Submitted:
11 February 2025
Posted:
12 February 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Study Design and Search Strategy
2.1.1. Systematic Search Phases
| KEY WORDS |
|---|
| Chronic obstructive pulmonary disease, COPD |
| Biomarker*, therapeutic target* |
| Epigenome-Wide Association*, sequencing, epigenetic*, DNA methylation*, long noncoding RNA*, circRNA*, miRNA*, histone* deacetylation*, histone* protein*, HDAC |
| Molecular mechanism*, extracellular vesicle* |
- 2. Systematic Search and Definition of PICOS
| Database | Steps | Query | Research in | Items found |
|---|---|---|---|---|
| PubMed | #1 | (((((((((((((Biomarker*[Title/Abstract]) OR ("therapeutic target*"[Title/Abstract])) OR ("Epigenome-Wide Association*"[Title/Abstract])) OR (sequencing[Title/Abstract])) OR (epigenetic*[Title/Abstract])) OR ("DNA methylation*"[Title/Abstract])) OR ("long noncoding RNA*"[Title/Abstract])) OR (circRNA*[Title/Abstract])) OR (miRNA*[Title/Abstract])) OR ("histone* deacetylation*"[Title/Abstract])) OR ("histone* protein*"[Title/Abstract])) OR (HDAC[Title/Abstract])) OR ("Molecular mechanism*"[Title/Abstract])) OR ("extracellular vesicle*"[Title/Abstract]) | Title/Abstract | 1,460,188 |
| #2 | "COPD"[Title/Abstract] OR "Chronic obstructive pulmonary disease"[Title/Abstract] | Title/Abstract | 83,977 | |
| #3 | Combine #1 AND #2 | 5,834 | ||
| #4 | Limit to (English) | 5,663 | ||
| #5 | Limit after 2020 | 2,509 | ||
| Scopus | #1 | TITLE-ABS-KEY ("Biomarker*" OR "therapeutic target*" OR "Epigenome-Wide Association*" OR "sequencing" OR "epigenetic*" OR " DNA methylation*" OR "long noncoding RNA*" OR "circRNA*" OR "miRNA*" OR "histone* deacetylation*" OR "histone* protein*" OR "HDAC" OR "molecular mechanism*" OR "extracellular vesicle*" ) | Title/Abstract/Keywords | 1,989,075 |
| #2 | TITLE-ABS-KEY (“COPD” OR "Chronic Obstructive Pulmonary Disease") | Title/Abstract/Keywords | 102,296 | |
| #3 | Combine #1 AND #2 | 7,433 | ||
| #4 | Limit to (English) and (Italian) | 7,078 | ||
| #5 | Limit after 2020 | 2,906 |
| Parameters | Inclusion Criteria | Exclusion Criteria |
|---|---|---|
| Participants | Studies in humans Studies including COPD patients |
In vitro and in vivo studies Participants with other malignancies |
| Interventions | Assessment of DNA methylation, Hystone modification, and ncRNA expression | Others |
| Comparisons | Control Group | Others |
| Outcomes | 1) to provide unbiased and exhaustive overview of the current knowledge on the epigenetic modification associated COPD; 2) to summarize the epigenetic modifications translated into clinical therapeutic interventions and biomarkers for COPD. |
Others |
| Study Design | Original studies in English | Review, Scoping Review, Narrative Review, Systematic Review, Meta-Analysis, Editorial, Book, Case Report, Conference Review, and Conference Paper |
- 3. Application of PICOS Study Design Exclusion Criteria
2.1.2. Title and Abstract Selection
2.1.3. Full-Text Selection According to PICOS Criteria
2.1.4. Synthesis Method
| Study | Country | Number of participants | Type of sample | Gene affected | Epigenetic alteration | Activity in COPD | Role of epigenetic mechanisms |
|---|---|---|---|---|---|---|---|
| Kachroo P, et al 2021 [16] | Boston (USA) | N=78 fetal N=160 adult COPD |
Lung tissue | Transcription factors, oxido-reductase, VEGFA-VEGFR2 | Hyper-/hypo-methylation | Air flow limitation, inflammation activation, lung remodeling | Fetal origin of COPD |
| Kachroo P, et al 2020 [17] | Boston (USA) | N=78 fetal N=160 adult COPD |
Lung tissue | Co-methylation: Wnt, Pi3K/AKT, MAPK, Hippo | DNA methylation imbalance | Low lung function | Fetal origin of COPD |
| Schwartz U, et al 2023 [18] | Heidelberg and Munich (Germany) Huston (USA) |
N=3 control N=3 COPD I N=5 COPD II-IV |
Parenchymal fibroblasts (lung tissue) |
3 cluster of genes involved in cell proliferation, DNA repair and extracellular matrix organization | Hyper-/hypo-methylation | Low lung function | Kinetics of DNA methylation in COPD |
