1. Introduction
Transcription factors (TFs) are DNA-binding proteins that interact with other transcriptional regulators, including chromatin remodelling/ modifying proteins, to either attract or prevent RNA polymerases from accessing the DNA template [
5,
53]. The complexity of 27 transcriptional regulation in plants is demonstrated by the fact that TFs make up around 7% of their coding sequence. As molecular switches, they respond to both internal and external stimuli by turning genes on or off, thereby ensuring precise control over cellular and physiological processes. The ability of TFs to modulate complex gene networks makes them indispensable for the proper functioning of plants [
35]. Plants are amazing organisms.
In addition to ensuring their growth and success in reproduction, they can construct intricate organic superstructures from basic inorganic molecules while stationary in space and exposed to harsh environmental conditions like water, light, temperature and nutrients, and to biological challenges from competitors, pests, and pathogens. Because of evolution, plants have a flexible developmental program that allows them to adapt reproduction to the environment and produce new vegetative organs. Short-term differentiation is another ability of plant cell to address more pressing environmental issues. The main mechanism governing plant development and differentiation is gene transcription, which is regulated by TFs and other proteins that either attract or prevent RNA polymerases from accessing the DNA template. TFs are usually defined as sequence-specific DNA-binding proteins that are capable of activating and/or repressing transcription. The genomes of plants seem to code many more TFs than those of animals, such as Drosophila melanogaster and Caenorhabditis elegans, which suggests that plants’ transcriptional control is at least as intricate as that of animals [
48]. Over 1,800 TF genes are found in Arabidopsis thaliana, accounting for over 7% of all protein-coding genes [
2].
Over the years, extensive studies has directed towards well-characterized TF families such as MYB, AP2/ERF, bZIP, and WRKY, which regulate various aspects of plant biology, from development and metabolism to stress responses [
20]. However, current progresses in genomic, transcriptomic, and proteomic technologies have unveiled a plethora of previously uncharacterized TFs. These novel TFs often exhibit unconventional structures, unique DNA-binding motifs, or distinct regulatory mechanisms, distinguishing them from classical TFs. Understanding these novel TFs is crucial, not only for unravelling the complexities of plant regulatory networks but also for addressing pressing agricultural challenges. With global food security threatened by climate change, resource limitations, and increasing population, identifying and leveraging these emerging TFs can pave the way for developing stress-resilient and high-yield crops. Furthermore, novel TFs hold promise for engineering plants with enhanced adaptability to diverse environments, improved nutritional content, and optimized secondary metabolite production.
In this review, we explore the latest discoveries in novel TFs, emphasizing their roles in plant development, stress responses, and metabolic regulation. We also discuss their impending applications in crop improvement, the challenges associated with their functional characterization, and future directions in this rapidly evolving field.
2. Novel Transcription Factor Families
2.1. GARP Family: New Insights into Photosynthesis Regulation
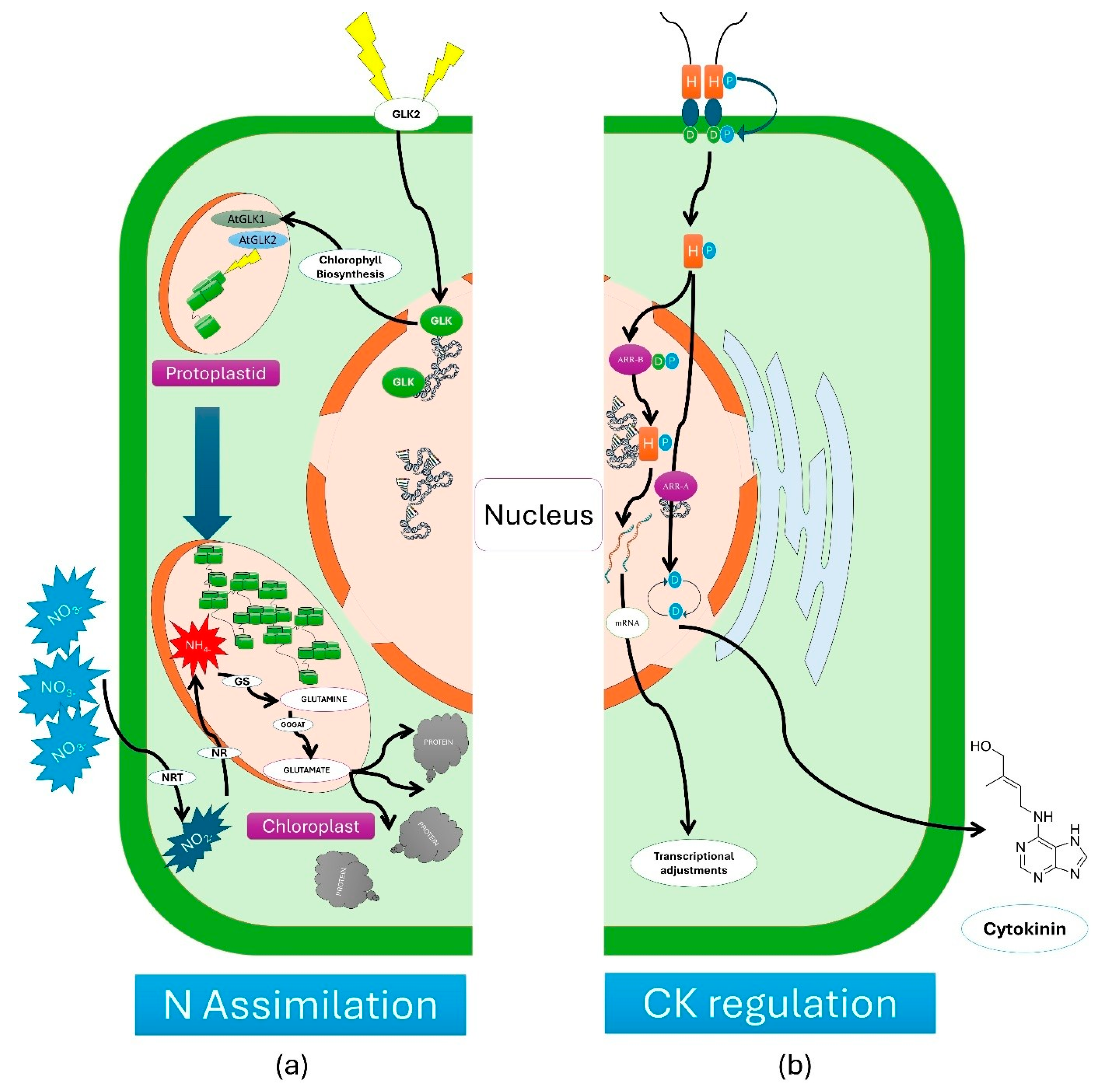
The GARP (Golden2, ARR-B, and Psr1) family of transcription factors plays an integral role in regulating chloroplast development and nutrient signalling pathways, particularly nitrogen assimilation [
7] (Figure1a.). These TFs bind to specific DNA sequences in photosynthesis-related and nutrient-responsive genes, modulating their expression to ensure optimal plant growth and development. Recent studies have proved the dual functionality of GARP TFs in both chloroplast biogenesis and metabolic adaptation. For instance, Golden2-like (GLK) transcription factors, a subgroup of GARP TFs, have been shown to directly regulate genes associated with chlorophyll biosynthesis, light harvesting, and thylakoid membrane assembly. This regulation is essential for efficient photosynthesis, especially under variable light conditions [
45]. Additionally, ARR-B proteins, another subgroup within the GARP family, serve as key mediators in the cytokinin signalling pathway. Cytokinins are plant hormones that control cell division, nutrient allocation, and chloroplast differentiation. ARR-B TFs help integrate cytokinin signals into the regulation of nitrogen uptake and assimilation, optimizing resource use efficiency [
42] (
Figure 1b).
