1. Introduction
Neurological diseases, including Alzheimer’s disease (AD), Parkinson’s disease (PD), amyotrophic lateral sclerosis (ALS), and multiple sclerosis (MS), pose substantial health and economic challenges worldwide. The increasing prevalence of these disorders, combined with their complex pathophysiology, necessitates the development of reliable molecular diagnostics [
1]. Traditional diagnostic methods often rely on clinical symptoms and imaging findings, which may not be sufficient for early disease detection or prognosis [
2]. Consequently, molecular biomarkers have emerged as powerful tools that provide insights into disease mechanisms, enhance diagnostic accuracy, and guide therapeutic interventions [
3].
Advancements in omics technologies, including genomics, transcriptomics, epigenomics, proteomics, and metabolomics, have led to the identification of novel biomarkers for neurological diseases [
4]. Genomic studies have uncovered key genetic risk factors and mutations associated with disease susceptibility, while transcriptomic analyses have highlighted dysregulated gene expression patterns [
5]. Epigenetic modifications, such as DNA methylation and histone modifications, provide insights into disease progression and gene regulation [
6]. Furthermore, proteomic and metabolomic approaches have refined our understanding of neurodegenerative and neuroinflammatory processes, identifying disease-specific proteins and metabolic alterations that may serve as potential biomarkers for early detection and prognosis [
7].
Recent technological developments, such as liquid biopsy and multi-omics data integration, have facilitated non-invasive biomarker detection and improved disease classification [
8]. Liquid biopsy techniques allow for the analysis of circulating nucleic acids, proteins, and extracellular vesicles, providing a minimally invasive approach to disease monitoring [
9]. Multi-omics integration, combining data from different molecular levels, has uncovered complex interactions underlying neurological conditions, paving the way for precision medicine strategies [
10].
This review explores the latest progress in molecular biomarker discovery, emphasizing its clinical applications in diagnosing and monitoring neurological diseases. By addressing the current challenges and future directions in the field, we aim to highlight the translational potential of molecular diagnostics in improving patient outcomes [
11].
2. Results
2.1. Genomic and Transcriptomic Biomarkers
Genetic mutations, single nucleotide polymorphisms (SNPs), and transcriptomic alterations have been implicated in various neurological diseases, emphasizing the role of genetic predisposition and gene expression dysregulation in neurodegeneration and neuroinflammation. Advances in next-generation sequencing (NGS) technologies have significantly improved our ability to detect rare and common genetic variations associated with neurological disorders. These technologies, including whole-exome sequencing (WES) and whole-genome sequencing (WGS), have facilitated the identification of pathogenic mutations in genes such as APP, PSEN1, and PSEN2 in Alzheimer’s disease (AD), as well as LRRK2, SNCA, and PINK1 in Parkinson’s disease (PD) [
12,
13].
Genome-wide association studies (GWAS) have identified numerous risk loci linked to AD, PD, and multiple sclerosis (MS), among others [
14]. For example, APOE ε4 remains a key genetic risk factor for AD, influencing amyloid-beta aggregation, tau pathology, and neuroinflammation through mechanisms involving lipid metabolism and synaptic dysfunction [
15]. Variants in TREM2 have also been associated with increased AD risk, affecting microglial response and neuroimmune regulation [
16]. In PD, LRRK2 mutations impact mitochondrial homeostasis and autophagy, while SNCA mutations contribute to alpha-synuclein aggregation, a hallmark of disease pathology [
17]. In MS, the HLA-DRB1 allele has been strongly associated with disease susceptibility, highlighting the role of immune system dysregulation in neuroinflammatory conditions [
18].
Beyond inherited genetic mutations, transcriptomic profiling has provided crucial insights into gene expression changes associated with neurological diseases [
19]. RNA sequencing (RNA-seq) studies have identified altered mRNA signatures in various neurodegenerative and neuroinflammatory disorders. Differential gene expression analysis has revealed upregulation of pro-inflammatory cytokines and downregulation of neuroprotective factors in AD and MS, supporting the role of immune activation and neuroinflammation in disease progression [
20]. Additionally, non-coding RNAs, including microRNAs (miRNAs) and long non-coding RNAs (lncRNAs), have emerged as critical regulators of neurodegenerative processes [
21]. miRNAs such as miR-155 and miR-146a have been implicated in neuroinflammation, while lncRNAs such as BACE1-AS modulate amyloid precursor protein (APP) processing and contribute to amyloid-beta plaque formation [
22].
