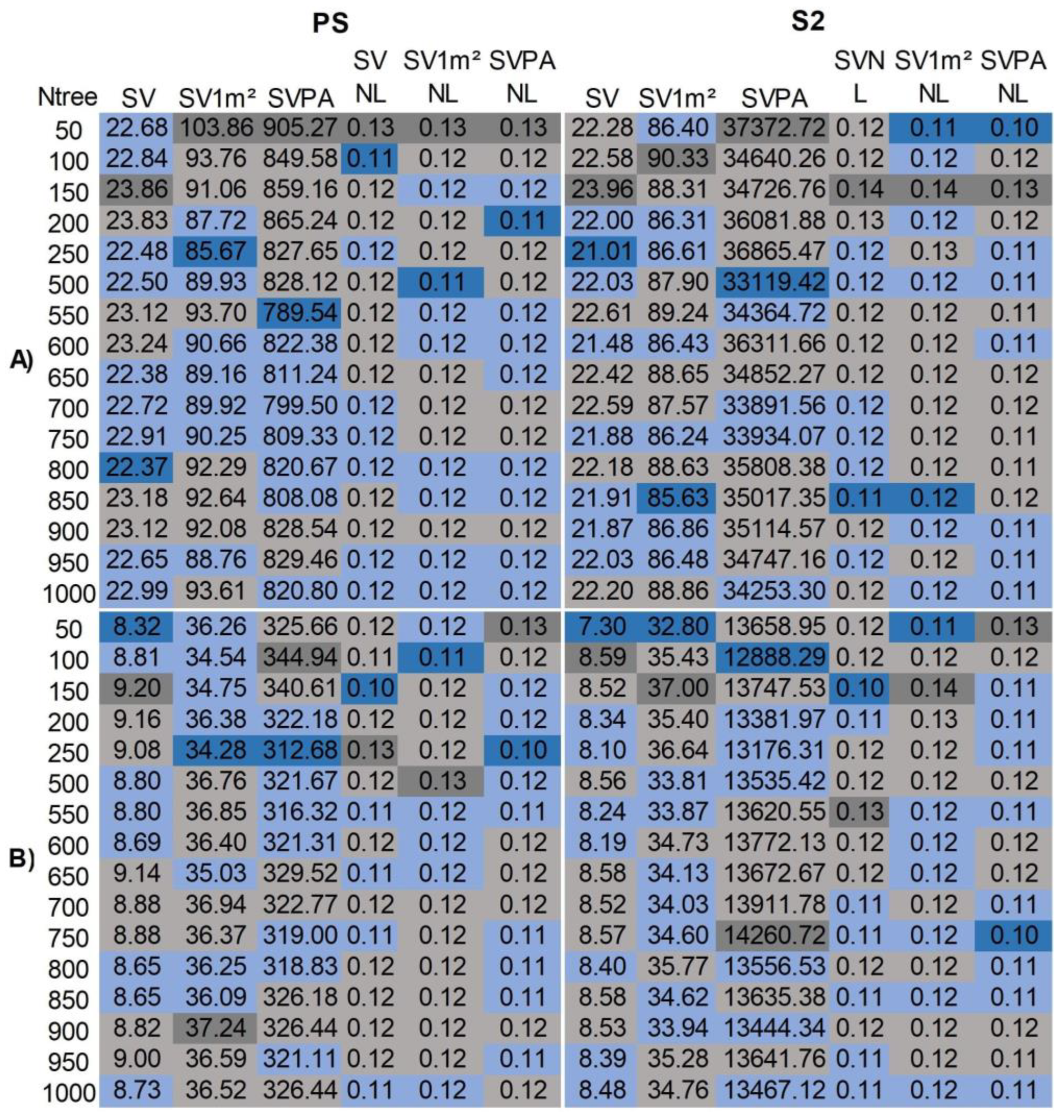
3.1. Optimization of Regression Model Parameters
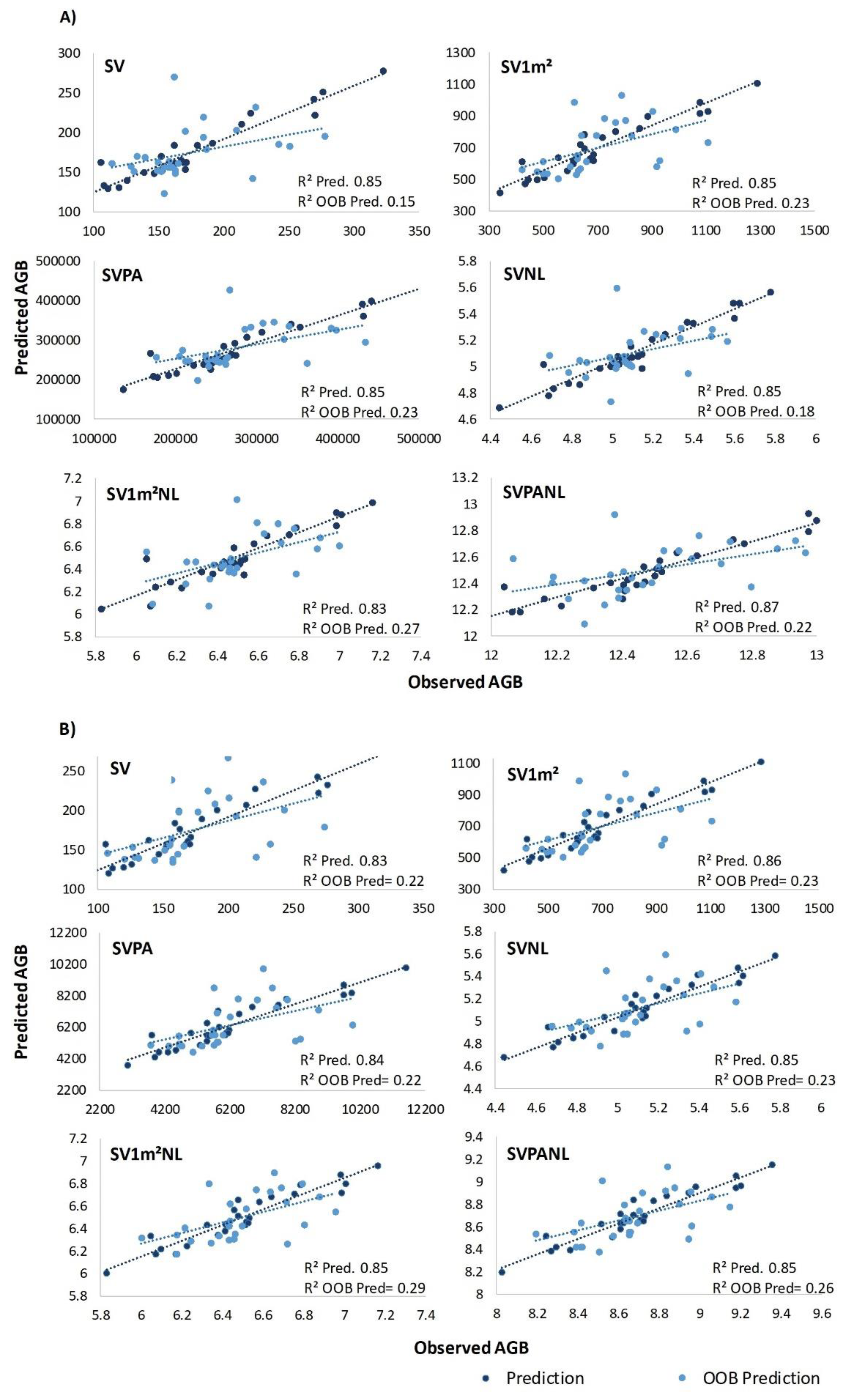
3.2. Predictive Performance of the RF Models
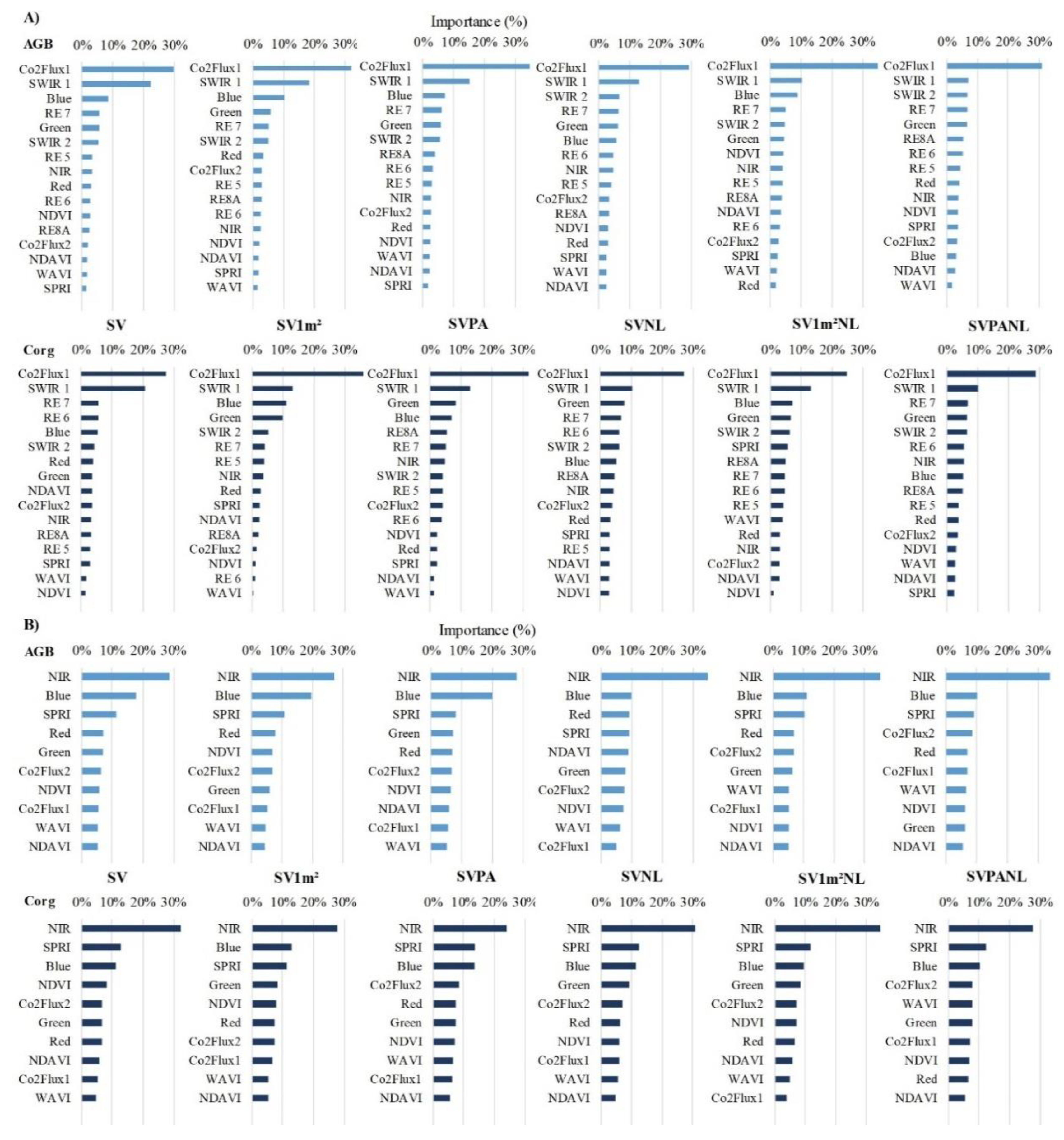
3.3. Importance of Predictor Variables
4. Discussion
4.1. AGB and Corg Estimation Accuracy and Efficiency of Sensors
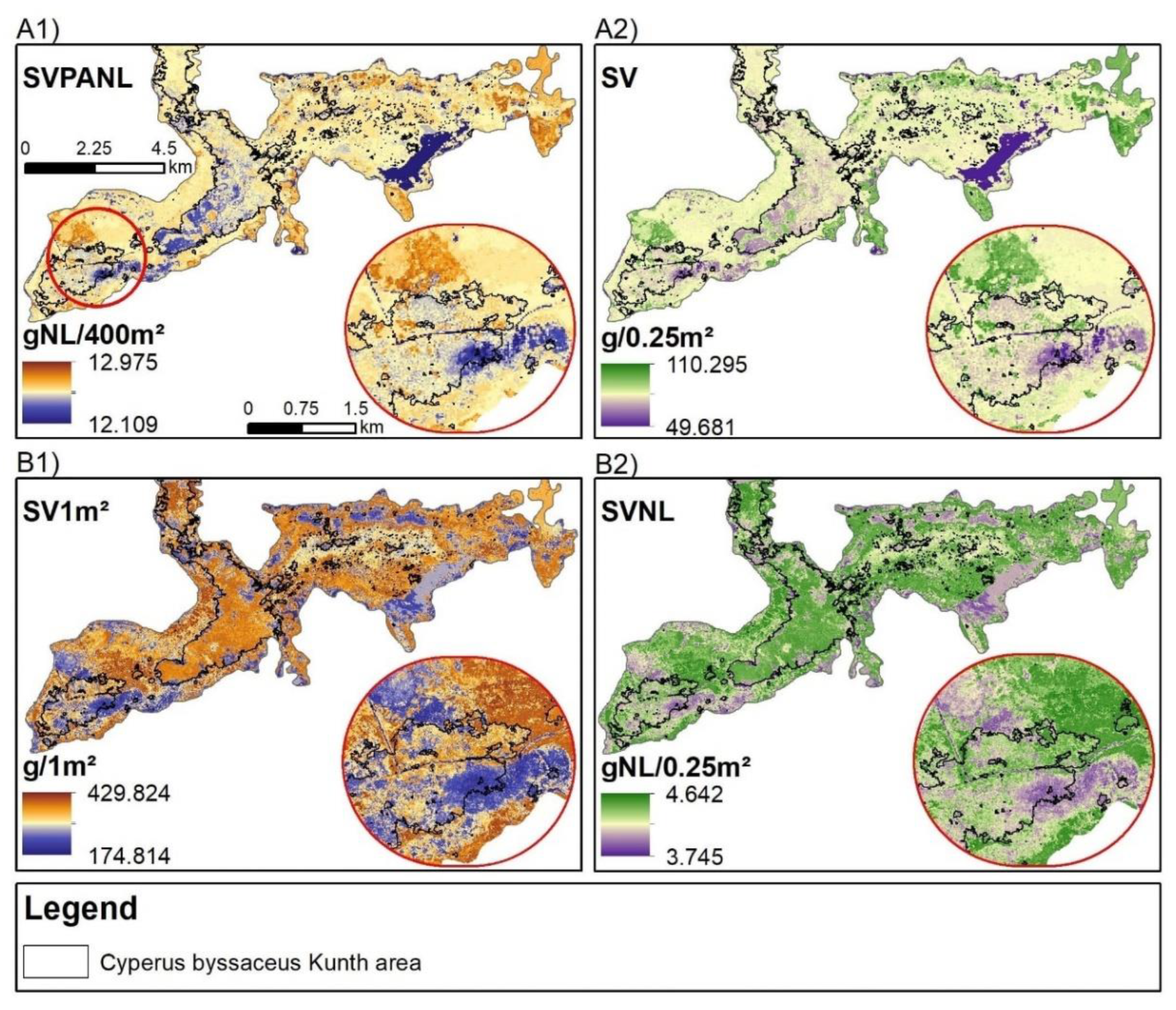
4.2. The Effect of Treatments and Plots Size on Model Performance and Importance of Spectral Variables
5. Conclusions
- Regarding RF parameters, different Ntrees impacted model errors, notably in non-normalized treatments, enhancing RF model precision. Thus, optimized RF models provide more accurate estimates. OOB estimates served effectively for validation, with average prediction errors within the limits found in validation sets in reference studies. This result is useful amidst wetland data collection challenges;