Figure 1.
1a. Regulation of Golden2-like (GLK) transcription factors on nitrogen assimilation a) Golden2-like2 (GLK2 TF) will get triggered through external factors and send a signal for chlorophyll biosynthesis to GLK in the nucleus, b) Arabidopsis thaliana GLK2 (GLK2) in protoplastid will increases the chloroplast for increasing the light use efficiency, c)applied nitrogen will be taken up by the plant as NO3
− (Nitrate) form and will be converted as Nitrite (NO2
−) through Nitrate reductase (NRT), d) the reduced NO2
− will be converted to ammonia (NH4) through nitrite reductase (NR), e) the accumulated NH4 with the help of chloroplast will be converted to Glutamine thorough Glutamine synthase (GS) e) Glutamine will be converted to glutamate with the help of glutamine and 2-oxoglutarate (GOGAT), further it will be converted into proteins.
Figure 2b. Regulation Cytokinin (CK) and transcriptional adjustments by type-A Arabidopsis response regulators (ARR-A) and type-B Arabidopsis response regulators (ARR-B) a) Activated CK will promote the ARR-A which regulated the proteins required for production of higher amount CK b) Activated CK will promote the ARR-B for synthesis of mRNA for transcriptional adjustments.
Figure 1.
1a. Regulation of Golden2-like (GLK) transcription factors on nitrogen assimilation a) Golden2-like2 (GLK2 TF) will get triggered through external factors and send a signal for chlorophyll biosynthesis to GLK in the nucleus, b) Arabidopsis thaliana GLK2 (GLK2) in protoplastid will increases the chloroplast for increasing the light use efficiency, c)applied nitrogen will be taken up by the plant as NO3
− (Nitrate) form and will be converted as Nitrite (NO2
−) through Nitrate reductase (NRT), d) the reduced NO2
− will be converted to ammonia (NH4) through nitrite reductase (NR), e) the accumulated NH4 with the help of chloroplast will be converted to Glutamine thorough Glutamine synthase (GS) e) Glutamine will be converted to glutamate with the help of glutamine and 2-oxoglutarate (GOGAT), further it will be converted into proteins.
Figure 2b. Regulation Cytokinin (CK) and transcriptional adjustments by type-A Arabidopsis response regulators (ARR-A) and type-B Arabidopsis response regulators (ARR-B) a) Activated CK will promote the ARR-A which regulated the proteins required for production of higher amount CK b) Activated CK will promote the ARR-B for synthesis of mRNA for transcriptional adjustments.
The third subgroup, Psr1 (Phosphorus Starvation Response 1), plays a critical role in phosphorus homeostasis. Psr1 TFs activate genes involved in phosphate transport and remobilization under nutrient-deprived conditions, ensuring continued photosynthetic activity despite environmental limitations [
9]. Collectively, these findings highlight the versatility of the GARP family in maintaining photosynthetic efficiency and nutrient balance, making them attractive targets for crop improvement. Genetic engineering approaches that enhance GARP TF expression or activity could lead to higher yields and better stress resilience in agricultural crops.
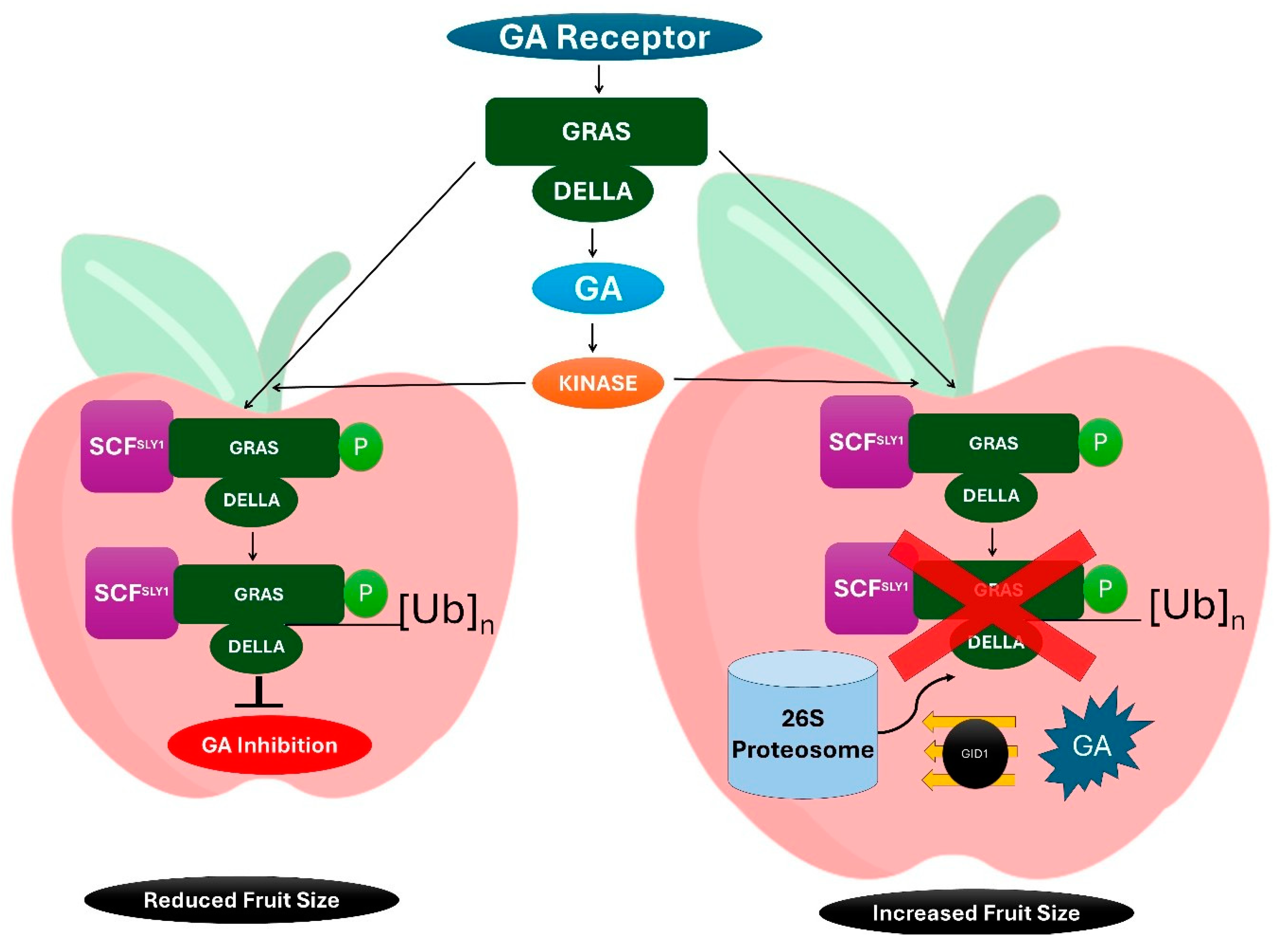
Figure 2.