The integration of multi-omics approaches, combining genomic, transcriptomic, and epigenomic data, has further enhanced our understanding of neurological disease mechanisms [
23]
Table 1. Recent studies have leveraged single-cell RNA sequencing (scRNA-seq) to uncover cell-type-specific transcriptional changes in brain tissues affected by neurodegenerative diseases. This high-resolution approach has provided new insights into the interactions between neurons, astrocytes, and microglia, revealing how genetic risk factors influence disease progression at the cellular level [
24].
2.2. Epigenomic and Proteomic Insights
Epigenetic modifications, including DNA methylation, histone acetylation, chromatin remodeling, and non-coding RNAs, play a critical role in neurological disease progression. These modifications influence gene expression without altering the DNA sequence, often in response to environmental factors, aging, and disease pathology. Aberrant DNA methylation has been observed in genes linked to neurodegeneration, including APP, PSEN1, and TREM2 in Alzheimer's disease (AD), as well as SNCA and LRRK2 in Parkinson's disease (PD) [
25,
26]. Specifically, hypermethylation of the APP promoter has been associated with reduced gene expression, potentially contributing to amyloid-beta (Aβ) dysregulation, while hypomethylation of PSEN1 may lead to increased amyloidogenic processing [
6]. In PD, aberrant methylation of SNCA influences α-synuclein aggregation, a hallmark of the disease [
27].
In addition to DNA methylation, histone modifications, such as acetylation and methylation, impact chromatin accessibility and gene transcription. Histone deacetylase (HDAC) dysregulation has been linked to cognitive decline in AD and synaptic dysfunction in PD [
28]. Pharmacological inhibition of HDACs has shown neuroprotective effects by enhancing memory and reducing tau pathology [
29]. Similarly, chromatin remodeling enzymes, including ATP-dependent chromatin remodelers, have been implicated in regulating neuronal plasticity and neuroinflammatory responses in multiple sclerosis (MS) [
30].
Beyond epigenetics, proteomic analyses have led to the discovery of cerebrospinal fluid (CSF) and blood-based biomarkers for neurodegenerative diseases. Advanced mass spectrometry and high-throughput proteomic techniques have enabled the identification of disease-specific proteins, facilitating early diagnosis and disease monitoring. Key CSF biomarkers include phosphorylated tau (p-tau), total tau (t-tau), amyloid-beta (Aβ42/Aβ40 ratio), and neurofilament light chain (NfL) [
7]. Elevated levels of p-tau and t-tau in AD correlate with tau aggregation and neuronal damage, while reduced CSF Aβ42 reflects amyloid deposition in the brain [
31].
In PD, proteomic studies have identified α-synuclein oligomers and DJ-1 protein levels as potential biomarkers [
10]. Additionally, NfL, a marker of axonal damage, is elevated in various neurodegenerative diseases, including AD, PD, and MS [
32]. Blood-based NfL measurements offer a promising non-invasive alternative for monitoring disease progression and therapeutic response [
33].