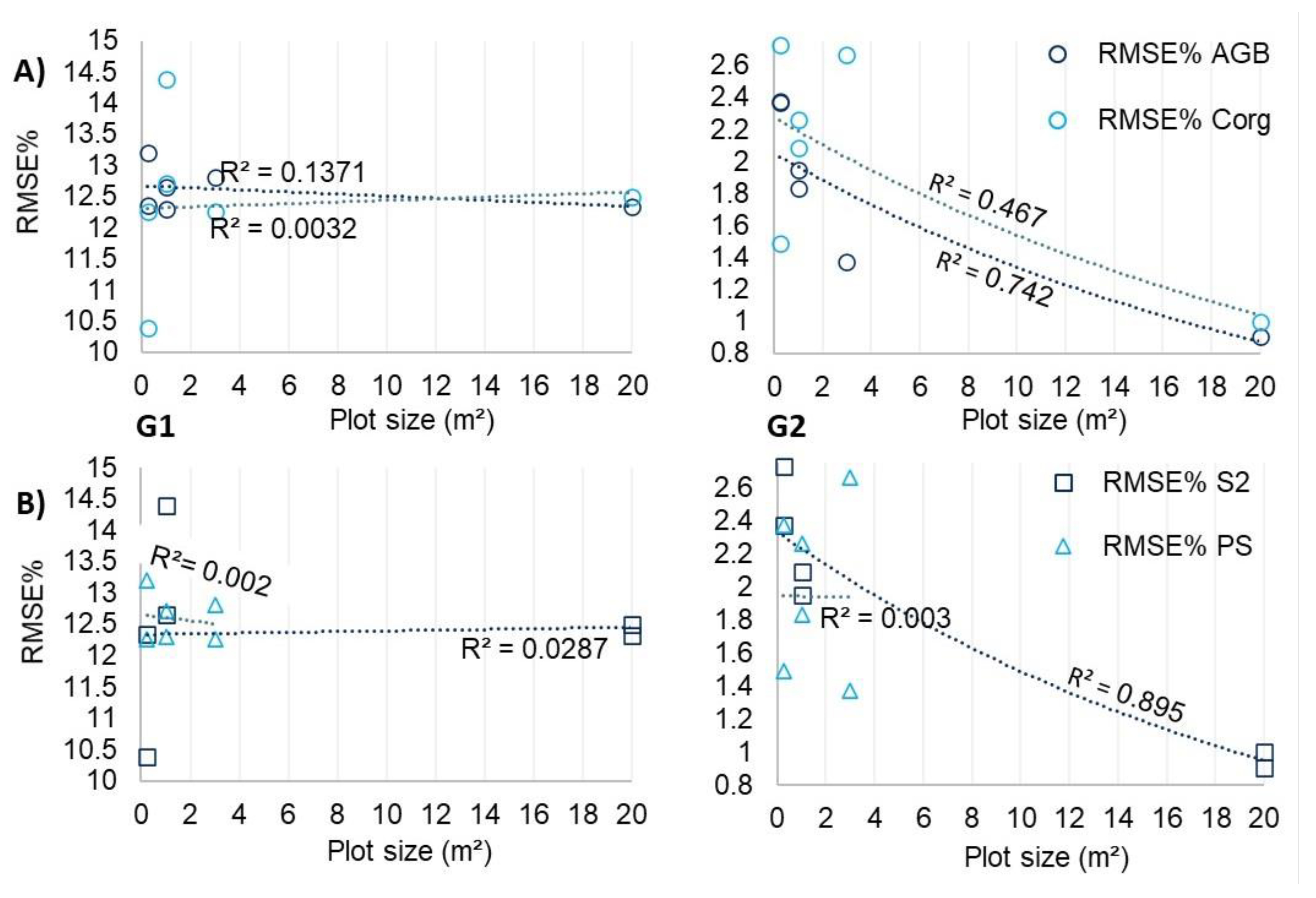
- Normalized sample data treatments enhanced RF model accuracy for AGB and Corg prediction. Estimation errors decrease as S2 model plot size increased, indicating smaller plots may compromise estimate reliability with S2;
- Utilizing S2 and PS sensors underscored, respectively, the value of medium spatial resolution satellite data for enhancing estimate accuracy and high-resolution data for delineating AGB and Corg spatial variability in wetlands. Sensors performance were close, however, S2 was more efficient;
- The RF method, employing the combination of VI CO2Flux and S2’s SWIR, Blue, Green, and RE bands 6 and 7 as predictors, excelled in AGB and Corg prediction. Leveraging an ML algorithm with VI and bands indicative of carbon fluxes and biomass changes proved beneficial, and these predictors serve as spectral indicators of these ecological functions;
- In addition to optimizing the parameters of the RF model, optimizing the input set of AGB and Corg collected in the field, i.e., evaluating normalization and plot sizes, has contributed to more accurate estimates. This approach holds promise for improved monitoring of the ecological processes of AGB and Corg storage in wetlands and for contributing to the understanding of these ecosystems as carbon sinks, vital for offsetting emissions and meeting national and global GHG reduction targets;
- We encourage future work that compares the effects of different plot sizes, sample data normalization methods, sensors, and VIs in RF models and other ML approaches on the accuracy of AGB and Corg estimates in marshes, as well as in other wetlands with emergent herbaceous vegetation, such as salt marshes and peatlands. This will contribute to the continued advancement of knowledge on improving the modeling of AGB and Corg in wetlands.