| Strom JE, et al 2022 [19] |
Northern Sweden | N=15 control N=18 COPD |
Macrophage from Broncho alveolar lavage (BAL) | DMPs co-localized with COPD-associated SNPs | DNA methylation imbalance | --- | Pathophysiology of COPD |
| Cordero AIH, et al, 2022 [10] | Vancouver (Canada) | N=27 control N=15 COPD |
Small airway epithelial brushings and buffy coat blood | DNAmGrimAge | DNA methylation imbalance | Biomarker for assessing accelerated aging in the airways of individuals with COPD | Biomarker |
| Morrow JD, et al 2020 [20] | Boston (USA) | N=336 control N=331 COPD |
Blood samples | Pi3KCD cg03971555 cg12033075 |
Hyper methylation | Predictive biomarker | Biomarker |
| Zhang Z, et al 2021 [21] | Wuxi (China) |
N=18/17 control N=8/16 COPD |
Lung tissue/bronchoscopies (bronco epithelial cells) | Nfr2 | Hyper methylation | Increased oxidative stress and cell death | Therapeutic target |
| Chen Q, et al 2022 [22] |
Groningen (Netherlands) |
N=966/8 control N=595/14 COPD |
whole blood/airway epithelial cells | AHRR cg05575921 cg21161138 |
Hypo methylation | Airway epithelial cell proliferation, dysregulate mitochondrial function, and reduce apoptotic processes | Therapeutic target |
| Study | Country | Number of participants | Type of sample | Gene affected | Epigenetic alteration | Activity in COPD | Role of epigenetic mechanisms |
|---|---|---|---|---|---|---|---|
| Günes GG, et al, 2022 [25] | Ghent (Belgium) |
N=40 control N=111 COPD |
Monocytes/lung tissue | PRMT7 | Histone methylation | Chronic inflammation | Pathophysiology of COPD /Therapeutic target |
| Study | Country | Number of participants | Type of sample | Gene affected | Epigenetic alteration | Activity in COPD | Role of epigenetic mechanisms |
|---|---|---|---|---|---|---|---|
| Duan R, et al, 2020 [28] | Beijing (China) |
N= 21 control N= 21 COPD |
Peripheral blood mononuclear cells | Gene involved in natural killer T cell activation and T-helper cell differentiation | Differential expression in COPD compared to control | Immune balance alteration | Pathophysiology of COPD /Therapeutic target |
| Xie T, et al, 2024 [29] | Hainan (China) |
N= 5 control N= 10 (acute and stable) COPD |
Peripheral blood mononuclear cells | Caspase 1, IL-18, IL-1b | UP-regulation hsa-circ_0008833-57aa |
Pyroptosis | Pathophysiology of COPD |
| Liu P, et al 2022 [27] | Anhui (China) |
N= 3 control N= 3 COPD |
Blood samples | miR-1273h-3p; miR-411-5p; miR-122-5p; miR-615-5p; miR-519d-3p; miR-485-3p; miR-3646; miR-4714-5p; miR-203b-5p; miR-193a-5p; miR-1261; miR-4690-5p; miR-939-5p; miR-9-5p; miR-2113; miR-7977 | UP-regulation circFCHO2; circMBOAT2, circPTPN22; circTBC1D22A; circACADM; circCKAP5 |
---- | Pathophysiology of COPD |
| Zhang C, et al, 20220[30] | Jiangsu (China) |
N= 17 COPD non-smoker N=23 COPD smoker |
Lung tissue | miR-24/PHPPL2 axis | Down regulation Circ_0006892 | Inflammatory injury | Pathophysiology of COPD |
| Wang Z, et al,2021 [31] | Hebei (China) |
N= 27 control N=21COPD |
Lung tissue | miR-145-5p/BRD4 axis | UP-regulation Circ_ANKRDII |
Inflammation, apoptosis and oxidative stress | Pathophysiology of COPD |
| Tang S, et al,2023 [32] | Hefei (China) |
N=30 control N=30 COPD |
Plasma samples | ---- | Differential expression circ_0008882; circ_00089763; circ_00062683; circ_00077607 | Immune balance alteration | Biomarkers |
| Shen X, et al, 2024 [33] | Jiangsu (China) |
N=29 control N=41 COPD |
Peripheral blood mononuclear cells | ---- | Differential expression circ_0049875 and circ_0042590 |
Acute exacerbation of COPD | Biomarkers |
| Study | Country | Number of participants | Type of sample | Gene affected | Epigenetic alteration | Activity in COPD | Role of epigenetic mechanisms |
|---|---|---|---|---|---|---|---|
| Wu S, et al, 2020 [37] | Taipei(Taiwan) | N=35 control N=64 COPD |
Peripheral blood mononuclear cells | IL-8, VCAM1, E-SEL | Down regulation lncRNA-IL7R | Inflammatory processes | Pathophysiology of COPD |
| Zhou Ai, et al, 2020 [38] | Xiangya (China) | N=3 control N=7COPD |
