Submitted:
03 February 2025
Posted:
05 February 2025
You are already at the latest version
Abstract
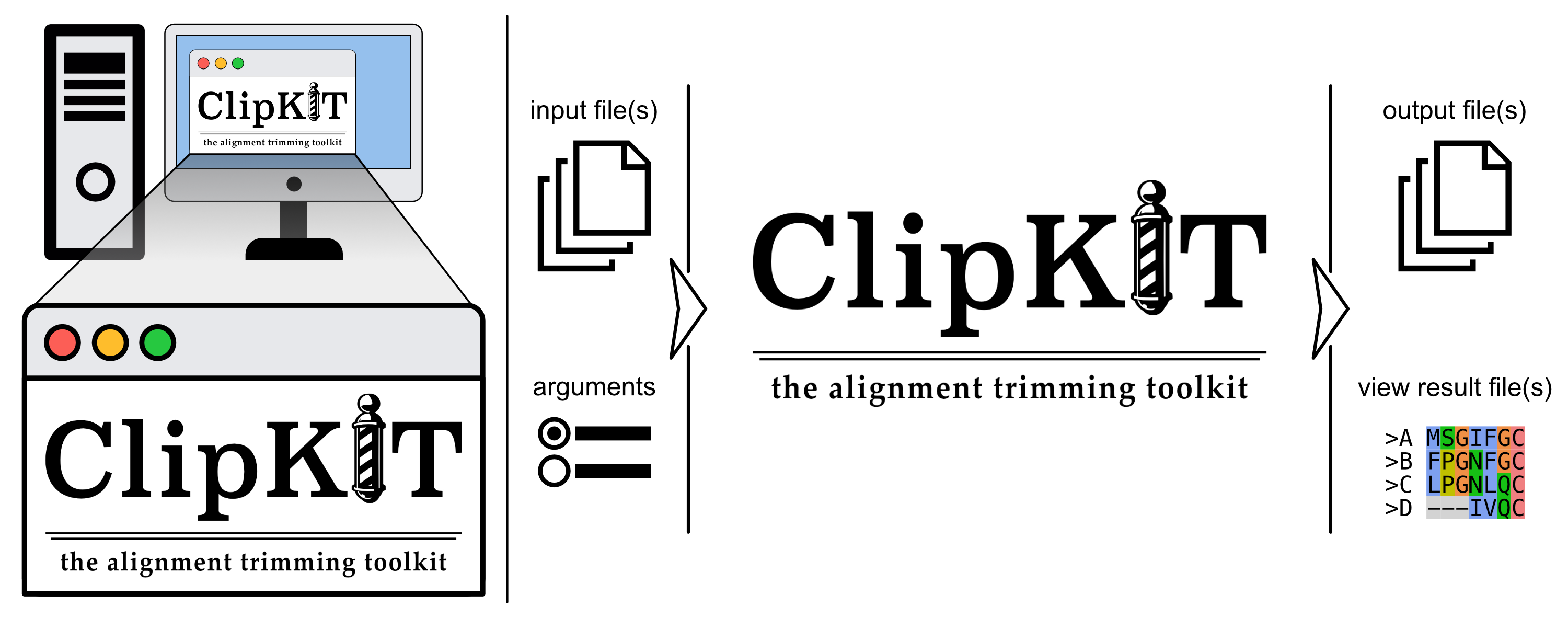
Keywords:
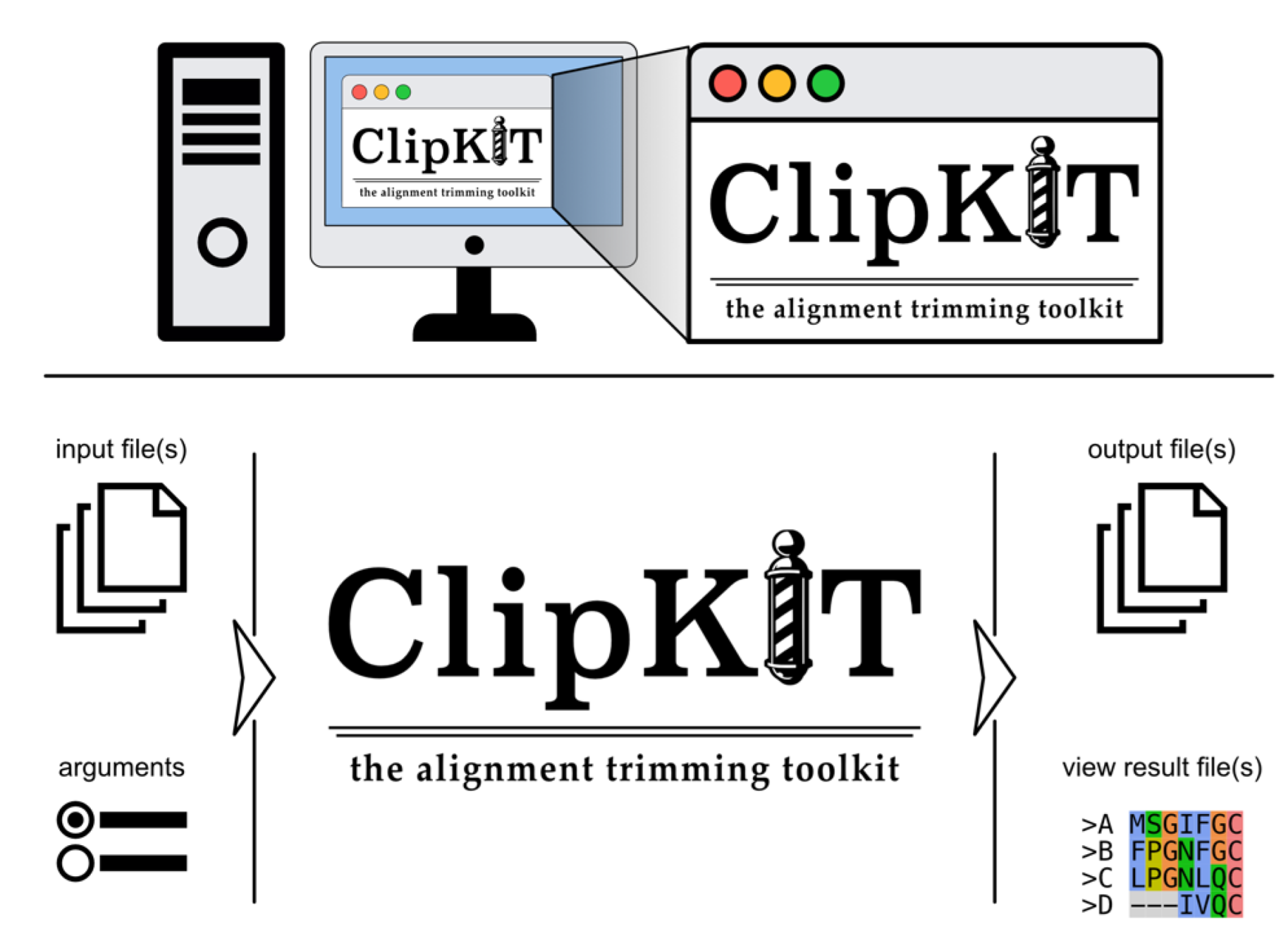
Introduction
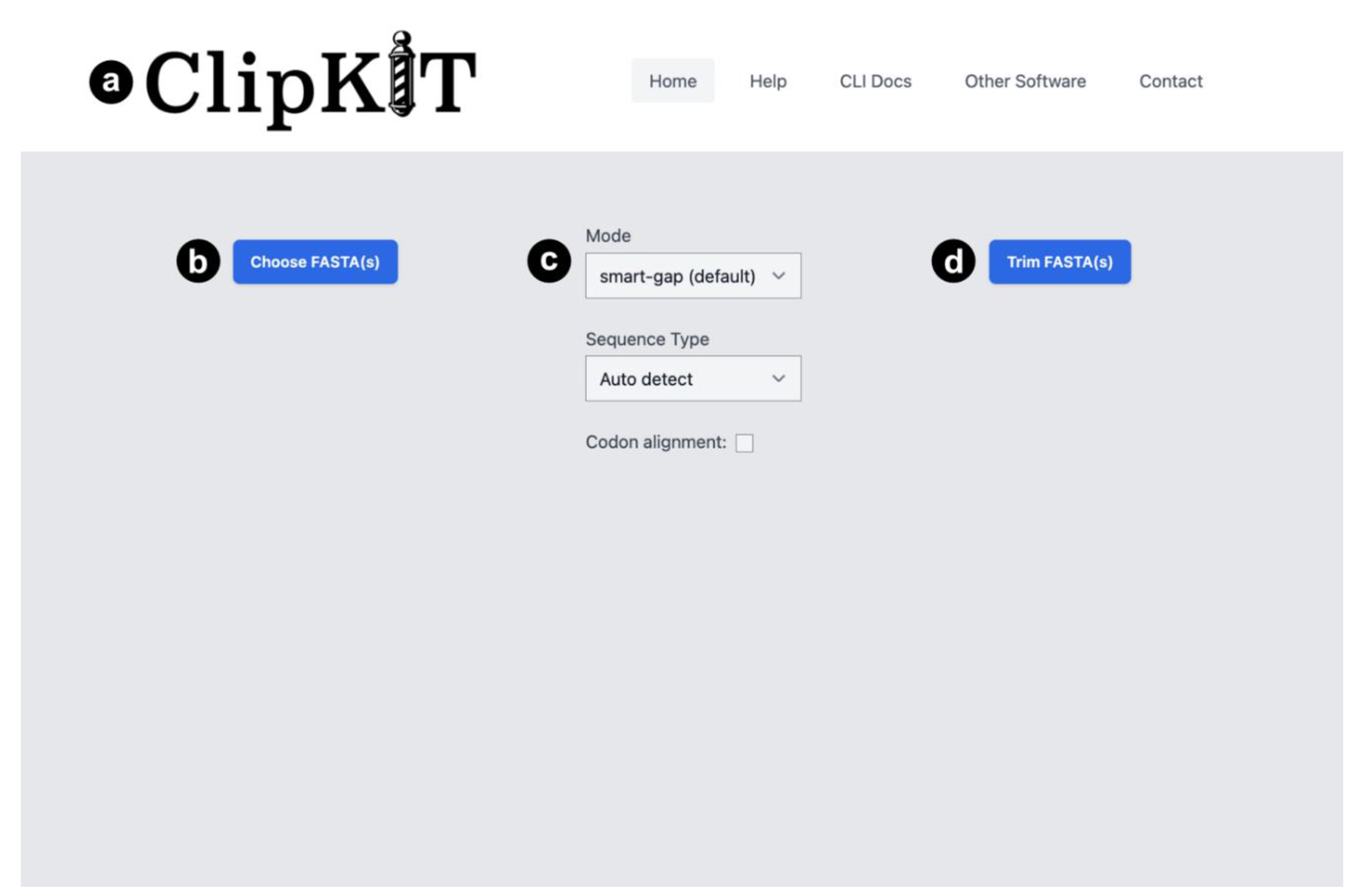
CLIPKIT WEB-APPLICATION
Input Data and Arguments
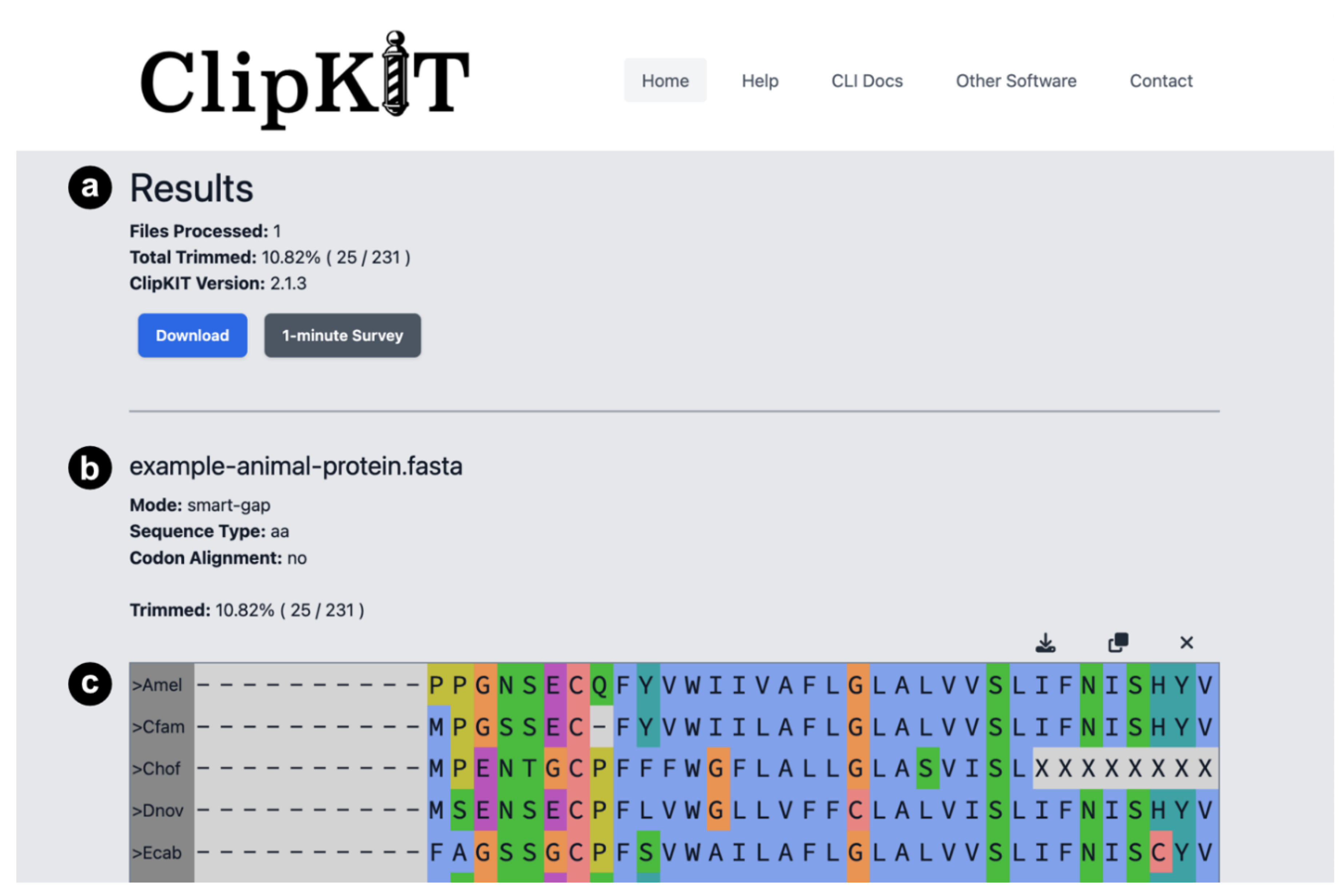
Results and Output Information
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Steenwyk, J.L.; Buida, T.J.; Li, Y.; Shen, X.-X.; Rokas, A. ClipKIT: A multiple sequence alignment trimming software for accurate phylogenomic inference. PLOS Biol. 2020, 18, e3001007. [Google Scholar] [CrossRef] [PubMed]