The integration of epigenomic and proteomic data into multi-omics analyses is enhancing biomarker discovery and precision medicine approaches. Combining epigenetic profiles with proteomic signatures may improve diagnostic accuracy, facilitate early disease detection, and guide personalized treatment strategies [
34]. Future research should focus on validating these biomarkers in large-scale longitudinal studies and exploring their potential as therapeutic targets.
2.3. Liquid Biopsy and Multi-Omics Integration
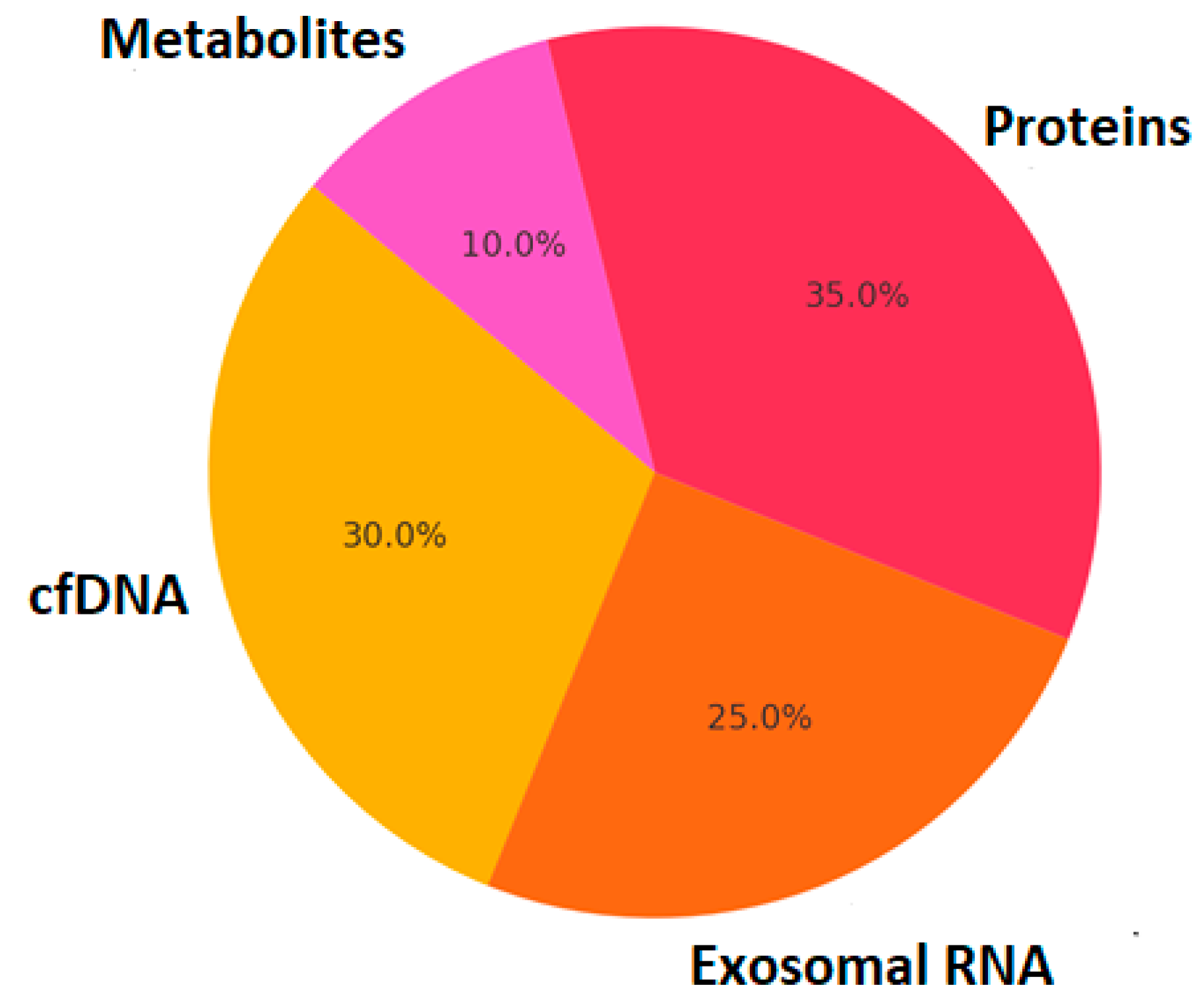
Liquid biopsy technologies have revolutionized the non-invasive detection of circulating biomarkers, offering new avenues for diagnosing and monitoring neurological diseases. Unlike traditional biopsies that require invasive sampling of brain tissue or cerebrospinal fluid (CSF), liquid biopsies analyze circulating molecular components in blood, plasma, saliva, or urine. These components include exosomal RNA, cell-free DNA (cfDNA), circulating tumor DNA (ctDNA), extracellular vesicles (EVs), proteins, and metabolites, providing a snapshot of disease pathophysiology in real time [
3].
Figure 1
One of the most promising aspects of liquid biopsy in neurology is its application in neurodegenerative disorders such as Alzheimer's disease (AD), Parkinson’s disease (PD), amyotrophic lateral sclerosis (ALS), and multiple sclerosis (MS). In AD, cell-free mitochondrial DNA (cf-mtDNA) levels have been correlated with disease progression, reflecting neuronal damage [
35]. Additionally, exosomal tau and amyloid-beta peptides in blood have been proposed as alternative biomarkers for early AD diagnosis [
36]. In PD, exosomal α-synuclein in plasma distinguishes PD patients from healthy individuals, serving as a potential diagnostic marker [
8].
Moreover, extracellular vesicles (EVs), which carry microRNAs (miRNAs), proteins, and lipids, play a pivotal role in disease pathogenesis and biomarker discovery. For example, miR-21 and miR-155 levels in circulating exosomes are elevated in MS patients, highlighting their role in neuroinflammation [
37]. Similarly, liquid biopsy analysis of circulating inflammatory cytokines and neurofilament light chain (NfL) provides valuable insights into disease severity and treatment response in neurodegenerative and neuroinflammatory conditions [
38].
Beyond individual biomarkers, multi-omics integration has emerged as a powerful approach for enhancing biomarker discovery and disease classification. By combining genomic, transcriptomic, proteomic, epigenomic, and metabolomic data, researchers can identify complex interactions that underlie neurological diseases. For example, in AD, integrating GWAS data with transcriptomic and proteomic datasets has led to the identification of novel risk genes and pathways involved in neurodegeneration [
33].
Multi-omics analyses have also been applied in drug development and precision medicine. Integrating metabolomics with proteomics has enabled the identification of metabolic disturbances in PD, providing potential therapeutic targets [
10]. Similarly, in MS, combining DNA methylation profiles with proteomic signatures has improved our understanding of immune dysregulation and personalized treatment approaches [
39].