Lung tissue | Notch1 | Down regulation lncRNA- HOXA-AS2 | Cell viability and Inflammatory processes | Pathophysiology of COPD |
| Wang Y, et al,2020 [39] | Wuhan (China) |
N=80 control N=80 stable COPD N=80 AECOPD |
Peripheral blood mononuclear cells | Mir-146a/TNF, IL-6, IL-8, IL-1IL-17 | Upregulation LncRNA-PVT1 |
Inflammatory processes | Prognostic biomarker |
| Liu S, et al, 2020 [42] | Wuhan (China) |
N=120control N=120 stable COPD N=120 AECOPD |
Blood samples | miR-125b, miR-133, miR-146a, miR-203/TNF, IL-6, IL-8, IL-1IL-17, IL-23 | Up regulation LncRNA-MALAT1 |
Inflammatory processes | Prognostic biomarker |
| Liu P, et al, 2021 [43] | Shanghai (China) |
N=90 control N=50 COPD |
Blood samples | Mir-18a-5p/TNF, IL-6, IL-8, IL-1 | Down regulation lncRNA-CASC2 | Inflammatory processes | Diagnostic biomarker |
| Zhao S, et al,2021 [44] | Jiangsu (China) |
N=150 control N=70 COPD |
Blood samples | Mir-181-5p/Wnt/b-catenin axis | Upregulation LncRNA-LUCAT1 |
Apoptotic/Inflammatory processes | Biomarker/therapeutic target |
| Dai Z, et al, 2022 [45] | Xiangya (China) | N=8 control N=5COPD |
Lung tissue | Bcl-2 | Up regulation lncRNA-HOTAIR | Apoptotic processes | Therapeutic target |
| Zong D, et al, 2022 [46] | Xiangya (China) | N=10 control N=10 COPD |
Lung tissue | Mir-152-3p/ERK | Up regulation lncRNA-CCT1 | Inflammatory processes | Therapeutic target |
| Study | Country | Number of participants | Type of sample | Gene affected | Epigenetic alteration | Activity in COPD | Role of epigenetic mechanisms |
|---|---|---|---|---|---|---|---|
| Wang L, et al 2022 [48] | Xiangya (China) |
N= 12 control N=12 COPD |
Peripheral blood mononuclear cells | IL-8 signaling; *********************** iCOS-iCOSL signaling |
Aberrant expression: miR-4453; miR-4736; miR-3118; miR-6967-5p; miR-132-3p; miR-96-5p; miR-4497 ***************** miR-16-5p; miR-1964-5p; miR-29b-3p; miR-2355-3p; miR-18a-5p; miR-1234-3p; miR-148-3p; miR-21-5p; miR-1184; miR-140-5p; miR-19b-3p; miR-223-3p; miR-1246; miR-130a-3p |
Inflammatory processes | Pathophysiology of COPD |
| Hu J, et al 2022 [49] | Wuhan (China) |
N= 3/9 control N=3/9 COPD |
BALF/ blood samples |
MAPK, RAS, FOXO | miR-129-5p; miR-3529-3p; miR-365b-3p; miR-6503-5p; miR-26-3p; miR-34b-5p; miR-4748; miR-491-5p; miR-158-3p |
Oxidative/inflammatory process | Pathophysiology of COPD |
| Zhang J, et al 2020 [50] | Xuzhou (China) |
N= 14/75 control N=36/53 COPD |
Alveolar macrophages/ Peripheral blood mononuclear cells | HAT1/TNF-IL-6-IL-8 | Up regulation miR-486-5p |
Inflammatory processes | Pathophysiology of COPD |
| Yang H, et al 2021 [52] | Xuzhou (China) |
N= 27 control N=21COPD |
Lung tissue | CDKN1B | Down regulation miR-221-3p |
Apoptotic and Inflammatory processes | Pathophysiology of COPD |
| Chang C, et al 2024 [54] | Beijing (China) |
N= 19 control N= 13 COPD |
Lung tissue | RAGE | Down regulation miR-23a-5p |
Oxidative/inflammatory process | Pathophysiology of COPD |
| Kim R, et al 2021 [55] | Newcastle, New South Wales (Australia) |
N= 5 control N= 10 COPD |
Lung tissue | SATB1/S100A9/NF-kB | Up regulation miR-21 |
Inflammatory processes | Pathophysiology of COPD |
| De Smet E, et al 2020 [56] | Ghent (Belgium) | N= 44 control N= 48 COPD |
Lung tissue | MMP12, ADAM19 | Up regulation miR-155 |
Inflammatory processes | Pathophysiology of COPD |
| Zhu Y, et al 2022 [57] | Brigham (USA) |
N= 17control N=8COPD |
Alveolar macrophages | LDLR | Down regulation miR-103a |
Oxidative/inflammatory process | Pathophysiology of COPD |
| Yang Z, et al 2023 [59] | Suzhou (China) |
N= 14 control N=14 COPD |
Lung tissue | NOS1 | Up regulation miR-4640-5p |
Pulmonary hypertension | Pathophysiology of COPD |
| Tasena H, et al 2022 [63] | Groningen (Netherlands) | N=6 control N=3 COPD |
Lung tissue | MICS5AC, COL4A1, COL5A1 | Up regulation miR-708-5p, let-7a-5p, miR-31-5p, miR-146a-5p |
Epithelial differentiation, chronic mucus hypersecretion (CMH) |
Pathophysiology of COPD |
| Singh P, et al 2024 [60] | Birmingham (USA) |
N= 9 control N=13 COPD |