Supplementary Materials
Author Contributions
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barbier, E.B.; Hacker, S.D.; Kennedy, C.; Koch, E.W.; Stier, A.C.; Silliman, B.R. The Value of Estuarine and Coastal Ecosystem Services. Ecol. Monogr. 2011, 81, 169–193. [Google Scholar] [CrossRef]
- Hiraishi, T.; Krug, T.; Tanabe, K.; Srivastava, N.; Baasansuren, J.; Fukuda, M.; Troxler, T.G. 2013 Supplement to the 2006 IPCC Guidelines for National Greenhouse Gas Inventories: Wetlands. IPCC Switz. 2014, Available online: https://tinyurl.com/5c4xm8rp.
- Webb, E.L.; Friess, D.A.; Krauss, K.W.; Cahoon, D.R.; Guntenspergen, G.R.; Phelps, J. A Global Standard for Monitoring Coastal Wetland Vulnerability to Accelerated Sea-Level Rise. Nat. Clim. Change 2013, 3, 458–465. [Google Scholar] [CrossRef]
- Rice, J.; Seixas, C.; Elena, M.; Bedoya, M.; Valderrama, N.; Anderson, C. Resumen Para Los Responsables de La Formulación de Políticas Del Informe de Evaluación Regional Sobre Diversidad Biológica y Servicios de Los Ecosistemas Para Las Américas de La Plataforma Intergubernamental Científico-Normativa Sobre Diversidad Biológica. Plataforma Intergub. Científico-Norm. Dobre Divers. Biológica Serv. Los Ecosistemas 2018, 34, Available online: https://tinyurl.com/4seyyrds.
- Poulter, B.; Fluet-Chouinard, E.; Hugelius, G.; Koven, C.; Fatoyinbo, L.; Page, S.E.; Rosentreter, J.A.; Smart, L.S.; Taillie, P.J.; Thomas, N.; et al. A Review of Global Wetland Carbon Stocks and Management Challenges. In Geophysical Monograph Series; Krauss, K.W., Zhu, Z., Stagg, C.L., Eds.; Wiley, 2021; pp. 1–20 ISBN 978-1-119-63928-2. [CrossRef]
- Byrd, K.B.; Ballanti, L.; Thomas, N.; Nguyen, D.; Holmquist, J.R.; Simard, M.; Windham-Myers, L. A Remote Sensing-Based Model of Tidal Marsh Aboveground Carbon Stocks for the Conterminous United States. ISPRS J. Photogramm. Remote Sens. 2018, 139, 255–271. [Google Scholar] [CrossRef]
- Ren, Y.; Mao, D.; Li, X.; Wang, Z.; Xi, Y.; Feng, K. Aboveground Biomass of Marshes in Northeast China: Spatial Pattern and Annual Changes Responding to Climate Change. Front. Ecol. Evol. 2022, 10, 1043811. [Google Scholar] [CrossRef]
- Dronova, I.; Taddeo, S.; Hemes, K.S.; Knox, S.H.; Valach, A.; Oikawa, P.Y.; Kasak, K.; Baldocchi, D.D. Remotely Sensed Phenological Heterogeneity of Restored Wetlands: Linking Vegetation Structure and Function. Agric. For. Meteorol. 2021, 296, 108215. [Google Scholar] [CrossRef]
- Campbell, A.D.; Temilola Fatoyinbo, L.; Charles, S.; Bourgeau-Chavez, L.L. A Review of Carbon Monitoring in Wet Carbon Systems Using Remote Sensing. Environ. Res. Lett. 2022, 17, 025009. [Google Scholar] [CrossRef]
- Requena-Mullor, J.M.; Reyes, A.; Escribano, P.; Cabello, J. Assessment of Ecosystem Functioning from Space: Advancements in the Habitats Directive Implementation. Ecol. Indic. 2018, 89, 893–902. [Google Scholar] [CrossRef]
- Cabello, J.; Fernández, N.; Alcaraz-Segura, D.; Oyonarte, C.; Piñeiro, G.; Altesor, A.; Delibes, M.; Paruelo, J.M. The Ecosystem Functioning Dimension in Conservation: Insights from Remote Sensing. Biodivers. Conserv. 2012, 21, 3287–3305. [Google Scholar] [CrossRef]
- Alcaraz-Segura, D.; Lomba, A.; Sousa-Silva, R.; Nieto-Lugilde, D.; Alves, P.; Georges, D.; Vicente, J.R.; Honrado, J.P. Potential of Satellite-Derived Ecosystem Functional Attributes to Anticipate Species Range Shifts. Int. J. Appl. Earth Obs. Geoinformation 2017, 57, 86–92. [Google Scholar] [CrossRef]
- Miller, G.J.; Morris, J.T.; Wang, C. Estimating Aboveground Biomass and Its Spatial Distribution in Coastal Wetlands Utilizing Planet Multispectral Imagery. Remote Sens. 2019, 11, 2020. [Google Scholar] [CrossRef]
- Warwick-Champion, E.; Davies, K.P.; Barber, P.; Hardy, N.; Bruce, E. Characterising the Aboveground Carbon Content of Saltmarsh in Jervis Bay, NSW, Using ArborCam and PlanetScope. Remote Sens. 2022, 14, 1782. [Google Scholar] [CrossRef]
- Zhao, Y.; Mao, D.; Zhang, D.; Wang, Z.; Du, B.; Yan, H.; Qiu, Z.; Feng, K.; Wang, J.; Jia, M. Mapping Phragmites Australis Aboveground Biomass in the Momoge Wetland Ramsar Site Based on Sentinel-1/2 Images. Remote Sens. 2022, 14, 694. [Google Scholar] [CrossRef]
- Li, C.; Zhou, L.; Xu, W. Estimating Aboveground Biomass Using Sentinel-2 MSI Data and Ensemble Algorithms for Grassland in the Shengjin Lake Wetland, China. Remote Sens. 2021, 13, 1595. [Google Scholar] [CrossRef]
- Cai, F.; Tang, B.-H.; Ji, X.; Huang, L.; Fu, Z.; Fan, D. Predicting Carbon Storage in the Yunnan-Kweichow Plateau Wetlands Using a Fusion of Multi-Source Remote Sensing Data and Machine Learning. In Proceedings of the IGARSS 2024 - 2024 IEEE International Geoscience and Remote Sensing Symposium; July 2024; pp. 4806–4809. [Google Scholar] [CrossRef]
- Wan, R.; Wang, P.; Wang, X.; Yao, X.; Dai, X. Modeling Wetland Aboveground Biomass in the Poyang Lake National Nature Reserve Using Machine Learning Algorithms and Landsat-8 Imagery. J. Appl. Remote Sens. 2018, 12, 046029. [Google Scholar] [CrossRef]
- Cutler, D.R.; Edwards Jr., T. C.; Beard, K.H.; Cutler, A.; Hess, K.T.; Gibson, J.; Lawler, J.J. Random Forests for Classification in Ecology. Ecology 2007, 88, 2783–2792. [Google Scholar] [CrossRef]
- Da Silva, J.V.S. Aprendizado de Máquinas Por “Random Forest” Para a Modelagem Da Altura de Árvores de Seringueira. Term paper, Universidade Federal de Mato Grosso do Sul: Mato Grosso do Sul, 2023, Available online:https://repositorio.ufms.br/handle/123456789/8264.