The application of artificial intelligence (AI) and machine learning in multi-omics research has further accelerated biomarker discovery. AI-driven algorithms can process large-scale datasets to uncover hidden patterns and predict disease risk, progression, and therapeutic responses [
40]. Future research should focus on standardizing liquid biopsy protocols, validating biomarkers in large-scale clinical trials, and integrating multi-omics data into clinical practice to advance precision neurology.
Additionally, regulatory approval and standardization remain critical challenges in translating biomarkers into clinical practice. While some biomarkers, such as CSF-based tau and Aβ42/Aβ40 ratio, have received regulatory validation (e.g., FDA-approved assays for Alzheimer’s disease diagnosis), others require further validation through multicenter clinical trials. The European Medicines Agency (EMA) and the U.S. Food and Drug Administration (FDA) emphasize the importance of reproducibility and large-scale cohort validation for biomarker approval. [
41]
Future research should prioritize:
- -
Large-scale multi-omics integration with well-characterized patient cohorts.
- -
Standardization of biomarker assessment protocols across different populations.
- -
AI-driven approaches to analyze high-dimensional biomarker data, improving diagnostic accuracy.
- -
Translation of biomarker panels into clinical decision-making tools through regulatory harmonization. [
5]
3. Discussion
The identification of molecular biomarkers has significantly improved our ability to diagnose, prognosticate, and monitor neurological diseases, allowing for earlier detection and more precise therapeutic interventions. The advent of multi-omics technologies has revolutionized the field by integrating genomic, transcriptomic, proteomic, metabolomic, and epigenomic data, leading to a more comprehensive understanding of disease mechanisms [
4].
One of the key challenges in biomarker research is ensuring clinical translation and standardization. While several promising biomarkers have been identified for Alzheimer’s disease (AD), Parkinson’s disease (PD), multiple sclerosis (MS), and amyotrophic lateral sclerosis (ALS), many remain in the experimental phase due to limited reproducibility, small sample sizes, and lack of validation in diverse populations [
3]. Large-scale, longitudinal studies with standardized protocols are essential for biomarker validation [
33].
Furthermore, liquid biopsy and non-invasive biomarker detection are emerging as critical components of precision neurology. Circulating biomarkers such as cell-free DNA (cfDNA), exosomal RNA, neurofilament light chain (NfL), and inflammatory cytokines have shown promise in tracking disease progression and response to treatment [
8]. However, the sensitivity and specificity of these biomarkers vary, necessitating multi-omics integration for improved diagnostic accuracy [
34].
Another transformative advancement is the application of artificial intelligence (AI) and machine learning (ML) algorithms in biomarker discovery and disease classification. AI-driven approaches can analyze vast amounts of multi-omics and imaging data to identify novel biomarker signatures, improve diagnostic predictions, and optimize personalized treatment strategies [
32]. In AD, for example, deep learning models have been used to integrate neuroimaging and molecular biomarker data, improving early diagnosis and differentiation from other dementias [
5]. In PD, AI-based analyses of blood-based biomarkers and voice patterns have enhanced early disease detection [
42].
Despite these advancements, several challenges remain:
Standardization and Validation: Biomarkers must undergo rigorous validation across independent cohorts before being implemented in clinical practice [
9].
Inter-individual Variability: Differences in genetics, lifestyle, and comorbidities influence biomarker expression, making it necessary to develop personalized reference ranges [
9].
Data Integration and Interpretation: Multi-omics datasets generate large volumes of information that require sophisticated computational tools and robust statistical frameworks to extract clinically relevant insights [
43].
Future research should focus on large-scale, multi-center collaborations to validate promising biomarkers, enhance AI-driven analytics, and integrate biomarker-based precision medicine approaches into routine neurological care. The ultimate goal is to develop non-invasive, cost-effective, and highly specific biomarkers that enable earlier intervention and improved patient outcomes [
11], [
44].
4. Materials and Methods
This review was conducted through a comprehensive literature search using the PubMed, Scopus, and Web of Science databases. The search strategy included the following keywords:
"molecular biomarkers"
"neurological diseases"
"genomics"
"proteomics"
"liquid biopsy"
"precision medicine"
Studies published between 2010 and 2024 were considered for inclusion. Articles were selected based on their relevance, methodological rigor, and impact in the field. Priority was given to systematic reviews, meta-analyses, clinical trials, and large-scale multi-omics studies. Exclusion criteria included small-scale studies with low statistical power, studies lacking appropriate validation cohorts, and those published in non-peer-reviewed sources.