Regulation of GRAS (GAI, RGA, and SCR) in GA (Gibberellic acid) production, a) the activation of ubiquitin ligase complex with GRAS-DELLA will lead to inhibition of GA production b) The GA inhibition will lead to reduced fruit size c) GA binds to GID1 which interacts with DELLA proteins d)26S proteasome degrades the DELLA protein through Ubiquitination e) degradation of DELLA proteins promote cell elongation and growth in response to gibberellin.
Figure 2.
Regulation of GRAS (GAI, RGA, and SCR) in GA (Gibberellic acid) production, a) the activation of ubiquitin ligase complex with GRAS-DELLA will lead to inhibition of GA production b) The GA inhibition will lead to reduced fruit size c) GA binds to GID1 which interacts with DELLA proteins d)26S proteasome degrades the DELLA protein through Ubiquitination e) degradation of DELLA proteins promote cell elongation and growth in response to gibberellin.
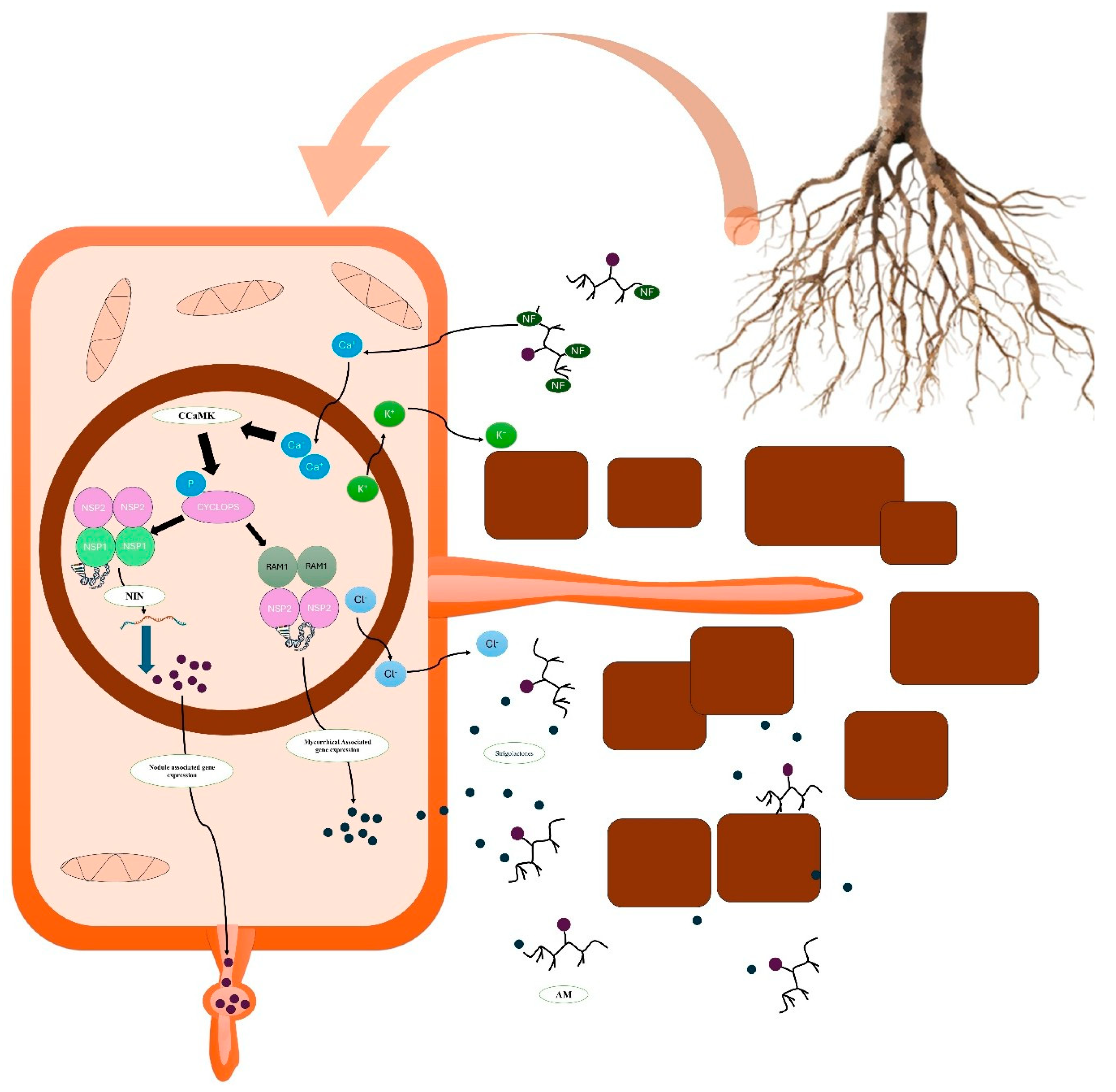
2.2. GRAS Proteins: Versatility in Symbiosis and Stress
GRAS (GAI, RGA, and SCR) proteins represent a diverse family of transcription factors involved in numerous plant processes, including gibberellin (GA) signaling, root development, symbiotic interactions, and abiotic stress responses. Initially identified for their roles in GA signaling, GRAS TFs have since been recognized for their multifaceted functions in plant biology [
28] (
Figure 2.). A key role of GRAS proteins lies in promoting symbiotic associations, particularly arbuscular mycorrhizal (AM) and rhizobial symbioses [
36]. For example, NSP1 (Nodulation Signalling Pathway 1) and NSP2 are indispensable for the establishment of nodules in leguminous plants. These nodules facilitate nitrogen fixation, a critical process for plant growth in nutrient-poor soils. NSP1 directly regulates the expression of nodulation genes, while NSP2 interacts with other regulatory components to fine-tune symbiotic responses [
44]. Similarly, RAM1 (Required for Arbuscular Mycorrhization 1), another GRAS TF, controls the expression of genes necessary for AM symbiosis, enhancing phosphorus uptake in plants [
24] (
Figure 2.).
GRAS proteins also play pivotal roles in abiotic stress responses, including drought, salinity, and heat stress. For instance, DELLA proteins, a subgroup of GRAS TFs, act as repressors in the gibberellin signalling pathway. Under stress conditions, reduced gibberellin levels stabilize DELLA proteins, which in turn activate stress-responsive genes (
Figure 3). This mechanism allows plants to prioritize survival over growth during adverse conditions [
8]. Another example is SCL14 (SCARECROW-like 14), which mediates plant responses to oxidative stress. SCL14 establishes a complex with TGA transcription factors to activate genes involved in detoxification pathways, protecting plants from reactive oxygen species (ROS) damage [
39]. In addition to stress and symbiosis, GRAS proteins are crucial for root and shoot development. SCR (SCARECROW) and SHR (SHORT-ROOT) are GRAS TFs that regulate root radial patterning and stem cell maintenance. These proteins form a regulatory cascade that ensures proper tissue differentiation and organ development [
16].