- Nishiwaki, A.A.M. Uso do LIDAR e a potencialidade de geração de renda mediante pagamento por serviço ambiental de sequestro de carbono em floresta tropical sazonalmente seca. Doctoral dissertation, Federal University of Pernambuco: Pernambuco, 2023, Available online: https://repositorio.ufpe.br/handle/123456789/52100.
- Rocha de Souza Pereira, F.; Kampel, M.; Gomes Soares, M.L.; Estrada, G.C.D.; Bentz, C.; Vincent, G. Reducing Uncertainty in Mapping of Mangrove Aboveground Biomass Using Airborne Discrete Return Lidar Data. Remote Sens. 2018, 10, 637. [Google Scholar] [CrossRef]
- Chan, C.K.; Gomez, C.A.; Kothikar, A.; Baiz-Villafranca, P.M. Satellite-Based Carbon Estimation in Scotland: AGB and SOC. Land 2023, 12, 818. [Google Scholar] [CrossRef]
- Mahmud Sujon, K.; Binti Hassan, R.; Tusnia Towshi, Z.; Othman, M.A.; Abdus Samad, M.; Choi, K. When to Use Standardization and Normalization: Empirical Evidence From Machine Learning Models and XAI. IEEE Access 2024, 12, 135300–135314. [Google Scholar] [CrossRef]
- Frazer, G.W.; Magnussen, S.; Wulder, M.A.; Niemann, K.O. Simulated Impact of Sample Plot Size and Co-Registration Error on the Accuracy and Uncertainty of LiDAR-Derived Estimates of Forest Stand Biomass. Remote Sens. Environ. 2011, 115, 636–649. [Google Scholar] [CrossRef]
- Dai, X.; Yang, G.; Liu, D.; Wan, R. Vegetation Carbon Sequestration Mapping in Herbaceous Wetlands by Using a MODIS EVI Time-Series Data Set: A Case in Poyang Lake Wetland, China. Remote Sens. 2020, 12, 3000. [Google Scholar] [CrossRef]
- Sun, S.; Wang, Y.; Song, Z.; Chen, C.; Zhang, Y.; Chen, X.; Chen, W.; Yuan, W.; Wu, X.; Ran, X.; et al. Modelling Aboveground Biomass Carbon Stock of the Bohai Rim Coastal Wetlands by Integrating Remote Sensing, Terrain, and Climate Data. Remote Sens. 2021, 13, 4321. [Google Scholar] [CrossRef]
- Adam, E.; Mutanga, O.; Abdel-Rahman, E.M.; Ismail, R. Estimating Standing Biomass in Papyrus (Cyperus Papyrus L.) Swamp: Exploratory of in Situ Hyperspectral Indices and Random Forest Regression. Int. J. Remote Sens. 2014, 35, 693–714. [Google Scholar] [CrossRef]
- Mutanga, O.; Adam, E.; Cho, M.A. High Density Biomass Estimation for Wetland Vegetation Using WorldView-2 Imagery and Random Forest Regression Algorithm. Int. J. Appl. Earth Obs. Geoinformation 2012, 18, 399–406. [Google Scholar] [CrossRef]
- Naidoo, L.; van Deventer, H.; Ramoelo, A.; Mathieu, R.; Nondlazi, B.; Gangat, R. Estimating above Ground Biomass as an Indicator of Carbon Storage in Vegetated Wetlands of the Grassland Biome of South Africa. Int. J. Appl. Earth Obs. Geoinformation 2019, 78, 118–129. [Google Scholar] [CrossRef]
- Zolkos, S.G.; Goetz, S.J.; Dubayah, R. A Meta-Analysis of Terrestrial Aboveground Biomass Estimation Using Lidar Remote Sensing. Remote Sens. Environ. 2013, 128, 289–298. [Google Scholar] [CrossRef]
- Wang, D.; Wan, B.; Liu, J.; Su, Y.; Guo, Q.; Qiu, P.; Wu, X. Estimating Aboveground Biomass of the Mangrove Forests on Northeast Hainan Island in China Using an Upscaling Method from Field Plots, UAV-LiDAR Data and Sentinel-2 Imagery. Int. J. Appl. Earth Obs. Geoinformation 2020, 85, 101986. [Google Scholar] [CrossRef]
- Qiu, B.; Li, S.; Cao, J.; Zhang, J.; Yang, K.; Luo, K.; Huang, K.; Jiang, X. Uncertainty Analysis of Forest Aboveground Carbon Stock Estimation Combining Sentinel-1 and Sentinel-2 Images. Forests 2024, 15, 2134. [Google Scholar] [CrossRef]
- Simioni, J.P.D.; Guasselli, L.A.; Etchelar, C.B. Connectivity among Wetlands of EPA of Banhado Grande, RS. RBRH 2017, 22. [Google Scholar] [CrossRef]
- Accordi, I.; Hartz, S.; ohlweiler, Adalberto, O. O Sistema Banhado Grande Como Uma Área Úmida de Importância Internacional.; Simpósio de áreas protegidas, Pelotas, Brazil, September 1 2003, Available online: https://tinyurl.com/79aeekcv.
- Serviço Geológico Do Brasil - CPRM - GeoSGB. Available online: https://geoportal.sgb.gov.br/geosgb/ (accessed on 17 January 2025).