To ensure the reliability and reproducibility of the findings, the literature review focused on studies that met the following criteria:
Genomic biomarkers: Studies investigating single nucleotide polymorphisms (SNPs), gene mutations, and genome-wide association studies (GWAS) in neurological diseases.
Transcriptomic biomarkers: RNA sequencing (RNA-seq) and differential gene expression studies.
Epigenomic biomarkers: DNA methylation, histone modifications, and chromatin remodeling studies.
Proteomic biomarkers: Studies identifying cerebrospinal fluid (CSF) and blood-based protein biomarkers.
Liquid biopsy: Analysis of exosomal RNA, circulating cfDNA, and extracellular vesicles as diagnostic tools.
Multi-omics approaches: Integration of genomics, transcriptomics, proteomics, and metabolomics in biomarker discovery.
AI and machine learning in biomarker analysis: Studies utilizing AI-driven approaches for biomarker identification and clinical applications.
Data extraction and synthesis followed PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analyses) guidelines, ensuring a transparent and reproducible selection process. Future systematic reviews may further benefit from network meta-analysis and real-world evidence from biobank datasets and electronic health records (EHRs).
5. Conclusions
The future of neurological disease diagnostics lies in integrating AI-enhanced multi-omics, liquid biopsy techniques, and standardized regulatory frameworks. By leveraging these advancements, precision neurology will facilitate earlier disease detection, personalized treatment strategies, and improved patient outcomes. Collaborative efforts between researchers, clinicians, and regulatory bodies are essential to bridge the gap between biomarker discovery and clinical implementation.
Molecular biomarkers are revolutionizing the landscape of neurological disease diagnostics and prognostics, providing deeper insights into disease mechanisms, early detection, and personalized therapeutic strategies. The integration of multi-omics approaches, incorporating genomics, transcriptomics, proteomics, metabolomics, and epigenomics, has enhanced our ability to identify novel disease-specific markers. This allows for greater diagnostic accuracy, improved patient stratification, and refined treatment response predictions.
One of the most transformative advancements in biomarker research is the use of multi-omics integration to uncover complex molecular interactions. Genome-wide association studies (GWAS) and transcriptomic profiling have helped elucidate the genetic basis of neurological diseases, while proteomic and metabolomic analyses have identified circulating biomarkers relevant for early disease detection. For instance, neurofilament light chain (NfL) has emerged as a robust biomarker of neurodegeneration, providing valuable insights into disease progression in Alzheimer’s disease (AD), Parkinson’s disease (PD), multiple sclerosis (MS), and amyotrophic lateral sclerosis (ALS).
In parallel, liquid biopsy techniques are reshaping non-invasive biomarker discovery, allowing for the detection of cell-free DNA (cfDNA), exosomal RNA, microRNAs (miRNAs), and extracellular vehicles (EVs) in various biofluids, including blood, cerebrospinal fluid (CSF), and plasma. These circulating biomarkers offer the advantage of real-time disease monitoring and have been successfully utilized in the early detection of AD, PD, and MS. For example, exosomal tau and amyloid-beta peptides have shown diagnostic potential for AD, while plasma α-synuclein and DJ-1 protein levels serve as promising biomarkers for PD.
Beyond individual biomarkers, the integration of artificial intelligence (AI) and machine learning (ML) algorithms has significantly enhanced biomarker discovery. AI-driven computational tools can analyze high-dimensional multi-omics datasets, enabling early disease prediction, biomarker validation, and precision medicine applications. In AD, deep learning models combining genetic, transcriptomic, and imaging data have been shown to improve diagnostic accuracy, while in PD, AI-based voice analysis has facilitated early disease detection.
Despite these advancements, several challenges remain:
Standardization and Validation: Large-scale multi-center studies are needed to validate molecular biomarkers across diverse populations before clinical implementation.
Inter-Individual Variability: Biomarker expression varies based on genetics, environment, and comorbidities, highlighting the need for personalized reference ranges.
Data Integration and Interpretation: The complexity of multi-omics datasets requires advanced bioinformatics tools and robust statistical models to extract meaningful insights.
Future research should focus on optimizing biomarker panels, refining AI-driven analytics, and establishing clinical guidelines for biomarker-based diagnostics. The convergence of multi-omics technologies, liquid biopsy innovations, and AI-driven analytics is transforming precision neurology, paving the way for earlier disease detection, improved monitoring, and more effective, individualized treatments.