Figure 3.
Role of nodulation signalling pathway 1(NSP1) and nodulation signalling pathway 2 (NSP2) in nodule and symbiotic association of mycorrhiza a) Node factor (NF) from mycorrhiza will trigger the calcium oscillation in the cells b) Calcium spiking will activate the nuclear-localized calcium- and calmodulin-dependent protein kinases (CCaMKs), c) CCaMK directly phosphorylates CYCLOPS, enabling its DNA-binding ability and transcriptional activity, d)CYCLOPS activates NPS 1 and NPS 2 leading to transcriptional activity through NIN (NODULE INCEPTION) triggering the nodule development d) CYCLOPS triggers the NSP1 and RAM1, helpful in formation of hyphopodia on the surface of the root and production of strigogalactone.
Figure 3.
Role of nodulation signalling pathway 1(NSP1) and nodulation signalling pathway 2 (NSP2) in nodule and symbiotic association of mycorrhiza a) Node factor (NF) from mycorrhiza will trigger the calcium oscillation in the cells b) Calcium spiking will activate the nuclear-localized calcium- and calmodulin-dependent protein kinases (CCaMKs), c) CCaMK directly phosphorylates CYCLOPS, enabling its DNA-binding ability and transcriptional activity, d)CYCLOPS activates NPS 1 and NPS 2 leading to transcriptional activity through NIN (NODULE INCEPTION) triggering the nodule development d) CYCLOPS triggers the NSP1 and RAM1, helpful in formation of hyphopodia on the surface of the root and production of strigogalactone.
Given their diverse roles, GRAS proteins offer immense potential for agricultural biotechnology. Genetic modifications that enhance GRAS protein activity, improve nutrient acquisition, stress tolerance, and overall plant resilience. For instance, overexpression of NSP1 and RAM1 enhances nodulation efficiency and phosphorus uptake, respectively, while manipulating DELLA proteins can optimize stress responses without compromising growth.
2.3. NAC with Unusual Regulatory Mechanisms
The NAC (NAM, ATAF, and CUC) family of transcription factors is renowned for its roles in plant development, stress responses, and senescence. However, recent discoveries have identified NAC subgroups with atypical regulatory mechanisms and DNA-binding properties, expanding our understanding of their functional diversity. Certain NAC proteins exhibit unusual DNA-binding motifs that deviate from the conventional NAC recognition sequences. These motifs enable them to target unique gene sets, particularly those involved in specialized secondary metabolism. For example, specific NAC TFs regulate genes responsible for flavonoid and lignin biosynthesis, which are critical for plant defence and structural integrity [
5]. Novel NAC TFs have been linked to enhanced drought and heat tolerance. Some of these TFs are capable of binding to promoter regions of abiotic stress-responsive genes and activating their expression under adverse conditions. They also interact with other regulatory proteins and signalling pathways, fine-tuning the plant’s adaptive responses [
14,
15,
32].
Recent data indicates that certain NAC TFs participate in chromatin remodelling by recruiting histone modifiers to stress-inducible genes [
31]. This epigenetic regulation ensures a rapid and sustained transcriptional response to environmental stimuli. A unique subgroup of NAC TFs has been implicated in regulating Programmed Cell Death (PCD) during senescence and pathogen attack. These TFs activate genes encoding proteases and other enzymes involved in cellular breakdown, contributing to nutrient remobilization and defence [
62]. The identification of NAC TFs with unusual regulatory properties opens new avenues for agricultural innovation. By engineering crops to overexpress stress-tolerant or metabolite-enhancing NAC TFs, researchers can develop plants with improved resilience and productivity. Furthermore, understanding their epigenetic roles could inform strategies for modulating gene expression in a stable yet flexible manner.
Beyond these major families, there are several other novel TFs such as bZIP, AITRs, TCP, CPP, NIN-like reported in different plant species (
Table 1). These novel TFs have a crucial part in their respective plant development and growth under some favourable or unfavourable conditions.
Table 1.
Novel transcription factors reported in various plant species and their functions.
Table 1.
Novel transcription factors reported in various plant species and their functions.
S.N. Tran-
Scription Factor (TF) |
TF Family |
Plant Species |
Function |
|
| SUSIBA2 |
WRKY |
Hordeum vulgare cv Pongo |
Binds to the Sugar-Responsive
Elements of the iso1 Promoter for participation in sugar signalling |
Sun et al., 2003 |
AtVOZ1
and AtVOZ2, |
|
Arabidopsis |
bind to the 38-bp cis-acting region of A. thaliana V-PPase gene, AVP1 |
Mit- suda et al., 2004 |
| NtWRKY12 |
WRKY |
Tobacco (Nicotiana tabacum ‘Samsun NN’ |
NtWRKY12 and TGA1a act synergistically in PR-1a expression induced by salicylic acid and bacterial elicitors. |
Van- verk et al., 2008 |
myb52,
myb-like
TF, hb5
hb15
showed |
|
Arabidopsis |
Hyper lignified SCW
Ectopic lignification |
Cassan-
Wang
et al.,
2013 |
| JcNAC1 |
NAC |
Jatropha curcas |
enhanced tolerance to
drought and increased
susceptibility to
pathogens |
Qin et
al.,
2014 |
Gb-
WRKY2 |
WRKY |
Ginkgo biloba |
flowers and strongly
induced by methyl
jasmonate |
Liao
et al.,
2015 |
NAC050
and
NAC052 |
NAC |
Arabidopsis |
involved in
transcriptional
repression and
flowering time control
by associating with the
histone demethylase
JMJ14. |
Ning
et al.,
2015 |
| GIP1 |
bZIP |
Arabidopsis |
early stages of
Arabidopsis
development |
Shaikhali,
2015 |
| AITR |
ABA-
induced
transcription
repressors
(AITRs) |
Arabidopsis |
6 Arabidopsis AITR
genes are induced by
exogenous ABA |
Tian
et al.,
2017 |
OsPCF2
(OsCPP5)
OsNIN-
like2, OsNIN-
like3 and OsNIN-
like4 |
TCP
CPP
NIN-like |
Oryza sativa L.)
genotype Hasawi |
regulators of OsNHX1
gene expression in a salt tolerant rice genotype |
Almeidaet al., 2017 |
| PvBMY1 |
APETALA2/Ethyle
Response Factor |
Pnaenicum virgatum L |
increase biomass yield in greenhouse-grown switchgrass by regulation of photosynthesis and related metabolism be-like |
AmHleyvpaotelidgne-
bavaram et al., 2018 |
| PvBMY3 |
Nuclear-
Factor Y |
|
|
|
| Bel-like |
|
|
|
|
3. Functional Insights and Mechanisms
3.1. Unique DNA-Binding Domains
Unique DNA-binding domains (DBDs) in plant novel transcription factors are specialized protein regions that enable precise regulation of gene expression by interacting with specific DNA sequences. These domains often exhibit distinctive structural features beyond classical motifs like homeodomains or zinc fingers, adapting to plant-specific regulatory needs. For instance, some plant transcription factors target DNA secondary structures such as G-quadruplexes or interact with cis-regulatory elements unique to plant genomes, enabling fine-tuned control over growth, development, and stress responses [
1,
19].