- Rossato, M.S. Os Climas Do Rio Grande Do Sul: Uma Proposta de Classificação Climática. Entre-Lugar 2020, 11, 57–85. [Google Scholar] [CrossRef]
- Pereira-Silva, L.; Trevisan, R.; Rodrigues, A.C.; Larridon, I. Combining the Small South American Genus Androtrichum into Cyperus (Cyperaceae). Plant Ecol. Evol. 2020, 153, 446–454. [Google Scholar] [CrossRef]
- Belloli, T.F.; Guasselli, L.A.; Kuplich, T.M.; Ruiz, L.F.C.; de Arruda, D.C.; Etchelar, C.B.; Simioni, J.D. Estimation of Aboveground Biomass and Carbon in Palustrine Wetland Using Bands and Multispectral Indices Derived from Optical Satellite Imageries PlanetScope and Sentinel-2A. J. Appl. Remote Sens. 2022, 16, 034516–034516. [Google Scholar] [CrossRef]
- Frantz, D.S.; Carraro, C.C.; Verdum, R.; Garcia, M. Caracterização de Ambientes Paludais Da Planície Costeira Do Rio Grande Do Sul Em Imagens Orbitais TM/Landsat 5.; Manaus, Brazil, 1990; Vol. 6, pp. 408–418. Available online: https://tinyurl.com/34hp6zej.
- Irgang, B.E.; Júnior, G.; de Senna Gastal Jr, C.V. Macrófitas Aquáticas Da Planície Costeira Do RS; UFRGS, 1996.
- Pratolongo, P.; Kandus, P. Dinámica Biomasa Aérea En Pajonales Scirpus Giganteus Juncales Schoenoplectus Californicus En Zona Front. Bajo Delta Río Paraná Argent. Ecotrópicos. 2005, 18, 30–37. [Google Scholar]
- Howard, J.; Hoyt, S.; Isensee, K; Telszewski, M; Pidgeon, E Coastal Blue Carbon: Methods for Assessing Carbon Stocks and Emissions Factors in Mangroves, Tidal Salt Marshes, and Seagrass Meadows; Arlington, Virginia, USA, 2014.
- Pompêo, M.L.M.; Moschini, C.V. Biomassa Das Macrófitas Aquáticas: O Método Do Quadro; RiMa São Carlos, SP, 2003.
- Sifleet, S.; Pendleton, L.; Murray, B.C. State of the Science on Coastal Blue Carbon: A Summary for Policy Makers. Available online: https://nicholasinstitute.duke.edu/sites/default/files/publications/state-of-science-coastal-blue-carbon-paper.pdf (accessed on 17 January 2025).
- Main-Knorn, M.; Pflug, B.; Louis, J.; Debaecker, V.; Müller-Wilm, U.; Gascon, F. Sen2Cor for Sentinel-2. Proc. SPIE 2017, 10427, 1042704. [Google Scholar] [CrossRef]
- Planet Imagery Product Specification. Available online: https://assets.planet.com/docs/Planet_Combined_Imagery_Product_Specs_letter_screen.pdf (accessed on 17 January 2025).
- Rouse, J.W.; Haas, R.H.; Deering, D.W.; Schell, J.A.; Harlan, J.C. Monitoring the Vernal Advancement and Retrogradation (Green Wave Effect) of Natural Vegetation; Texas A&M Univ. College Station, TX, United States, 1974; Available online: https://ntrs.nasa.gov/citations/19750020419.
- Villa, P.; Laini, A.; Bresciani, M.; Bolpagni, R. A Remote Sensing Approach to Monitor the Conservation Status of Lacustrine Phragmites Australis Beds. Wetl. Ecol. Manag. 2013, 21, 399–416. [Google Scholar] [CrossRef]
- Villa, P.; Bresciani, M.; Braga, F.; Bolpagni, R. Comparative Assessment of Broadband Vegetation Indices Over Aquatic Vegetation. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 3117–3127. [Google Scholar] [CrossRef]
- Gamon, J.A.; Serrano, L.; Surfus, J.S. The Photochemical Reflectance Index: An Optical Indicator of Photosynthetic Radiation Use Efficiency across Species, Functional Types, and Nutrient Levels. Oecologia 1997, 112, 492–501. [Google Scholar] [CrossRef]
- Rahman, A.F.; Gamon, J.A.; Fuentes, D.A.; Roberts, D.A.; Prentiss, D. Modeling Spatially Distributed Ecosystem Flux of Boreal Forest Using Hyperspectral Indices from AVIRIS Imagery. J. Geophys. Res. Atmospheres 2001, 106, 33579–33591. [Google Scholar] [CrossRef]
- Baptista, G. Validação Da Modelagem de Seqüestro de Carbono Para Ambientes Tropicais de Cerrado, Por Meio de Dados AVIRIS e Hyperion. In Proceedings of the Anais XI SBSR; INPE: Belo Horizonte, 2003; pp. 1037–1044. [Google Scholar]
- Zhu, X.; Song, L.; Weng, Q.; Huang, G. Linking In Situ Photochemical Reflectance Index Measurements With Mangrove Carbon Dynamics in a Subtropical Coastal Wetland. J. Geophys. Res. Biogeosciences 2019, 124, 1714–1730. [Google Scholar] [CrossRef]
- Mohanty, P.C.; Shetty, S.; Mahendra, R.S.; Nayak, R.K.; Sharma, L.K.; Rama Rao, E.P. Spatio-Temporal Changes of Mangrove Cover and Its Impact on Bio-Carbon Flux along the West Bengal Coast, Northeast Coast of India. Eur. J. Remote Sens. 2021, 54, 525–537. [Google Scholar] [CrossRef]
- Liu, Y.; Wu, C.; Tian, F.; Wang, X.; Gamon, J.A.; Wong, C.Y.S.; Zhang, X.; Gonsamo, A.; Jassal, R.S. Modeling Plant Phenology by MODIS Derived Photochemical Reflectance Index (PRI). Agric. For. Meteorol. 2022, 324, 109095. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Pedregosa, F.; Pedregosa, F.; Varoquaux, G.; Varoquaux, G.; Org, N.; Gramfort, A.; Gramfort, A.; Michel, V.; Michel, V.; Fr, L.; et al. Scikit-Learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Guerini Filho, M.; Kuplich, T.M.; Quadros, F.L.F.D. Estimating Natural Grassland Biomass by Vegetation Indices Using Sentinel 2 Remote Sensing Data. Int. J. Remote Sens. 2020, 41, 2861–2876. [Google Scholar] [CrossRef]
- Morris, J.D.; Daood, S.S.; Nimmo, W. Machine Learning Prediction and Analysis of Commercial Wood Fuel Blends Used in a Typical Biomass Power Station. Fuel 2022, 316, 123364. [Google Scholar] [CrossRef]
- Xing, J.; Luo, K.; Wang, H.; Fan, J. Estimating Biomass Major Chemical Constituents from Ultimate Analysis Using a Random Forest Model. Bioresour. Technol. 2019, 288, 121541. [Google Scholar] [CrossRef]
- Valbuena, R.; Hernando, A.; Manzanera, J.A.; Görgens, E.B.; Almeida, D.R.A.; Silva, C.A.; García-Abril, A. Evaluating Observed versus Predicted Forest Biomass: R-Squared, Index of Agreement or Maximal Information Coefficient? Eur. J. Remote Sens. 2019, 52, 345–358. [Google Scholar] [CrossRef]