Table 2
Author Contributions
Conceptualization, A.M.; methodology, A.M.; software, A.M. and K.M..; validation, A.M..; formal analysis, A.M.; investigation, A.M. and A.I.; resources, K.M.; A.I..; data curation, A.M..; writing—original draft preparation, A.M..; writing—review and editing, A.M. and A.I.; visualization, K.M.; supervision, A.M. and C.S.; project administration, A.M.; funding acquisition, A.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| MS |
multiple sclerosis |
| ALS |
amyotrophic lateral sclerosis |
| AD |
Alzheimer’s disease |
| PD |
Parkinson’s disease |
References
- GBD 2021 Nervous System Disorders Collaborators. Global, regional, and national burden of disorders affecting the nervous system, 1990-2021: a systematic analysis for the Global Burden of Disease Study 2021. Lancet Neurol. 2024 Apr;23(4):344-381. Erratum in: Lancet Neurol. 2024 May;23(5): e9. doi: 10.1016/S1474-4422(24)00114-5. Erratum in: Lancet Neurol. 2024 Jul;23(7): e11. doi: 10.1016/S1474-4422(24)00231-X. [CrossRef] [PubMed] [PubMed Central]
- Jack CR Jr, Bennett DA, Blennow K, Carrillo MC, Dunn B, Haeberlein SB, Holtzman DM, Jagust W, Jessen F, Karlawish J; Contributors. NIA-AA Research Framework: Toward a biological definition of Alzheimer's disease. Alzheimers Dement. 2018 Apr;14(4):535-562. [CrossRef] [PubMed] [PubMed Central]
- Hampel H, O'Bryant SE, Molinuevo JL, Zetterberg H, Masters CL, Lista S, Kiddle SJ, Batrla R, Blennow K. Blood-based biomarkers for Alzheimer disease: mapping the road to the clinic. Nat Rev Neurol. 2018 Nov;14(11):639-652. [CrossRef] [PubMed] [PubMed Central]
- Hasin Y, Seldin M, Lusis A. Multi-omics approaches to disease. Genome Biol. 2017 May 5;18(1):83. [CrossRef] [PubMed] [PubMed Central]
- Zhang S.W., Xu J.Y., Zhang T. DGMP: identifying cancer driver genes by jointing DGCN and MLP from multiomics genomic data. Genomics Proteomics Bioinformatics. 2022;20:928–938. [CrossRef]
- De Jager PL, Srivastava G, Lunnon K, Burgess J, Schalkwyk LC, Yu L, Eaton ML, Keenan BT, Ernst J, McCabe C. Alzheimer's disease: early alterations in brain DNA methylation at ANK1, BIN1, RHBDF2 and other loci. Nat Neurosci. 2014 Sep;17(9):1156-63. [CrossRef] [PubMed] [PubMed Central]
- Olsson B, Lautner R, Andreasson U, Öhrfelt A, Portelius E, Bjerke M, Hölttä M, Rosén C, Olsson C, Strobel G. CSF and blood biomarkers for the diagnosis of Alzheimer's disease: a systematic review and meta-analysis. Lancet Neurol. 2016 Jun;15(7):673-684. [CrossRef] [PubMed]
- Gulisano W, Maugeri D, Baltrons MA, Fà M, Amato A, Palmeri A, D'Adamio L, Grassi C, Devanand DP, Honig LS. Role of Amyloid-β and Tau Proteins in Alzheimer's Disease: Confuting the Amyloid Cascade. J Alzheimers Dis. 2018;64(s1): S611-S631. Erratum in: J Alzheimers Dis. 2019;68(1):415. doi: 10.3233/JAD-189015. [CrossRef] [PubMed] [PubMed Central]
- Teunissen CE, Verberk IMW, Thijssen EH, Vermunt L, Hansson O, Zetterberg H, van der Flier WM, Mielke MM, Del Campo M. Blood-based biomarkers for Alzheimer's disease: towards clinical implementation. Lancet Neurol. 2022 Jan;21(1):66-77. [CrossRef] [PubMed]
- Kodam P, Sai Swaroop R, Pradhan SS, Sivaramakrishnan V, Vadrevu R. Integrated multi-omics analysis of Alzheimer's disease shows molecular signatures associated with disease progression and potential therapeutic targets. Sci Rep. 2023 Mar 6;13(1):3695. [CrossRef] [PubMed] [PubMed Central]
- Hampel H, Vergallo A, Afshar M, Akman-Anderson L, Arenas J, Benda N, Batrla R, Broich K, Caraci F, Cuello AC. Blood-based systems biology biomarkers for next-generation clinical trials in Alzheimer's diseases. Dialogues Clin Neurosci. 2019;21(2):177-191. [CrossRef] [PubMed] [PubMed Central]