The identification of these novel DBDs is motivated by developments in genetics and bioinformatics. Techniques like RNA sequencing, ChIP-seq, and CRISPR-based screens help uncover transcription factors and their DNA-binding preferences [
56]. Protein structure 162 prediction tools, such as AlphaFold, provide insights into the structural basis of DNA 163 recognition in novel plant transcription factors [
17]. Biochemical methods, including 164 electrophoretic mobility shift assays (EMSAs), further validate DNA-binding specificity 165 and unravel functional interactions [
52].
Functionally, plant transcription factors with unique DBDs play critical roles in diverse processes such as seed development, flowering, and response to environmental cues like drought, salinity, and pathogens. They often act as master regulators, integrating signals from phytohormones like auxins, gibberellins, or abscisic acid, to control large networks of target genes. Recent discoveries include TFs with atypical DNA-binding domains. Examples include: TIFY proteins, which interact with jasmonic acid signalling components to regulate stress responses. RWP-RK TFs, pivotal in nitrogen-related signaling pathways.
3.2. Post-Translational Modifications (PTMs)
Novel TFs often exhibit complex regulation through PTMs such as phosphorylation, ubiquitination, and SUMOylation. For example: The ubiquitin-mediated degradation of certain GRAS TFs fine-tunes their activity under fluctuating environmental conditions.
PTMs of plant novel TFs are critical regulatory mechanisms that fine-tune their activity, stability, localization, and interactions, enabling plants to respond dynamically to internal and external stimuli. These chemical modifications alter the functional properties of TFs after protein synthesis, providing an additional layer of control over gene expression. Phosphorylation, mediated by kinases, is among the most common PTMs, modulating the DNA-binding activity, nuclear localization, or protein-protein interactions of TFs in response to environmental signals like light, drought, or pathogens [
58]. Ubiquitination often targets TFs for proteasomal degradation, ensuring timely downregulation of transcriptional activity, while sumoylation can stabilize TFs or alter their subcellular distribution [
66,
67]. Acetylation and methylation affect TF stability and DNA-binding efficiency, linking transcriptional regulation to broader epigenetic contexts [
12].
PTMs also enable plant TFs to serve as switches for molecules in signaling cascades. For example, phosphorylation of TFs downstream of hormone signalling pathways, such as those involving auxins, abscisic acid, or jasmonates, fine-tunes gene expression for stress responses, growth, and development. Crosstalk between different PTMs, such as phosphorylation and ubiquitination, further enhances the specificity and adaptability of TF-mediated transcriptional regulation.
3.3. Non-Coding RNA Interactions
Non-coding RNAs (ncRNAs) regulate novel transcription factors (TFs) in plants, influencing their stability, activity, and specificity to control gene expression during growth, development, and stress responses. Among ncRNAs, microRNAs (miRNAs) are well-studied and regulate TFs post-transcriptionally by targeting their mRNAs for degradation or translational repression. For instance, in Arabidopsis thaliana, miR156 targets SPL (SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE) TFs, modulating vegetative phase transitions and flowering. Overexpression of miR156 delays flowering, while its suppression accelerates it [
18]. Long non-coding RNAs (lncRNAs) interact with TFs at various levels, often serving as scaffolds, decoys, or guides to regulate transcription [
33]. In rice, the lncRNA LAIR interacts with NAC TFs to control leaf senescence during stress, enhancing the expression of stress-responsive genes and promoting survival [
43]. Small interfering RNAs (siRNAs) guide chromatin-level regulation by recruiting TFs to specific genomic loci, often silencing transposable elements or stress-responsive genes. For example, in maize, siRNAs guide TFs to ensure genome stability by suppressing transposable elements during development [
65]. Circular RNAs (circRNAs), though less explored in plants, act as sponges or decoys for miRNAs or TFs, indirectly modulating transcriptional networks [
7,
67]. These diverse interactions provide plants with flexible regulatory mechanisms to adapt to environmental challenges and developmental cues. Leveraging ncRNA-TF interactions can revolutionize agriculture by enabling precise control of traits. For example, engineering miR156 or lncRNAs like LAIR can optimize flowering time, improve stress tolerance, or enhance productivity, offering sustainable solutions for crop improvement in changing climate conditions [
37].
4. Biotechnological Applications
4.1. Enhancing Stress Tolerance
Novel TFs have been harnessed to develop stress-resilient crops. These TFs activate or repress stress-responsive pathways, enabling plants to adapt to environmental challenges such as drought, salinity, cold, and pathogen attacks. For instance, overexpression of GARP TFs enhances photosynthetic efficiency under nutrient-limited conditions, while manipulation of TIFY proteins confers drought and pest resistance [
50]. The DREB (Dehydration-Responsive Element Binding) family of TFs, which belong to the AP2/ERF superfamily. These TFs bind to dehydration-responsive elements (DREs) in the promoters of stress-responsive genes. Overexpression of DREB2A in crops like rice and wheat has been shown to improve drought and heat tolerance by activating genes involved in osmo-protection and water-use efficiency [
49].
Another example is the NAC (ATAF1/2, CUC2, NAM,) family of TFs, which regulate multiple stress-response pathways. For instance, the overexpression of SNAC1 in rice enhances tolerance to drought and salinity by improving root architecture and osmotic balance, resulting in better water absorption and retention [
41]. MYB transcription factors also play a significant role in stress tolerance. AtMYB44, for example, enhances tolerance to drought and salinity in Arabidopsis by modulating abscisic acid (ABA)-responsive pathways [
29]. WRKY transcription factors, such as WRKY28 in Arabidopsis, are key regulators in pathogen defense by modulating salicylic acid and jasmonic acid signaling pathways [
27].
4.2. Boosting Yield and Quality
By targeting GRAS and NAC TFs, researchers have achieved improvements in biomass, seed quality, and secondary metabolite production. The integration of synthetic biology approaches with novel TFs has accelerated trait optimization. GRAS TFs, named after GAI, RGA, and SCR, are involved in hormonal signaling, particularly gibberellins (GA), and root development [
21]. For example, the GRAS TF DRO1 in rice regulates root angle, enhancing deep rooting and improving drought tolerance, which directly contributes to higher yields under water-limited conditions (Batool et al., 2024). Similarly, SlGRAS40 in tomatoes influences fruit ripening and quality by regulating sugar accumulation and stress resistance [
47].
NAC TFs (NAM, ATAF, and CUC) are known for their role in stress tolerance and senescence, which are crucial for crop productivity and quality. The rice NAC TF OsNAC67 improves grain yield and quality under drought by enhancing photosynthesis and reducing oxidative damage [
57]. AtNAP, a NAC TF in Arabidopsis, regulates senescence, ensuring nutrient remobilization to seeds, thereby improving seed quality [
30].
4.3. Precision Breeding Tools
CRISPR-Cas9 technology has been employed to modify novel TFs and their target genes, enabling precise control over regulatory networks. This approach holds promise for tailoring crops to specific environmental challenges.