Lung tissue | ANO1 | Down regulation miR-381 |
Mucus production and secretion | Pathophysiology /therapeutic target |
| Zheng L, et al 2021 [61] | Hefei (China) | N= 400 control N=400 COPD N=50 control+COPD |
Blood samples/ Lung tissue |
E-cadherin, α-SMA,Vimentin,N-cadherin | Down regulation miR-30 |
Epithelial-mesenchymal transition | Pathophysiology of COPD |
| Shi X, et al 2022 [51] | Qinghai (China) |
N= 40 control N=40 COPD |
Blood samples | ------ | Up regulation miR-486-5p; miR-106b-5p |
Hypoxia/Pulmonary Hypertension | Biomarkers |
| Shen Y, et al 2021[ 53] | Nanjing (China) |
N=77 control N=155 COPD |
Blood samples | TNF, IL-6, IL-8, IL-1 | Up regulation miR-221-3p; miR-92a-3p |
Inflammatory processes | Biomarkers |
| Zhang L, et al 2022 [64] | Guizhou (China) |
N= 6 control N=6 COPD |
Blood samples | SLC17A9 | Down regulation miR-548ar-3p |
------ | Biomarkers |
| Nadi E, et al 2022 [65] | Hamadan (Iran) |
N=60 control N=60 COPD |
Blood samples | ------ | Down regulation miR-146a;miR-216 |
Oxidative/inflammatory process | Biomarkers |
| Ding Y, et al 2023 [66] | Hefei (China) |
N=26 control N=59 COPD |
Blood samples | ------ | Down regulation miR-150-5p |
Inflammatory processes | Biomarkers |
| Zhang X, et al 2022 [67] | ChongQing (China) |
N=33 control N=36 COPD |
Blood samples | ------ | Down regulation miR-423-5p |
------ | Biomarkers |
| Tao S, et al 2024 [68] | Xiangya (China) |
N= 23 control N= 240 COPD |
Blood samples | ------ ***** PIK3R2 |
Down regulation miR-1290; miR-1246 ****** Up regulation miR-4433a-5p |
------ ***** Apoptotic and Inflammatory processes |
Biomarkers |
| Cazola-Rivero S, et al 2020 [69] | Tenerife (Spain) |
N=13 control N=24 COPD |
Blood samples | MAPK, chemokines, Wnt | Down regulation miR-1246 |
Emphysema development | Biomarkers |
| Wang C, et al 2021 [70] | Jiaozuo (China) |
N=70 control N=140 COPD |
Blood samples | TNF, IL-1, IL-6 | Up regulation miR-126 |
Inflammatory processes | Biomarkers |
| Huang H, et al 2020 [71] | Kunshan (China) |
N=80 control N=160 COPD |
Blood samples | ------ | Up regulation miR-210 |
Pulmonary hypertension | Biomarkers |
| Burke H, et al 2022 [72] | Southampton (United Kingdom) |
N=20 control N=24 COPD |
BALF/Extra vesicles | ------ | Down regulation miR-338-3p; miR-204-5p ****** Up regulation miR-223-3p; miR-182-5p; miR-2110 |
Inflammatory patterns | Biomarkers |
| Wang F, et al 2023 [73] | Peking (China) |
N=42 control N=111COPD |
Blood samples/Exosome | ------ | Up regulation miR-1258 |
Inflammatory processes | Biomarkers |
| Shen H, et al 2021 [74] | Binzhou (China) |
N=20 control N=20 COPD |
Blood samples | ARHGEF12, BCAT1 |
Up regulation miR-196-5p Down regulation miR-361-5p |
Epithelial hyperplasia | Therapeutic target |
| Study | Country | Number of participants | Type of sample | Gene affected | Epigenetic alteration | Activity in COPD | Role of epigenetic mechanisms |
|---|---|---|---|---|---|---|---|
| Li B, et al 2023 [76] | Ningxia (China) |
N=6 control N=14 COPD |
Lung tissue | 18 hub genes | ceRNA aberrant expression | Immune cells infiltration/differentiation; cell proliferation | Pathophysiology of COPD |
| Feng X, et al 2023 [77] | Ningxia (China) |
N=6 control N=7COPD |
Lung tissue | TNF/NF-kb; IL-6/JAK/STAT3 | ceRNA aberrant expression | Inflammatory processes | Pathophysiology of COPD |
2.1.5. Study Risk of Bias Assessment
3. Results
3.1. Flow Diagram
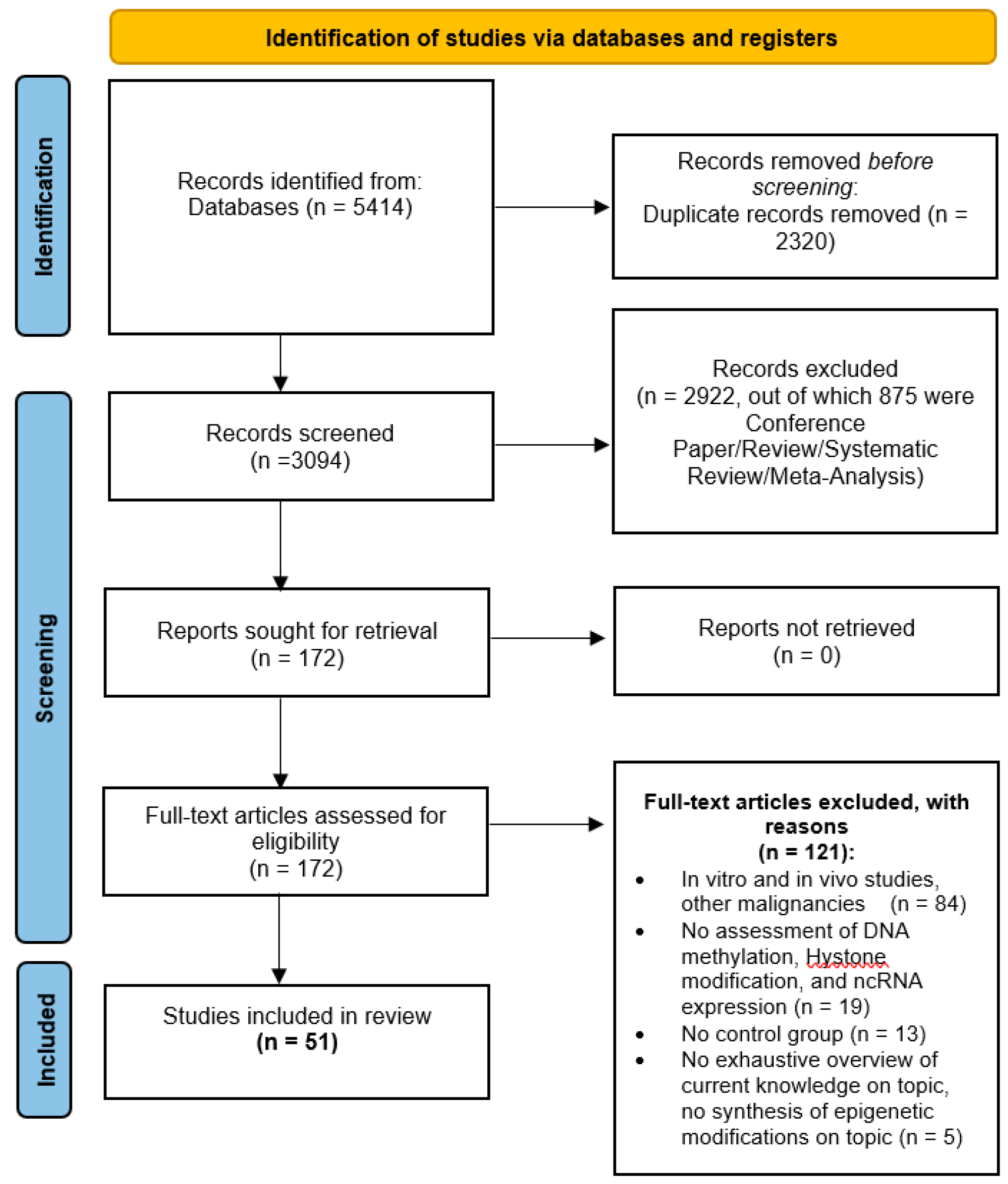
3.2. Study Selection and Characteristics
3.3. Synthesized Findings
3.4. DNA Methylation
3.5. Histone Modifications
3.6. Non-Coding RNA
3.6.1. Circular RNA
3.6.2. Long Non-Coding RNA
3.6.3. Micro RNA
3.6.4. Competing Endogenus RNA
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Disclaimer
References
- Barnes, P.J. Inflammatory Mechanisms in Patients with Chronic Obstructive Pulmonary Disease. Journal of Allergy and Clinical Immunology, Mosby Inc. July 1, 2016, pp 16–27. [CrossRef]
- WHO. Https://Www.Who.Int/News-Room/Fact-Sheets/Detail/Chronic-Obstructive-Pulmonary-Disease-(Copd); 2023.