- Mauya, E.W.; Madundo, S. Modelling Above Ground Biomass Using Sentinel 2 and Planet Scope Data in Dense Tropical Montane Forests of Tanzania. Tanzan. J. For. Nat. Conserv. 2022, 91, 132–153. [Google Scholar]
- Lawrence, R.L.; Wood, S.D.; Sheley, R.L. Mapping Invasive Plants Using Hyperspectral Imagery and Breiman Cutler Classifications (randomForest). Remote Sens. Environ. 2006, 100, 356–362. [Google Scholar] [CrossRef]
- Ghosh, S.M.; Behera, M.D.; Jagadish, B.; Das, A.K.; Mishra, D.R. A Novel Approach for Estimation of Aboveground Biomass of a Carbon-Rich Mangrove Site in India. J. Environ. Manage. 2021, 292, 112816. [Google Scholar] [CrossRef]
- Baloloy, A.B.; Blanco, A.C.; Candido, C.G.; Argamosa, R.J.L.; Dumalag, J.B.L.C.; Dimapilis, L.L.C.; Paringit, E.C. Estimation of Mangrove Forest Aboveground Biomass Using Multispectral Bands, Vegetation Indices and Biophysical Variables Derived from Optical Satellite Imageries: Rapideye, Planetscope and Sentinel-2. ISPRS Ann. Photogramm. Remote Sens. Spat. Inf. Sci. 2018, IV–3, 29–36. [Google Scholar] [CrossRef]
- Han, X.; Wang, Y.; Ke, Y.; Liu, T.; Zhou, D. Phenological Heterogeneities of Invasive Spartina Alterniflora Salt Marshes Revealed by High-Spatial-Resolution Satellite Imagery. Ecol. Indic. 2022, 144, 109492. [Google Scholar] [CrossRef]
- Swoish, M.; Da Cunha Leme Filho, J.F.; Reiter, M.S.; Campbell, J.B.; Thomason, W.E. Comparing Satellites and Vegetation Indices for Cover Crop Biomass Estimation. Comput. Electron. Agric. 2022, 196, 106900. [Google Scholar] [CrossRef]
- Padró, J.-C.; Muñoz, F.-J.; Ávila, L.Á.; Pesquer, L.; Pons, X. Radiometric Correction of Landsat-8 and Sentinel-2A Scenes Using Drone Imagery in Synergy with Field Spectroradiometry. Remote Sens. 2018, 10, 1687. [Google Scholar] [CrossRef]
- Naik, P.; Dalponte, M.; Bruzzone, L. Prediction of Forest Aboveground Biomass Using Multitemporal Multispectral Remote Sensing Data. Remote Sens. 2021, 13, 1282. [Google Scholar] [CrossRef]
- Mao, P.; Ding, J.; Jiang, B.; Qin, L.; Qiu, G.Y. How Can UAV Bridge the Gap between Ground and Satellite Observations for Quantifying the Biomass of Desert Shrub Community? ISPRS J. Photogramm. Remote Sens. 2022, 192, 361–376. [Google Scholar] [CrossRef]
- Rohlf, F.J.; Sokal, R.R. Biometry : The Principles and Practice of Statistics in Biological Research; 2nd ed.; W. H. Freeman, 1981.
- Santos, H.V.S. Estimativa de biomassa aérea e teor de carbono da espécie Rhizophora mangle L. Thesis dissertation, Universidade Federal de Sergipe, 2012. Available online: https://ri.ufs.br/handle/riufs/6642.
- Hossain, M.; Othman, S.; Bujang, J.S.; Kusnan, M. Net Primary Productivity of Bruguiera Parviflora (Wight & Arn.) Dominated Mangrove Forest at Kuala Selangor, Malaysia. For. Ecol. Manag. 2008, 255, 179–182. [Google Scholar] [CrossRef]
- Goetz, S.; Dubayah, R. Advances in Remote Sensing Technology and Implications for Measuring and Monitoring Forest Carbon Stocks and Change. Carbon Manag. 2011, 2, 231–244. [Google Scholar] [CrossRef]
- Hosseini, Z.; Latifi, H.; Naghavi, H.; Bakhtiarvand Bakhtiari, S.; Fassnacht, F.E. Influence of Plot and Sample Sizes on Aboveground Biomass Estimations in Plantation Forests Using Very High Resolution Stereo Satellite Imagery. For. Int. J. For. Res. 2021, 94, 278–291. [Google Scholar] [CrossRef]
- Silva, M.A.S. da; Faria, A.L.L. de Índice CO2 flux para avaliar perdas de serviços ecossistêmicos em mangues impactados por tempestade de granizo no Sudeste do Brasil. GEOUSP 2023, 27, e. [Google Scholar] [CrossRef]
- Della-Silva, J.L.; da Silva Junior, C.A.; Lima, M.; Teodoro, P.E.; Nanni, M.R.; Shiratsuchi, L.S.; Teodoro, L.P.R.; Capristo-Silva, G.F.; Baio, F.H.R.; de Oliveira, G.; et al. CO2Flux Model Assessment and Comparison between an Airborne Hyperspectral Sensor and Orbital Multispectral Imagery in Southern Amazonia. Sustainability 2022, 14, 5458. [Google Scholar] [CrossRef]
- Rossi, F.S.; de Araújo Santos, G.A.; de Souza Maria, L.; Lourençoni, T.; Pelissari, T.D.; Della-Silva, J.L.; Oliveira Júnior, J.W.; Silva, A. de A. e; Lima, M.; Teodoro, P.E.; et al. Carbon Dioxide Spatial Variability and Dynamics for Contrasting Land Uses in Central Brazil Agricultural Frontier from Remote Sensing Data. J. South Am. Earth Sci. 2022, 116, 103809. [Google Scholar] [CrossRef]
- Lu, Y.; Zhu, X. Response of Mangrove Carbon Fluxes to Drought Stress Detected by Photochemical Reflectance Index. Remote Sens. 2021, 13, 4053. [Google Scholar] [CrossRef]
- Pham, T.D.; Yokoya, N.; Xia, J.; Ha, N.T.; Le, N.N.; Nguyen, T.T.T.; Dao, T.H.; Vu, T.T.P.; Pham, T.D.; Takeuchi, W. Comparison of Machine Learning Methods for Estimating Mangrove Above-Ground Biomass Using Multiple Source Remote Sensing Data in the Red River Delta Biosphere Reserve, Vietnam. Remote Sens. 2020, 12, 1334. [Google Scholar] [CrossRef]
- Byrd, K.B.; O’Connell, J.L.; Di Tommaso, S.; Kelly, M. Evaluation of Sensor Types and Environmental Controls on Mapping Biomass of Coastal Marsh Emergent Vegetation. Remote Sens. Environ. 2014, 149, 166–180. [Google Scholar] [CrossRef]
- Chen, J.M.; Pavlic, G.; Brown, L.; Cihlar, J.; Leblanc, S.G.; White, H.P.; Hall, R.J.; Peddle, D.R.; King, D.J.; Trofymow, J.A.; et al. Derivation and Validation of Canada-Wide Coarse-Resolution Leaf Area Index Maps Using High-Resolution Satellite Imagery and Ground Measurements. Remote Sens. Environ. 2002, 80, 165–184. [Google Scholar] [CrossRef]
- Nandy, S.; Singh, R.; Ghosh, S.; Watham, T.; Kushwaha, S.P.S.; Kumar, A.S.; Dadhwal, V.K. Neural Network-Based Modelling for Forest Biomass Assessment. Carbon Manag. 2017, 8, 305–317. [Google Scholar] [CrossRef]
- Ponzoni, F.J.; Shimabukuro, Y.E.; Kuplich, T.M. Sensoriamento remoto da vegetação; Oficina de Textos, 2015; ISBN 978-85-7975-211-7.