5. Challenges and Future Directions
Despite notable advancements in describing novel transcription factors (TFs), several challenges remain. Functional redundancy among TF families complicates the identification of distinct roles, and limited knowledge of TF interactomes hinders a comprehensive understanding of their involvement in complex regulatory networks. Additionally, ethical and regulatory challenges surrounding the deployment of genetically modified crops need to be addressed. Future research should focus on utilizing high-throughput functional screens to discover more novel TFs, integrating multi-omics approaches to map TF-mediated regulatory networks, and developing robust biotechnological applications that can be effectively deployed in the field to increase the yield of crops and resilience.
6. Conclusions
The discovery of novel transcription factors has revolutionized our understanding of plant gene regulation. These TFs offer unprecedented opportunities to address global challenges such as food security and climate resilience. By leveraging advanced genetic and biotechnological tools, we are able to realize the maximum potential of these molecular regulators to create sustainable agricultural systems.
Acknowledgments
This publication is connected to the Interreg VI-A IPA Hungary-Serbia Interreg Programme (grant number HUSRB/23S/11/027). Also, Authors are thankful for their logistic support of Department of Agriculture, Koneru Lakshmaiah Education Foundation Green Fileds, Vaddeswaram, A.P.
References
- Ferrero, L.; Zhang, W.; Benhamed, M.; Crespi, M.; Ariel, F. Non-B DNA in plant genomes: prediction, mapping, and emerging roles. Trends in Plant Science 2024. [Google Scholar] [CrossRef] [PubMed]
- Udvardi, M.K.; Kakar, K.; Wandrey, M.; Montanari, O.; Murray, J.; Andriankaja, A.; Zhang, J.Y.; Benedito, V.; Hofer, J.M.; Chueng, F.; et al. Legume transcription factors: global regulators of plant development and response to the environment. Plant Physiology 2007, 144, 538–549. [Google Scholar] [CrossRef] [PubMed]
- Menconi, J.; Perata, P.; Gonzali, S. Novel R2R3 MYB transcription factors regulate anthocyanin synthesis in Aubergine tomato plants. BMC Plant Biology 2023, 23, 148–148. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.; Ling, J.; Song, L.; Zhao, L.; Wang, Y.; Zhao, T. Transcriptomic profiling of tomato leaves identifies novel transcriptionfactors responding to dehydration stress. International Journal of Molecular Sciences 2023, 24, 9725. [Google Scholar] [CrossRef]
- Chen, L.; Wu, F.; Zhang, J. NAC and MYB families and lignin biosynthesis-related members identification and expression analysis in Melilotus albus. Plants 2021, 10, 303–303. [Google Scholar] [CrossRef]
- Liao, Y.L.; Shen, Y.B.; Chang, J.; Zhang, W.W.; Cheng, S.Y.; Xu, F. Isolation, expression, and promoter analysis of GbWRKY2: a novel transcription factor gene from Ginkgo biloba. International Journal of Genomics, 2015; 607185. [Google Scholar]
- Zhao, X.; Yang, J.; Li, X.; Li, G.; Sun, Z.; Chen, Y.; Chen, Y.; Xia, M.; Li, Y.; Yao, L.; et al. Identification and expression analysis of GARP superfamily genes in response to nitrogen and phosphorus stress in Spirodela polyrhiza. BMC Plant Biology 2022, 22, 308. [Google Scholar] [CrossRef]
- Alabadí, D.; Sun, T.P. Green Revolution DELLA Proteins: Functional Analysis and Regulatory Mechanisms. Annual Review of Plant Biology, 2025; 76. [Google Scholar]
- Puga, M.I.; Poza-Carrión, C.; Martinez-Hevia, I.; Perez-Liens, L.; Paz-Ares, J. Recent advances in research on phosphate starvation signaling in plants. Journal of plant research, 2024; 1–16. [Google Scholar]
- Chen, H.; Pugh, B.F. What do transcription factors interact with? Journal of molecular biology 2021, 433, 166883. [Google Scholar] [CrossRef]
- Sun, C.; Palmqvist, S.; Olsson, H.; Borén, M.; Ahlandsberg, S.; Jansson, C. A novel WRKY transcription factor, SUSIBA2, participates in sugar signaling in barley by binding to the sugar-responsive elements of the iso1 promoter. The Plant Cell 2003, 15, 2076–2092. [Google Scholar] [CrossRef]
- Sharma, M.; Sidhu, A.K.; Samota, M.K.; Gupta, M.; Koli, P.; Choudhary, M. Post-Translational Modifications in Histones and Their Role in Abiotic Stress Tolerance in Plants. Proteomes 2023, 11, 38–38. [Google Scholar] [CrossRef]
- Mitsuda, N.; Hisabori, T.; Takeyasu, K.; Sato, M.H. , 2004.
- Ou, X.; Sun, L.; Chen, Y.; Zhao, Z.; Jian, W. Characteristics of NAC transcription factors in Solanaceae crops and their roles in responding to abiotic and biotic stresses. Biochemical and Biophysical Research Communications, 2024; 149840. [Google Scholar]
- Bo, C.; Liu, D.; Yang, J.; Ji, M.; Li, Z.; Zhu, Y.; Duan, Y.; Xue, J.; Xue, T. Comprehensive in silico characterization of NAC transcription factor family of Pinellia ternata and functional analysis of PtNAC66 under high-temperature tolerance in transgenic Arabidopsis thaliana. Plant Physiology and Biochemistry 2024, 208, 108539. [Google Scholar] [CrossRef]
- Waseem, M.; Nkurikiyimfura, O.; Niyitanga, S.; Jakada, B.H.; Shaheen, I.; Aslam, M.M. GRAS transcription factors emerging regulator in plants growth, development, and multiple stresses. Molecular Biology Reports 2022, 49, 9673–9685. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Sun, G.; Xiao, J.; He, X.; Jiang, H.; Zhang, Z.; Zhang, Q.; Li, K.; Zhang, S.; Shi, X.; et al. AlphaFold-guided redesign of a plant pectin methylesterase inhibitor for broad-spectrum disease resistance. Molecular Plant 2024, 17, 1344–1368. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.S.; Ali, A.; Abbas, M.; Sun, Y.; Li, Y.; Li, Q.; Ragauskas, A.J. Harnessing miRNA156: A molecular Toolkit for reshaping plant development and achieving ideal architecture. Plant Physiology and Biochemistry 2024, 215, 109071. [Google Scholar] [CrossRef] [PubMed]
- Suhorukova, A.V.; Sobolev, D.S.; Milovskaya, I.G.; Fadeev, V.S.; Goldenkova-Pavlova, I.V.; Tyurin, A.A. A Molecular Orchestration of Plant Translation under Abiotic Stress. Cells 2023, 12, 2445. [Google Scholar] [CrossRef]
- Kumari, P.; Ojha, R.; Varshney, V.; Gupta, V.; Salvi, P. Transcription Factors and Their Regulatory Role in Plant Defence Response. In Proceedings of the Biotechnological Advances for Disease Tolerance in Plants. Springer Nature; 2024; pp. 337–362. [Google Scholar]
- Dutta, M.; Saha, A.; Moin, M.; Kirti, P.B. Genome-wide identification, transcript profiling and bioinformatic analyses of GRAS transcription factor genes in rice. Frontiers in Plant Science 2021, 12, 777285. [Google Scholar] [CrossRef]
- Qin, X.; Zheng, X.; Huang, X.; Lii, Y.; Shao, C.; Xu, Y.; Chen, F. A novel transcription factor JcNAC1 response to stress in newmodel woody plant Jatropha curcas. Planta 2014, 239, 511–520. [Google Scholar] [CrossRef]
- Méndez, T.; Guajardo, J.; Cruz, N.; Gutiérrez, R.A.; Norambuena, L.; Vega, A.; Moya-León, M.A.; Herrera, R. The Characterization of a Novel PrMADS11 Transcription Factor from Pinus radiata Induced Early in Bent Pine Stem. International Journal of Molecular Sciences 2024, 25, 7245–7245. [Google Scholar] [CrossRef]
- Hartmann, R.M.; Schaepe, S.; Nübel, D.; Petersen, A.C.; Bertolini, M.; Vasilev, J.; Küster, H.; Hohnjec, N. Insights into the complex role of GRAS transcription factors in the arbuscular mycorrhiza symbiosis. Scientific Reports 2019, 9, 3360. [Google Scholar] [CrossRef]
- Ning, Y.Q.; Ma, Z.Y.; Huang, H.W.; Mo, H.; Zhao, T.T.; Li, L.; Cai, T.; Chen, S.; Ma, L.; He, X.J. Two novel NAC transcription factors regulate gene expression and flowering time by associating with the histone demethylase JMJ14. Nucleic acids research 2015, 43, 1469–1484. [Google Scholar] [CrossRef]
- Zhang, X.; Xu, Z.; Yu, X.; Zhao, L.; Zhao, M.; Han, X.; Qi, S. Identification of two novel R2R3-MYB transcription factors, 333 PsMYB114L and PsMYB12L, related to anthocyanin biosynthesis in Paeonia suffruticosa. International journal of molecular sciences 334 2019, 20, 1055. [Google Scholar] [CrossRef]
- Mishra, S.; Roychowdhury, R.; Ray, S.; Hada, A.; Kumar, A.; Sarker, U.; Aftab, T.; Das, R. , 2024.