- Agustí, A.; Celli, B.R.; Criner, G.J.; Halpin, D.; Anzueto, A.; Barnes, P.; Bourbeau, J.; Han, M.L.K.; Martinez, F.J.; de Oca, M.M.; et al. Global Initiative for Chronic Obstructive Lung Disease 2023 Report: GOLD Executive Summary. European Respiratory Journal 2023, 61. [Google Scholar] [CrossRef] [PubMed]
- Safiri, S.; Carson-Chahhoud, K.; Noori, M.; Nejadghaderi, S.A.; Sullman, M.J.M.; Ahmadian Heris, J.; Ansarin, K.; Mansournia, M.A.; Collins, G.S.; Kolahi, A.A.; et al. Burden of Chronic Obstructive Pulmonary Disease and Its Attributable Risk Factors in 204 Countries and Territories, 1990-2019: Results from the Global Burden of Disease Study 2019. The BMJ 2022. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Aarsand, R.; Schotte, K.; Han, J.; Lebedeva, E.; Tsoy, E.; Maglakelidze, N.; Soriano, J.B.; Bill, W.; Halpin, D.M.G.; et al. Tobacco and COPD: Presenting the World Health Organization (WHO) Tobacco Knowledge Summary. Respiratory Research, BioMed Central Ltd December 1, 2024. [CrossRef]
- Lee, Y.J.; Choi, S.; Kwon, S.Y.; Lee, Y.; Lee, J.K.; Heo, E.Y.; Chung, H.S.; Kim, D.K. A Genome-Wide Association Study in Early Copd: Identification of One Major Susceptibility Loci. International Journal of COPD 2020, 15, 2967–2975. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Bakshi, A.; Zhu, Z.; Hemani, G.; Vinkhuyzen, A.A.E.; Lee, S.H.; Robinson, M.R.; Perry, J.R.B.; Nolte, I.M.; Van Vliet-Ostaptchouk, J.V.; et al. Genetic Variance Estimation with Imputed Variants Finds Negligible Missing Heritability for Human Height and Body Mass Index. Nat Genet 2015, 47, 1114–1120. [Google Scholar] [CrossRef]
- Cho, M.H.; Hobbs, B.D.; Silverman, E.K. Genetics of Chronic Obstructive Pulmonary Disease: Understanding the Pathobiology and Heterogeneity of a Complex Disorder. The Lancet Respiratory Medicine, Elsevier Ltd May 1, 2022, pp 485–496. [CrossRef]
- Feng, X.; Dong, H.; Li, B.; Yu, L.; Zhu, J.; Lou, C.; Zhang, J. Integrative Analysis of the Expression Profiles of Whole Coding and Non-Coding RNA Transcriptomes and Construction of the Competing Endogenous RNA Networks for Chronic Obstructive Pulmonary Disease. Front Genet 2023, 14. [Google Scholar] [CrossRef]
- Hernandez Cordero, A.I.; Yang, C.X.; Li, X.; Yang, J.; Shaipanich, T.; MacIsaac, J.L.; Lin, D.T.S.; Kobor, M.S.; Horvath, S.; Man, S.F.P.; et al. The Blood DNA Methylation Clock GrimAge Is a Robust Surrogate for Airway Epithelia Aging. Biomedicines 2022, 10. [Google Scholar] [CrossRef]
- Li, B.; Zhang, J.; Dong, H.; Feng, X.; Yu, L.; Zhu, J.; Zhang, J. Systematic Analysis of Various RNA Transcripts and Construction of Biological Regulatory Networks at the Post-Transcriptional Level for Chronic Obstructive Pulmonary Disease. J Transl Med 2023, 21. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 Statement: An Updated Guideline for Reporting Systematic Reviews. Syst Rev 2021, 10. [Google Scholar] [CrossRef]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G.; Antes, G.; Atkins, D.; Barbour, V.; Barrowman, N.; Berlin, J.A.; Clark, J.; et al. Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. PLoS Medicine, Public Library of Science July 1, 2009. [CrossRef]
- Downes, M.J.; Brennan, M.L.; Williams, H.C.; Dean, R.S. Development of a Critical Appraisal Tool to Assess the Quality of Cross-Sectional Studies (AXIS).
- Moore, L.D.; Le, T.; Fan, G. DNA Methylation and Its Basic Function. Neuropsychopharmacology, January 2013, pp 23–38. [CrossRef]
- Kachroo, P. Priyadarshini Kachroo 2021.
- Kachroo, P.; Morrow, J.D.; Kho, A.T.; Vyhlidal, C.A.; Silverman, E.K.; Weiss, S.T.; Tantisira, K.G.; DeMeo, D.L. Co-Methylation Analysis in Lung Tissue Identifies Pathways for Fetal Origins of COPD. European Respiratory Journal 2020, 56. [Google Scholar] [CrossRef]
- Schwartz, U.; Llamazares Prada, M.; Pohl, S.T.; Richter, M.; Tamas, R.; Schuler, M.; Keller, C.; Mijosek, V.; Muley, T.; Schneider, M.A.; et al. High-resolution Transcriptomic and Epigenetic Profiling Identifies Novel Regulators of COPD. EMBO J 2023, 42. [Google Scholar] [CrossRef]
- Strom, J.E.; Merid, S.K.; Pourazar, J.; Blomberg, A.; Lindberg, A.; Ringh, M.V.; Hagemann-Jensen, M.; Ekstrom, T.J.; Behndig, A.F.; Melen, E. Chronic Obstructive Pulmonary Disease Is Associated with Epigenome-Wide Differential Methylation in BAL Lung Cells. Am J Respir Cell Mol Biol 2022, 66, 638–647. [Google Scholar] [CrossRef] [PubMed]