- Hardy J. The genetic causes of neurodegenerative diseases. J Alzheimers Dis. 2001 Feb;3(1):109-116. [CrossRef] [PubMed]
- Van der Brug MP, Singleton A, Gasser T. Parkinson's disease: FROM HUMAN GENETICS TO CLINICAL TRIALS. SCIENCE TRANSLATIONAL MEDICINE 2015;7(305) p.305ps20 . [CrossRef]
- Jansen IE, Savage JE, Watanabe K, Bryois J, Williams DM, Steinberg S, Sealock J, Karlsson IK, Hägg S, Athanasiu L. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer's disease risk. Nat Genet. 2019 Mar;51(3):404-413. Erratum in: Nat Genet. 2020 Mar;52(3):354. doi: 10.1038/s41588-019-0573-x. [CrossRef] [PubMed] [PubMed Central]
- Mahley RW. Apolipoprotein E: from cardiovascular disease to neurodegenerative disorders. J Mol Med (Berl). 2016 Jul;94(7):739-46. [CrossRef] [PubMed] [PubMed Central]
- Ulrich JD, Ulland TK, Colonna M, Holtzman DM. Elucidating the Role of TREM2 in Alzheimer's Disease. Neuron. 2017 Apr 19;94(2):237-248. [CrossRef] [PubMed]
- Brás J, Guerreiro R, Hardy J. SnapShot: Genetics of Parkinson's disease. Cell. 2015 Jan 29;160(3):570-570.e1. [CrossRef] [PubMed]
- International Multiple Sclerosis Genetics Consortium. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011 Aug 10;476(7359):214-9. [CrossRef] [PubMed] [PubMed Central]
- Zhang B, Gaiteri C, Bodea LG, Wang Z, McElwee J, Podtelezhnikov AA, Zhang C, Xie T, Tran L, Dobrin R. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer's disease. Cell. 2013 Apr 25;153(3):707-20. [CrossRef] [PubMed] [PubMed Central]
- Prinz M, Priller J. The role of peripheral immune cells in the CNS in steady state and disease. Nat Neurosci. 2017 Feb;20(2):136-144. [CrossRef] [PubMed]
- Guo C, Jeong HH, Hsieh YC, Klein HU, Bennett DA, De Jager PL, Liu Z, Shulman JM. Tau Activates Transposable Elements in Alzheimer's Disease. Cell Rep. 2018 Jun 5;23(10):2874-2880. [CrossRef] [PubMed] [PubMed Central]
- Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE, St Laurent G 3rd, Kenny PJ, Wahlestedt C. Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of beta-secretase. Nat Med. 2008 Jul;14(7):723-30. [CrossRef] [PubMed] [PubMed Central]
- Baranzini SE, Oksenberg JR. The Genetics of Multiple Sclerosis: From 0 to 200 in 50 Years. Trends Genet. 2017 Dec;33(12):960-970. [CrossRef] [PubMed] [PubMed Central]
- Habib N, McCabe C, Medina S, Varshavsky M, Kitsberg D, Dvir-Szternfeld R, Green G, Dionne D, Nguyen L, Marshall JL. Disease-associated astrocytes in Alzheimer's disease and aging. Nat Neurosci. 2020 Jun;23(6):701-706. [CrossRef] [PubMed] [PubMed Central]
- Lardenoije R, Iatrou A, Kenis G, Kompotis K, Steinbusch HW, Mastroeni D, Coleman P, Lemere CA, Hof PR, van den Hove DL. The epigenetics of aging and neurodegeneration. Prog Neurobiol. 2015 Aug;131:21-64. [CrossRef] [PubMed] [PubMed Central]
- Watson CT, Roussos P, Garg P, Ho DJ, Azam N, Katsel PL, Haroutunian V, Sharp AJ. Genome-wide DNA methylation profiling in the superior temporal gyrus reveals epigenetic signatures associated with Alzheimer's disease. Genome Med. 2016 Jan 19;8(1):5. [CrossRef] [PubMed] [PubMed Central]
- Masliah E, Dumaop W, Galasko D, Desplats P. Distinctive patterns of DNA methylation associated with Parkinson disease: identification of concordant epigenetic changes in brain and peripheral blood leukocytes. Epigenetics. 2013 Oct;8(10):1030-8. [CrossRef] [PubMed] [PubMed Central]
- Fischer A. Epigenetic memory: the Lamarckian brain. EMBO J. 2014 May 2;33(9):945-67. [CrossRef] [PubMed] [PubMed Central]
- Govindarajan N, Rao P, Burkhardt S, Sananbenesi F, Schlüter OM, Bradke F, Lu J, Fischer A. Reducing HDAC6 ameliorates cognitive deficits in a mouse model for Alzheimer's disease. EMBO Mol Med. 2013 Jan;5(1):52-63. [CrossRef] [PubMed] [PubMed Central]