- Khan, Y.; Xiong, Z.; Zhang, H.; Liu, S.; Yaseen, T.; Hui, T. Expression and roles of GRAS gene family in plant growth, signal transduction, biotic and abiotic stress resistance and symbiosis formation-A review. Plant Biology 2022, 24, 404–416. [Google Scholar] [CrossRef] [PubMed]
- Duan, B.; Xie, X.; Jiang, Y.; Zhu, N.; Zheng, H.; Liu, Y.; Hua, X.; Zhao, Y.; Sun, Y. GhMYB44 enhances stomatal closure to confer drought stress tolerance in cotton and Arabidopsis. Plant Physiology and Biochemistry 2023, 198, 107692. [Google Scholar] [CrossRef]
- Kim, H.J.; Hong, S.H.; Kim, Y.W.; Lee, I.H.; Jun, J.H.; Phee, B.K.; Rupak, T.; Jeong, H.; Lee, Y.; Hong, B.S.; et al. Gene regulatory cascade of senescence-associated NAC transcription factors activated by ETHYLENE-INSENSITIVE2-mediated leaf senescence signalling in Arabidopsis. Journal of experimental botany 2014, 65, 4023–4036. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Tan, N.W.K.; Chung, F.Y.; Yamaguchi, N.; Gan, E.S.; Ito, T. Transcriptional regulators of plant adaptation to heat stress. International Journal of Molecular Sciences 2023, 24, 13297. [Google Scholar] [CrossRef]
- Xia, L.; Sun, S.; Han, B.; Yang, X. NAC domain transcription factor gene GhNAC3 confers drought tolerance in plants. Plant Physiology and Biochemistry 2023, 195, 114–123. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Cui, Y.; Feng, Y.; Hu, Y.; Liu, L.; Duan, L. Long non-coding RNAs of plants in response to abiotic stresses and their regulating roles in promoting environmental adaption. Cells 2023, 12, 729. [Google Scholar] [CrossRef]
- Singh, P.; Mukhopadhyay, K. Comprehensive molecular dissection of TIFY Transcription factors reveal their dynamic responses to biotic and abiotic stress in wheat. Triticum aestivum L.). Scientific Reports 2021, 11, 9739. [Google Scholar] [CrossRef]
- Rushton, P.J.; Bokowiec, M.T.; Laudeman, T.W.; Brannock, J.F.; Chen, X.; Timko, M.P. TOBFAC: the database of tobacco transcription factors. BMC bioinformatics 2008, 9, 1–7. [Google Scholar] [CrossRef]
- Ho-Plágaro, T.; García-Garrido, J.M. Multifarious and interactive roles of GRAS transcription factors during arbuscular mycorrhiza development. Frontiers in Plant Science 2022, 13, 836213. [Google Scholar] [CrossRef]
- Chorostecki, U.; Bologna, N.G.; Ariel, F. The plant noncoding transcriptome: a versatile environmental sensor. The EMBO Journal 2023, 42, 114400. [Google Scholar] [CrossRef]
- Deng, C.; Wang, J.; Lu, C.; Li, Y.; Kong, D.; Hong, Y.; Huang, H.; Dai, S. CcMYB6-1 and CcbHLH1, two novel transcription factors synergistically involved in regulating anthocyanin biosynthesis in cornflower. Plant physiology and biochemistry 2020, 151, 271–283. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.P.; Kim, C. Photosynthetic ROS and retrograde signaling pathways. New Phytologist 2024, 244, 1183–1198. [Google Scholar] [CrossRef]
- Ambavaram, M.M.; Ali, A.; Ryan, K.P.; Peoples, O.; Snell, K.D.; Somleva, M.N. Novel transcription factors PvBMY1 and PvBMY3increase biomass yield in greenhouse-grown switchgrass. Panicum virgatum L.). Plant Science 2018, 273, 100–109. [Google Scholar] [CrossRef] [PubMed]
- Pooam, M.; El-Ballat, E.M.; Jourdan, N.; Ali, H.M.; Hano, C.; Ahmad, M.; El-Esawi, M.A. SNAC3 transcription factor enhances arsenic stress tolerance and grain yield in rice (Oryza sativa L.) through regulating physio-biochemical mechanisms, stress responsive genes, and cryptochrome 1b. Plants 2023, 12, 2731. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Zhang, L.; Yu, Q.; Zhang, J.; Li, P.; Zhang, Y.; Xing, X.; Ding, L.; Fang, W.; Chen, F.; et al. Integrated signals of jasmonates, sugars, cytokinins and auxin influence the initial growth of the second buds of chrysanthemum after decapitation. Biology 2021, 10, 440–440. [Google Scholar] [CrossRef]
- Panchal, A.; Maurya, J.; Seni, S.; Singh, R.K.; Prasad, M. An insight into the roles of regulatory ncRNAs in plants: An abiotic stress and developmental perspective. Plant Physiology and Biochemistry 2023, 201, 107823. [Google Scholar] [CrossRef]
- Chakraborty, S.; Harris, J.M. At the crossroads of salinity and rhizobium-legume symbiosis. Molecular Plant-Microbe Interactions 2022, 35, 540–553. [Google Scholar] [CrossRef]
- Liu, Z.; Xiong, T.; Zhao, Y.; Qiu, B.; Chen, H.; Kang, X.; Yang, J. Genome-wide characterization and analysis of golden 2-like transcription factors related to leaf chlorophyll synthesis in diploid and triploid Eucalyptus urophylla. Frontiers in Plant Science 2022, 13, 952877. [Google Scholar] [CrossRef]
- Verk, M.C.V.; Pappaioannou, D.; Neeleman, L.; Bol, J.F.; Linthorst, H.J. A novel WRKY transcription factor is required for induction of PR-1a gene expression by salicylic acid and bacterial elicitors. Plant Physiology 2008, 146, 1983–1995. [Google Scholar] [CrossRef]
- Chen, H.; Li, H.; Lu, X.; Chen, L.; Liu, J.; Wu, H. Identification and expression analysis of GRAS transcription factors to elucidatecandidate genes related to stolons, fruit ripening and abiotic stresses in woodland strawberry (Fragaria vesca). International journal of molecular sciences 2019, 20, 4593. [Google Scholar] [CrossRef]