- Morrow, J.D.; Make, B.; Regan, E.; Han, M.L.; Hersh, C.P.; Tal-Singer, R.; Quackenbush, J.; Choi, A.M.K.; Silverman, E.K.; DeMeo, D.L. DNA Methylation Is Predictive of Mortality in Current and Former Smokers. Am J Respir Crit Care Med 2020, 201, 1099–1109. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Fu, C.; Liu, J.; Sai, X.; Qin, C.; Di, T.; Yang, Y.; Wu, Y.; Bian, T. Hypermethylation of the Nrf2 Promoter Induces Ferroptosis by Inhibiting the Nrf2-GPX4 Axis in COPD. International Journal of COPD 2021, 16, 3347–3362. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Nwozor, K.O.; van den Berge, M.; Slebos, D.J.; Faiz, A.; Jonker, M.R.; Boezen, H.M.; Heijink, I.H.; de Vries, M. From Differential DNA Methylation in COPD to Mitochondria: Regulation of AHRR Expression Affects Airway Epithelial Response to Cigarette Smoke. Cells 2022, 11. [Google Scholar] [CrossRef]
- Kodal, J.B.; Kobylecki, C.J.; Vedel-Krogh, S.; Nordestgaard, B.G.; Bojesen, S.E. AHRR Hypomethylation, Lung Function, Lung Function Decline and Respiratory Symptoms. European Respiratory Journal 2018, 51. [Google Scholar] [CrossRef]
- Yao, H.; Rahman, I. Role of Histone Deacetylase 2 in Epigenetics and Cellular Senescence: Implications in Lung Inflammaging and COPD. Am J Physiol Lung Cell Mol Physiol 2012, 303, 557–566. [Google Scholar] [CrossRef]
- Günes Günsel, G.; Conlon, T.M.; Jeridi, A.; Kim, R.; Ertüz, Z.; Lang, N.J.; Ansari, M.; Novikova, M.; Jiang, D.; Strunz, M.; et al. The Arginine Methyltransferase PRMT7 Promotes Extravasation of Monocytes Resulting in Tissue Injury in COPD. Nat Commun 2022, 13. [Google Scholar] [CrossRef]
- Santer, L.; Bär, C.; Thum, T. Circular RNAs: A Novel Class of Functional RNA Molecules with a Therapeutic Perspective. Molecular Therapy 2019, 27, 1350–1363. [Google Scholar] [CrossRef]
- Liu, P.; Wang, Y.; Zhang, N.; Zhao, X.; Li, R.; Wang, Y.; Chen, C.; Wang, D.; Zhang, X.; Chen, L.; et al. Comprehensive Identification of RNA Transcripts and Construction of RNA Network in Chronic Obstructive Pulmonary Disease. Respir Res 2022, 23. [Google Scholar] [CrossRef]
- Duan, R.; Niu, H.; Yu, T.; Cui, H.; Yang, T.; Hao, K.; Wang, C. Identification and Bioinformatic Analysis of Circular RNA Expression in Peripheral Blood Mononuclear Cells from Patients with Chronic Obstructive Pulmonary Disease. International Journal of COPD 2020, 15, 1391–1401. [Google Scholar] [CrossRef]
- Xie, T.; Yang, Z.; Xian, S.; Lin, Q.; Huang, L.; Ding, Y. Hsa_circ_0008833 Promotes COPD Progression via Inducing Pyroptosis in Bronchial Epithelial Cells. Exp Lung Res 2024, 50, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Gu, S.; Kang, X. CircRNA Circ_0006892 Regulates MiR-24/PHLPP2 Axis to Mitigate Cigarette Smoke Extract-Induced Bronchial Epithelial Cell Injury. Biotechnol Appl Biochem 2022, 69, 735–748. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zuo, Y.; Gao, Z. CircANKRD11 Knockdown Protects HPMECs from Cigarette Smoke Extract-Induced Injury by Regulating MiR-145-5p/BRD4 Axis. International Journal of COPD 2021, 16, 887–899. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Ding, Y.; Zhou, Z.; Yang, W. Identification and Bioinformatic Analysis of CircRNAs in the Plasma of Patients with Very Severe Chronic Obstructive Pulmonary Disease. BMC Pulm Med 2023, 23. [Google Scholar] [CrossRef]
- Shen, X.R.; Liu, Y.Y.; Qian, R.Q.; Zhang, W.Y.; Huang, J.A.; Zhang, X.Q.; Zeng, D.X. Circular RNA Expression of Peripheral Blood Mononuclear Cells Associated with Risk of Acute Exacerbation in Smoking Chronic Obstructive Pulmonary Disease. International Journal of COPD 2024, 19, 789–797. [Google Scholar] [CrossRef]
- Mattick, J.S.; Amaral, P.P.; Carninci, P.; Carpenter, S.; Chang, H.Y.; Chen, L.L.; Chen, R.; Dean, C.; Dinger, M.E.; Fitzgerald, K.A.; et al. Long Non-Coding RNAs: Definitions, Functions, Challenges and Recommendations. Nat Rev Mol Cell Biol 2023, 24, 430–447. [Google Scholar] [CrossRef]
- Tang, W.; Shen, Z.; Guo, J.; Sun, S. Screening of Long Non-Coding RNA and TUG1 Inhibits Proliferation with TGF-β Induction in Patients with COPD. International Journal of COPD 2016, 11, 2951–2964. [Google Scholar] [CrossRef]
- Devadoss, D.; Long, C.; Langley, R.J.; Manevski, M.; Nair, M.; Campos, M.A.; Borchert, G.; Rahman, I.; Chand, H.S. Long Noncoding Transcriptome in Chronic Obstructive Pulmonary Disease. American Journal of Respiratory Cell and Molecular Biology, American Thoracic Society 2019, pp 678–688. [CrossRef]
- Wu, S.M.; Feng, P.H.; Chuang, H.C.; Ho, S.C.; Fan Chung, K.; Chen, K.Y.; Wu, G.S.; Chen, T.T.; Tseng, C.H.; Liu, W.T.; et al. Impaired Lnc-IL7R Modulatory Mechanism of Toll-like Receptors Is Associated with an Exacerbator Phenotype of Chronic Obstructive Pulmonary Disease. FASEB Journal 2020, 34, 13317–13332. [Google Scholar] [CrossRef]