- Huynh JL, Casaccia P. Epigenetic mechanisms in multiple sclerosis: implications for pathogenesis and treatment. Lancet Neurol. 2013 Feb;12(2):195-206. [CrossRef] [PubMed] [PubMed Central]
- Blennow K, Zetterberg H. Biomarkers for Alzheimer's disease: current status and prospects for the future. J Intern Med. 2018 Dec;284(6):643-663. [CrossRef] [PubMed]
- Mollenhauer B, Locascio JJ, Schulz-Schaeffer W, Sixel-Döring F, Trenkwalder C, Schlossmacher MG. α-Synuclein and tau concentrations in cerebrospinal fluid of patients presenting with parkinsonism: a cohort study. Lancet Neurol. 2011 Mar;10(3):230-40. Erratum in: Lancet Neurol. 2011 Apr;10(4):297. [CrossRef] [PubMed]
- Khalil M, Teunissen CE, Otto M, Piehl F, Sormani MP, Gattringer T, Barro C, Kappos L, Comabella M, Fazekas F. Neurofilaments as biomarkers in neurological disorders. Nat Rev Neurol. 2018 Oct;14(10):577-589. [CrossRef] [PubMed]
- Mattsson N, Andreasson U, Zetterberg H, Blennow K. Alzheimer’s Disease Neuroimaging Initiative. Association of Plasma Neurofilament Light With Neurodegeneration in Patients With Alzheimer Disease. JAMA Neurol. 2017 May 1;74(5):557-566. [CrossRef] [PubMed] [PubMed Central]
- Chen Y, He Y, Han J, Wei W, Chen F. Blood-brain barrier dysfunction and Alzheimer's disease: associations, pathogenic mechanisms, and therapeutic potential. Front Aging Neurosci. 2023 Nov 13;15:1258640. [CrossRef] [PubMed] [PubMed Central]
- Podlesniy P, Llorens F, Puigròs M, Serra N, Sepúlveda-Falla D, Schmidt C, Hermann P, Zerr I, Trullas R. Cerebrospinal Fluid Mitochondrial DNA in Rapid and Slow Progressive Forms of Alzheimer's Disease. Int J Mol Sci. 2020 Aug 31;21(17):6298. [CrossRef] [PubMed] [PubMed Central]
- Shi M, Liu C, Cook TJ, Bullock KM, Zhao Y, Ginghina C, Li Y, Aro P, Dator R, He C. Plasma exosomal α-synuclein is likely CNS-derived and increased in Parkinson's disease. Acta Neuropathol. 2014 Nov;128(5):639-650. [CrossRef] [PubMed] [PubMed Central]
- Selmaj I, Cichalewska M, Namiecinska M, Galazka G, Horzelski W, Selmaj KW, Mycko MP. Global exosome transcriptome profiling reveals biomarkers for multiple sclerosis. Ann Neurol. 2017 May;81(5):703-717. [CrossRef] [PubMed]
- Shao, Y., Le, W. Recent advances and perspectives of metabolomics-based investigations in Parkinson’s disease. Mol Neurodegeneration 14, 3 (2019). https://. [CrossRef]
- Baranzini SE, Mudge J, van Velkinburgh JC, Khankhanian P, Khrebtukova I, Miller NA, Zhang L, Farmer AD, Bell CJ, Kim RW. Genome, epigenome and RNA sequences of monozygotic twins discordant for multiple sclerosis. Nature. 2010 Apr 29;464(7293):1351-6. [CrossRef] [PubMed] [PubMed Central]
- Barthélemy NR, Salvadó G, Schindler SE, He Y, Janelidze S, Collij LE, Saef B, Henson RL, Chen CD, Gordon BA. Highly accurate blood test for Alzheimer's disease is similar or superior to clinical cerebrospinal fluid tests. Nat Med. 2024 Apr;30(4):1085-1095. [CrossRef] [PubMed] [PubMed Central]
- Antonio Suppa, Giovanni Costantini, Francesco Asci, Pietro Di Leo, Mohammad Sami Al-Wardat, Giulia Di Lazzaro, Simona Scalise, Antonio Pisani, Giovanni Saggio.Voice in Parkinson's Disease: A Machine Learning Study. Front. Neurol., 2022 Sec. Movement Disorders Volume 13 – 2022. https://. [CrossRef]
- Maher MC, Uricchio LH, Torgerson DG, Hernandez RD. Population genetics of rare variants and complex diseases. Hum Hered. 2012;74(3-4):118-28. [CrossRef] [PubMed] [PubMed Central]
- Robinson WH, Fontoura P, Lee BJ, de Vegvar HE, Tom J, Pedotti R, DiGennaro CD, Mitchell DJ, Fong D, Ho PP. Protein microarrays guide tolerizing DNA vaccine treatment of autoimmune encephalomyelitis. Nat Biotechnol. 2003 Sep;21(9):1033-9. [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).