- Lehti-Shiu, M.D.; Panchy, N.; Wang, P.; Uygun, S.; Shiu, S.H. Diversity, expansion, and evolutionary novelty of plant DNA-binding transcription factor families. Biochimica et Biophysica Acta (BBA)-Gene Regulatory Mechanisms 2017, 1860, 3–20. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, F.; Shah, S.H.; Jan, A. Overexpression of the DREB1A gene under stress-inducible promoter delays leaf senescence and improves drought tolerance in rice. Cereal Research Communications 2023, 51, 851–857. [Google Scholar] [CrossRef]
- Cai, L.; Guo, Z.; Ding, J.; Gai, Z.; Liu, J.; Meng, Q.; Yang, X.; Zhang, N.; Wang, Q. Genome-Wide Identification and Exogenous Hormone and Stress Response Expression Analysis of the GARP Gene Family in Soybean (Glycine max). Agriculture, 2024; 2109. [Google Scholar]
- Shaikhali, J. GIP1 protein is a novel cofactor that regulates DNA-binding affinity of redox-regulated members of bZIP transcription factors involved in the early stages of Arabidopsis development. Protoplasma 2015, 252, 867–883. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Yang, Y.Y.; Chen, J.Y.; Lakshmanan, P.; Kuang, J.F.; Lu, W.J.; Shan, W. MaNAC029 modulates ethylene biosynthesis and 390 fruit quality and undergoes MaXB3-mediated proteasomal degradation during banana ripening. Journal of Advanced Research 2023, 53, 33–47. [Google Scholar] [CrossRef]
- Sutradhar, M.; Singh, B.K.; Samanta, S.; Ali, M.N.; Mandal, N. The overexpression of OsMed 37_6, a mediator complex subunit enhances salt stress tolerance in rice. Biocatalysis and Agricultural Biotechnology 2024, 58, 103212. [Google Scholar] [CrossRef]
- Liu, Y.; Zhu, P.; Cai, S.; Haughn, G.; Page, J.E. Three novel transcription factors involved in cannabinoid biosynthesis in Cannabis sativa L. Plant Molecular Biology 2021, 106, 49–65. [Google Scholar] [CrossRef]
- Almeida, D.M.; Gregorio, G.B.; Oliveira, M.M.; Saibo, N.J. Five novel transcription factors as potential regulators of OsNHX1 gene expression in a salt tolerant rice genotype. Plant molecular biology 2017, 93, 61–77. [Google Scholar] [CrossRef]
- Hecker, D.; Lauber, M.; Ardakani, F.B.; Ashrafiyan, S.; Manz, Q.; Kersting, J.; Hoffmann, M.; Schulz, M.H.; List, M. Computational tools for inferring transcription factor activity. Proteomics 2023, 23, 2200462. [Google Scholar] [CrossRef]
- Rahman, H.; Ramanathan, V.; Nallathambi, J.; Duraialagaraja, S.; Muthurajan, R. Over-expression of a NAC 67 transcription factor from finger millet (Eleusine coracana L.) confers tolerance against salinity and drought stress in rice. BMC biotechnology 2016, 16, 7–20. [Google Scholar] [CrossRef]
- Baoxiang, W.; Zhiguang, S.; Yan, L.; Bo, X.; Jingfang, L.; Ming, C.; Yungao, X.; Bo, Y.; Jian, L.; Jinbo, L.; et al. A pervasive phosphorylation cascade modulation of plant transcription factors in response to abiotic stress. Planta 2023, 258, 73. [Google Scholar] [CrossRef]
- Iannelli, M.A.; Nicolodi, C.; Coraggio, I.; Fabriani, M.; Baldoni, E.; Frugis, G. A Novel Role of Medicago truncatula KNAT3/4/5- like Class 2 KNOX Transcription Factors in Drought Stress Tolerance. International Journal of Molecular Sciences 2023, 24, 12668. [Google Scholar] [CrossRef] [PubMed]
- Cassan-Wang, H.; Goué, N.; Saidi, M.N.; Legay, S.; Sivadon, P.; Goffner, D.; Grima-Pettenati, J. , 2013.
- Batool, A.; Zahra, N.; Naseer, R.; Shahzad, S.; Iqbal, S.; Kausar, A.; Raza, A. Harnessing the role of genes involved in plant architectural changes. Plant Growth Regulation 2023, 101, 15–34. [Google Scholar] [CrossRef]
- Vranic´, M.; Perochon, A.; Doohan, F.M. Transcriptional Profiling Reveals the Wheat Defences against Fusarium Head Blight Disease Regulated by a NAC Transcription Factor. Plants 2023, 12, 2708. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Lv, F.; Gao, L.; Gu, J.; Yang, R.; Li, S.; Li, Y.; Li, S.; Wang, P. Novel R2R3-MYB transcription factor LiMYB75 enhances leaf callus regeneration efficiency in Lagerstroemia indica. Forests 2023, 14, 517. [Google Scholar] [CrossRef]
- Tian, H.; Chen, S.; Yang, W.; Wang, T.; Zheng, K.; Wang, Y.; Cheng, Y.; Zhang, N.; Liu, S.; Li, D.; et al. A novel family of transcription factors conserved in angiosperms is required for ABA signalling. Plant, cell & environment 2017, 40, 2958–2971. [Google Scholar]
- Cahn, J.; Regulski, M.; Lynn, J.; Ernst, E.; Alves, C.D.S.; Ramakrishnan, S.; Chougule, K.; Wei, S.; Lu, Z.; Xu, X.; et al. MaizeCODE reveals bi-directionally expressed enhancers that harbor molecular signatures of maize domestication. Nature Communications 2024, 15, 10854. [Google Scholar] [CrossRef]
- Singh, M.; Singh, A.; Yadav, N.; Yadav, D.K. Current perspectives of ubiquitination and SUMOylation in abiotic stress tolerance in plants. Frontiers in Plant Science 2022, 13, 993194. [Google Scholar] [CrossRef]
- Zhang, D.; Ma, Y.; Naz, M.; Ahmed, N.; Zhang, L.; Zhou, J.J.; Yang, D.; Chen, Z. Advances in CircRNAs in the Past Decade: Review of CircRNAs Biogenesis, Regulatory Mechanisms, and Functions in Plants 2024, 15, 958–958.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).