- Zhou, A.Y.; Zhao, Y.Y.; Zhou, Z.J.; Duan, J.X.; Zhu, Y.Z.; Cai, S.; Chen, P. Microarray Analysis of Long Non-Coding RNAs in Lung Tissues of Patients with Copd and Hoxa-As2 Promotes Hpmecs Proliferation via Notch1. International Journal of COPD 2020, 15, 2449–2460. [Google Scholar] [CrossRef]
- Wang, Y.; Lyu, X.; Wu, X.; Yu, L.; Hu, K. Long Non-Coding RNA PVT1, a Novel Biomarker for Chronic Obstructive Pulmonary Disease Progression Surveillance and Acute Exacerbation Prediction Potentially through Interaction with MicroRNA-146a. J Clin Lab Anal 2020, 34. [Google Scholar] [CrossRef]
- Feng, F.; Qi, Y.; Dong, C.; Yang, C. Pvt1 Regulates Inflammation and Cardiac Functiovia the Mapk/Nf-Κb Pathway in a Sepsis Model. Exp Ther Med 2018, 16, 4471–4478. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zeng, Z.; Shen, X.; Wu, Z.; Dong, Y.; Cheng, J.C.H. MicroRNA-146a-5p Negatively Regulates pro-Inflammatory Cytokine Secretion and Cell Activation in Lipopolysaccharide Stimulated Human Hepatic Stellate Cells through Inhibition of Toll-like Receptor 4 Signaling Pathways. Int J Mol Sci 2016, 17. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Liu, M.; Dong, L. The Clinical Value of LncRNA MALAT1 and Its Targets MiR-125b, MiR-133, MiR-146a, and MiR-203 for Predicting Disease Progression in Chronic Obstructive Pulmonary Disease Patients. J Clin Lab Anal 2020, 34. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Zhang, H.; Zeng, H.; Meng, Y.; Gao, H.; Zhang, M.; Zhao, L. LncRNA CASC2 Is Involved in the Development of Chronic Obstructive Pulmonary Disease via Targeting MiR-18a-5p/IGF1 Axis. Ther Adv Respir Dis 2021, 15. [Google Scholar] [CrossRef]
- Zhao, S.; Lin, C.; Yang, T.; Qian, X.; Lu, J.; Cheng, J. Expression of Long Non-Coding RNA LUCAT1 in Patients with Chronic Obstructive Pulmonary Disease and Its Potential Functions in Regulating Cigarette Smoke Extract-Induced 16HBE Cell Proliferation and Apoptosis. J Clin Lab Anal 2021, 35. [Google Scholar] [CrossRef]
- Dai, Z.; Liu, X.; Zeng, H.; Chen, Y. Long Noncoding RNA HOTAIR Facilitates Pulmonary Vascular Endothelial Cell Apoptosis via DNMT1 Mediated Hypermethylation of Bcl-2 Promoter in COPD. Respir Res 2022, 23. [Google Scholar] [CrossRef]
- Zong, D.; Liu, X.; Li, J.; Long, Y.; Ouyang, R.; Chen, Y. LncRNA-CCAT1/MiR-152-3p Is Involved in CSE-Induced Inflammation in HBE Cells via Regulating ERK Signaling Pathway. Int Immunopharmacol 2022, 109. [Google Scholar] [CrossRef]
- Billi, M.; De Marinis, E.; Gentile, M.; Nervi, C.; Grignani, F. Nuclear MiRNAs: Gene Regulation Activities. International Journal of Molecular Sciences, Multidisciplinary Digital Publishing Institute (MDPI) June 1, 2024. [CrossRef]
- Wang, L.; Zhao, H.; Raman, I.; Yan, M.; Chen, Q.; Li, Q.Z. Peripheral Blood Mononuclear Cell Gene Expression in Chronic Obstructive Pulmonary Disease: MiRNA and MRNA Regulation. J Inflamm Res 2022, 15, 2167–2180. [Google Scholar] [CrossRef]
- Hu, J.; Wang, W.; Lu, Q.; Du, L.; Qin, T. Differential Expression of MiRNAs in Bronchoalveolar Lavage Fluid and Plasma from Patients with Chronic Obstructive Pulmonary Disease. Medicine (United States) 2022, 101. [Google Scholar] [CrossRef]
- Zhang, J.; Xu, Z.; Kong, L.; Gao, H.; Zhang, Y.; Zheng, Y.; Wan, Y. Mirna-486-5p Promotes Copd Progression by Targeting Hat1 to Regulate the Tlr4-Triggered Inflammatory Response of Alveolar Macrophages. International Journal of COPD 2020, 15, 2991–3001. [Google Scholar] [CrossRef]
- Shi, X. feng; He, X.; Sun, Z. rui; Wang, J. xiang; Gu, Y. hai; Xie, Y. bang; Duo, J. Different Expression of Circulating MicroRNA Profile and Plasma SP-D in Tibetan COPD Patients. Sci Rep 2022, 12. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Zhang, L.; Wang, Q. MicroRNA-221-3p Alleviates Cell Apoptosis and Inflammatory Response by Targeting Cyclin Dependent Kinase Inhibitor 1B in Chronic Obstructive Pulmonary Disease. Bioengineered 2021, 12, 5705–5715. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Lu, H.; Song, G. MiR-221-3p and MiR-92a-3p Enhances Smoking-Induced Inflammation in COPD. J Clin Lab Anal 2021, 35. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.; Huang, K.; Xu, X.; Duan, R.; Yu, T.; Chu, X.; Chen, C.; Li, B.; Yang, T. MiR-23a-5p Alleviates Chronic Obstructive Pulmonary Disease through Targeted Regulation of RAGE-ROS Pathway. Respir Res 2024, 25. [Google Scholar] [CrossRef]
- Kim, R.Y.; Sunkara, K.P.; Bracke, K.R.; Jarnicki, A.G.; Donovan, C.; Hsu, A.C.; Ieni, A.; Beckett, E.L.; Galvão, I.; Wijnant, S.; et al. A MicroRNA-21-Mediated SATB1/S100A9/NF-κB Axis Promotes Chronic Obstructive Pulmonary Disease Pathogenesis; 2021; Vol. 13. https://www.science.org.