- Talavera, G.; Castresana, J. Improvement of Phylogenies after Removing Divergent and Ambiguously Aligned Blocks from Protein Sequence Alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef] [PubMed]
- Criscuolo, A.; Gribaldo, S. BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol. Biol. 2010, 10, 210. [Google Scholar] [CrossRef] [PubMed]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef] [PubMed]
- Dress, A.W.; Flamm, C.; Fritzsch, G.; Grünewald, S.; Kruspe, M.; Prohaska, S.J.; Stadler, P.F. Noisy: Identification of problematic columns in multiple sequence alignments. Algorithms Mol. Biol. 2008, 3, 7. [Google Scholar] [CrossRef] [PubMed]
- Dereeper, A.; Guignon, V.; Blanc, G.; Audic, S.; Buffet, S.; Chevenet, F.; Dufayard, J.-F.; Guindon, S.; Lefort, V.; Lescot, M.; et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008, 36, W465–W469. [Google Scholar] [CrossRef] [PubMed]
- Steenwyk, J.L.; Buida, T.J.; Labella, A.L.; Li, Y.; Shen, X.-X.; Rokas, A. PhyKIT: a broadly applicable UNIX shell toolkit for processing and analyzing phylogenomic data. Bioinformatics 2021, 37, 2325–2331. [Google Scholar] [CrossRef] [PubMed]
- Steenwyk, J.L.; Martínez-Redondo, G.I.; Buida, T.J.; Gluck-Thaler, E.; Shen, X.; Gabaldón, T.; Rokas, A.; Fernández, R. PhyKIT: A Multitool for Phylogenomics. Curr. Protoc. 2024, 4, e70016. [Google Scholar] [CrossRef] [PubMed]
- Steenwyk, J.L.; Buida, T.J.; Gonçalves, C.; Goltz, D.C.; Morales, G.; Mead, M.E.; LaBella, A.L.; Chavez, C.M.; Schmitz, J.E.; Hadjifrangiskou, M.; et al. BioKIT: a versatile toolkit for processing and analyzing diverse types of sequence data. Genetics 2022, 221, iyac079. [Google Scholar] [CrossRef] [PubMed]
- Steenwyk, J.L.; Buida, T.J.; Rokas, A.; King, N. OrthoHMM: Improved Inference of Ortholog Groups using Hidden Markov Models 2024. [CrossRef]
- Steenwyk, J.L.; Goltz, D.C.; Buida, T.J.; Li, Y.; Shen, X.-X.; Rokas, A. OrthoSNAP: A tree splitting and pruning algorithm for retrieving single-copy orthologs from gene family trees. PLOS Biol. 2022, 20, e3001827. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