- Vorster, A.G.; Evangelista, P.H.; Stovall, A.E.L.; Ex, S. Variability and Uncertainty in Forest Biomass Estimates from the Tree to Landscape Scale: The Role of Allometric Equations. Carbon Balance Manag. 2020, 15, 8. [Google Scholar] [CrossRef]

| Sensor | March/2018 | August/2018 | November/2018 |
|---|---|---|---|
| Sentinel-2A | Mar 11 | Aug 28 | Nov 16 |
| PlanetScope | Mar 13 | Aug 17 | Nov 21 |
| Field data collection | Mar 14 | Aug 17 | Nov 22 |
| Vegetation Indices | Equation | References |
|---|---|---|
| NDVI – Normalized Difference | [48] | |
| NDAVI – Aquatic by Normalized Difference | [49] | |
| WAVI – Adjusted to Water | [50] | |
| sPRI – Photochemical Reflectance |
|
[51] |
| CO²Flux1– Integrated | [52,53] | |
| CO²Flux2 – Integrated NDAVI | [39] |
| Treatments | Legend | |
|---|---|---|
| Group 1 | Group 2 | |
| SV | SVNL | Sample Values obtained with a 50x50 cm sampler (SV). Plot area equal to the sampler (0.25m²); the same in NL |
| SV1m² | SV1m²NL | Sample Values estimated to the plot area of 1m² (SV1m²); the same in NL |
| SVPA | SVPANL | Sample Values estimated to plot area equal to the sensor pixel (SVPA), PS (3m²) and S2 (20m²); the same in NL |
| AGB | ||||||||
|---|---|---|---|---|---|---|---|---|
| Group | Sensor | Treatment | R2 | RMSE | RMSE% | RMSE OOB | RMSE OOB% | |
| G1 | S2 | SV | 0.85 | 21.46 | 12.35 | 39.60 | 22.75 | |
| SV1m² | 0.85 | 87.55 | 12.65 | 157.26 | 22.58 | |||
| SVPA | 0.85 | 34246.41 | 12.33 | 58938.28 | 20.98 | |||
| PS | SV | 0.83 | 22.89 | 13.19 | 62.74 | 35.93 | ||
| SV1m² | 0.86 | 85.19 | 12.31 | 163.32 | 23.49 | |||
| SVPA | 0.84 | 804.33 | 12.81 | 1502.88 | 23.67 | |||
| G2 | S2 | SVNL | 0.85 | 0.12 | 2.37 | 0.21 | 4.04 | |
| SV1m²NL | 0.83 | 0.13 | 1.95 | 0.22 | 3.34 | |||
| SVPANL | 0.87 | 0.11 | 0.91 | 0.21 | 1.71 | |||
| PS | SVNL | 0.85 | 0.12 | 2.37 | 0.22 | 4.24 | ||
| SV1m²NL | 0.85 | 0.12 | 1.83 | 0.21 | 3.17 | |||
| SVPANL | 0.85 | 0.12 | 1.37 | 0.21 | 2.41 | |||
| Corg | ||||||||
| G1 | S2 | SV | 0.89 | 7.41 | 10.39 | 16.17 | 19.71 | |
| SV1m² | 0.79 | 41.83 | 14.39 | 57.38 | 22.41 | |||
| SVPA | 0.84 | 14228.77 | 12.50 | 24846.43 | 21.73 | |||
| PS | SV | 0.85 | 8.79 | 12.26 | 16.54 | 21.83 | ||
| SV1m² | 0.84 | 36.51 | 12.71 | 63.21 | 21.88 | |||
| SVPA | 0.84 | 318.91 | 12.27 | 573.27 | 23.02 | |||
| G2 | S2 | SVNL | 0.86 | 0.12 | 2.73 | 0.21 | 5.08 | |
| SV1m²NL | 0.85 | 0.12 | 2.09 | 0.23 | 4.06 | |||
| SVPANL | 0.86 | 0.12 | 1.00 | 0.20 | 1.70 | |||
| PS | SVNL | 0.86 | 0.11 | 1.49 | 0.21 | 2.72 | ||
| SV1m²NL | 0.83 | 0.13 | 2.26 | 0.21 | 3.69 | |||
| SVPANL | 0.85 | 0.12 | 2.67 | 0.21 | 5.02 | |||
| AGB | |||||
|---|---|---|---|---|---|
| Sensor | Treatment. | μObs | μPred | μOOB | |
| G1 | S2 | SV | 172.61 | 173.78 | 174.05 |
| SV1m² | 658.32 | 660.43 | 664.10 | ||
| SVPA | 276178.96 | 277714.74 | 280909.88 | ||
| PS | SV | 172.61 | 173.51 | 174.64 | |
| SV1m² | 658.32 | 660.09 | 663.16 | ||
| SVPA | 6214.03 | 6278.38 | 6349.69 | ||
| G2 | S2 | SVNL | 5.102 | 5.103 | 5.106 |
| SV1m²NL | 6.488 | 6.502 | 6.504 | ||
| SVPANL | 12.480 | 12.489 | 12.500 | ||
| PS | SVNL | 5.102 | 5.107 | 5.116 | |
| SV1m²NL | 6.488 | 6.493 | 6.497 | ||
| SVPANL | 8.686 | 8.687 | 8.693 | ||
| Corg | |||||
| G1 | S2 | SV | 71.54 | 71.31 | 72.15 |
| SV1m² | 273.82 | 278.26 | 278.69 | ||
| SVPA | 114456.71 | 113874.3 | 114321.65 | ||
| PS | SV | 71.54 | 71.69 | 71.87 | |
| SV1m² | 273.82 | 274.76 | 276.37 | ||
| SVPA | 2575.28 | 2599.82 | 2625.71 | ||
| G2 | S2 | SVNL | 4.223 | 4.222 | 4.221 |
| SV1m²NL | 5.61 | 5.616 | 5.634 | ||
| SVPANL | 11.601 | 11.605 | 11.616 | ||
| PS | SVNL | 4.223 | 4.236 | 4.253 | |
| SV1m²NL | 5.61 | 5.618 | 5.636 | ||
| SVPANL | 7.807 | 7.814 | 7.824 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