- De Smet, E.G.; Van Eeckhoutte, H.P.; Avila Cobos, F.; Blomme, E.; Verhamme, F.M.; Provoost, S.; Verleden, S.E.; Venken, K.; Maes, T.; Joos, G.F.; et al. The Role of MiR-155 in Cigarette Smoke-Induced Pulmonary Inflammation and COPD. Mucosal Immunol 2020, 13, 423–436. [Google Scholar] [CrossRef]
- Zhu, Y.; Han, Y.; Almuntashiri, S.; Dutta, S.; Wang, X.; Owen, C.A.; Zhang, D. Dysregulation of MiR-103a Mediates Cigarette Smoking–Induced Lipid-Laden Macrophage Formation. Am J Respir Cell Mol Biol 2022, 67, 695–707. [Google Scholar] [CrossRef]
- Arif, R.; Pandey, A.; Zhao, Y.; Arsenault-Mehta, K.; Khoujah, D.; Mehta, S. Treatment of Pulmonary Hypertension Associated with COPD: A Systematic Review. ERJ Open Research, European Respiratory Society January 1, 2022. [CrossRef]
- Yang, Z.; Li, P.; Yuan, Q.; Wang, X.; Ma, H.H.; Zhuan, B. Inhibition of MiR-4640-5p Alleviates Pulmonary Hypertension in Chronic Obstructive Pulmonary Disease Patients by Regulating Nitric Oxide Synthase 1. Respir Res 2023, 24. [Google Scholar] [CrossRef]
- Singh, P.; Li, F.J.; Dsouza, K.; Stephens, C.T.; Zheng, H.; Kumar, A.; Dransfield, M.T.; Antony, V.B. Low Dose Cadmium Exposure Regulates MiR-381–ANO1 Interaction in Airway Epithelial Cells. Sci Rep 2024, 14. [Google Scholar] [CrossRef]
- Zheng, L.; Jiang, Y.L.; Fei, J.; Cao, P.; Zhang, C.; Xie, G.F.; Wang, L.X.; Cao, W.; Fu, L.; Zhao, H. Circulatory Cadmium Positively Correlates with Epithelial-Mesenchymal Transition in Patients with Chronic Obstructive Pulmonary Disease. Ecotoxicol Environ Saf 2021, 215. [Google Scholar] [CrossRef]
- Tasena, H.; Faiz, A.; Timens, W.; Noordhoek, J.; Hylkema, M.N.; Gosens, R.; Hiemstra, P.S.; Spira, A.; Postma, D.S.; Tew, G.W.; et al. MicroRNA–MRNA Regulatory Networks Underlying Chronic Mucus Hypersecretion in COPD. European Respiratory Journal 2018, 52. [Google Scholar] [CrossRef]
- Tasena, H.; Timens, W.; Van Den Berge, M.; Van Broekhuizen, J.; Kennedy, B.K.; Hylkema, M.N.; Brandsma, C.A.; Heijink, I.H. MicroRNAs Associated with Chronic Mucus Hypersecretion in COPD Are Involved in Fibroblast–Epithelium Crosstalk. Cells 2022, 11. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liu, X.; Zheng, Y.; Du, F.; He, G. MiR-548ar-3p Increases Cigarette Smoke Extract-Induced Chronic Obstructive Pulmonary Disease (COPD) Injury through Solute Carrier Family 17 Member 9 (SLC17A9). Arch Biol Sci 2022, 74, 97–105. [Google Scholar] [CrossRef]
- Nadi, E.; Geramirad, G.; Kahramfar, Z.; Rasouli-Saravani, A.; Solgi, G. Peripheral Blood Expressions of MicroRNA-146a and MicroRNA-218 in Chronic Obstructive Pulmonary Disease with/without Cigarette Smoke Exposure. Iran J Allergy Asthma Immunol 2022, 21, 399–406. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Tang, S.; Zhou, Z.; Wei, H.; Yang, W. Plasma MiR-150-5p as a Biomarker for Chronic Obstructive Pulmonary Disease. International Journal of COPD 2023, 18, 399–406. [Google Scholar] [CrossRef]
- Zhang, X.; Shi, Q.; Xiong, L.; Shi, S.; Li, Y.; Wang, Y.; Zhang, M. Clinical Relevance of MiR-423-5p Levels in Chronic Obstructive Pulmonary Disease Patients. Clinics 2022, 77. [Google Scholar] [CrossRef]
- Tao, S.; Liao, C.; Wang, Y.; Xu, D.; Li, Z.; Li, F. Differential MiRNA Profiling Reveals MiR-4433a-5p as a Key Regulator of Chronic Obstructive Pulmonary Disease Progression via PIK3R2- Mediated Phenotypic Modulation. Comb Chem High Throughput Screen 2023, 27, 2323–2334. [Google Scholar] [CrossRef]
- Cazorla-Rivero, S.; Mura-Escorche, G.; Gonzalvo-Hernández, F.; Mayato, D.; Córdoba-Lanús, E.; Casanova, C. Circulating Mir-1246 in the Progression of Chronic Obstructive Pulmonary Disease (Copd) in Patients from the Bode Cohort. International Journal of COPD 2020, 15, 2727–2737. [Google Scholar] [CrossRef]
- Wang, C.; Feng, D.; Dong, S.; He, R.; Fan, B. Dysregulated Circulating MicroRNA-126 in Chronic Obstructive Pulmonary Disease: Linkage with Acute Exacerbation Risk, Severity Degree, and Inflammatory Cytokines. J Clin Lab Anal 2022, 36. [Google Scholar] [CrossRef]
- Huang, H.; Wu, F.; Yang, J.; Li, H.; Cai, M.; Shan, K.; Shi, S. Increased Plasma Level of MiR-210 as a Potential Diagnostic Marker for Chronic Obstructive Pulmonary Disease Induced Pulmonary Hypertension. Clin Lab 2020, 66, 971–977. [Google Scholar] [CrossRef]
- Burke, H.; Cellura, D.; Freeman, A.; Hicks, A.; Ostridge, K.; Watson, A.; Williams, N.P.; Spalluto, C.M.; Staples, K.J.; Wilkinson, T.M.A. Pulmonary EV MiRNA Profiles Identify Disease and Distinct Inflammatory Endotypes in COPD. Front Med (Lausanne) 2022, 9. [Google Scholar] [CrossRef]
- Wang, F.; Yang, B.; Qiao, J.; Bai, L.; Li, Z.; Sun, W.; Liu, Q.; Yang, S.; Cui, L. Serum Exosomal MicroRNA-1258 May as a Novel Biomarker for the Diagnosis of Acute Exacerbations of Chronic Obstructive Pulmonary Disease. Sci Rep 2023, 13. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.F.; Liu, Y.; Qu, P.P.; Tang, Y.; Li, B.B.; Cheng, G.L. Mir-361-5p/Abca1 and Mir-196-5p/Arhgef12 Axis Involved in γ-Sitosterol Inducing Dual Anti-Proliferative Effects on Bronchial Epithelial Cells of Chronic Obstructive Pulmonary Disease. International Journal of COPD 2021, 16, 2741–2753. [Google Scholar] [CrossRef]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A CeRNA Hypothesis: The Rosetta Stone of a Hidden RNA Language? Cell, Elsevier B.V. August 5, 2011, pp 353–358. [CrossRef]
- Li, B.; Zhang, J.; Dong, H.; Feng, X.; Yu, L.; Zhu, J.; Zhang, J. Systematic Analysis of Various RNA Transcripts and Construction of Biological Regulatory Networks at the Post-Transcriptional Level for Chronic Obstructive Pulmonary Disease. J Transl Med 2023, 21. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Dong, H.; Li, B.; Yu, L.; Zhu, J.; Lou, C.; Zhang, J. Integrative Analysis of the Expression Profiles of Whole Coding and Non-Coding RNA Transcriptomes and Construction of the Competing Endogenous RNA Networks for Chronic Obstructive Pulmonary Disease. Front Genet 2023, 14. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





