Submitted:
03 February 2025
Posted:
04 February 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Production of Fermentation Broth
2.2 Feed Solutions for Filtration with the Ceramic Membrane
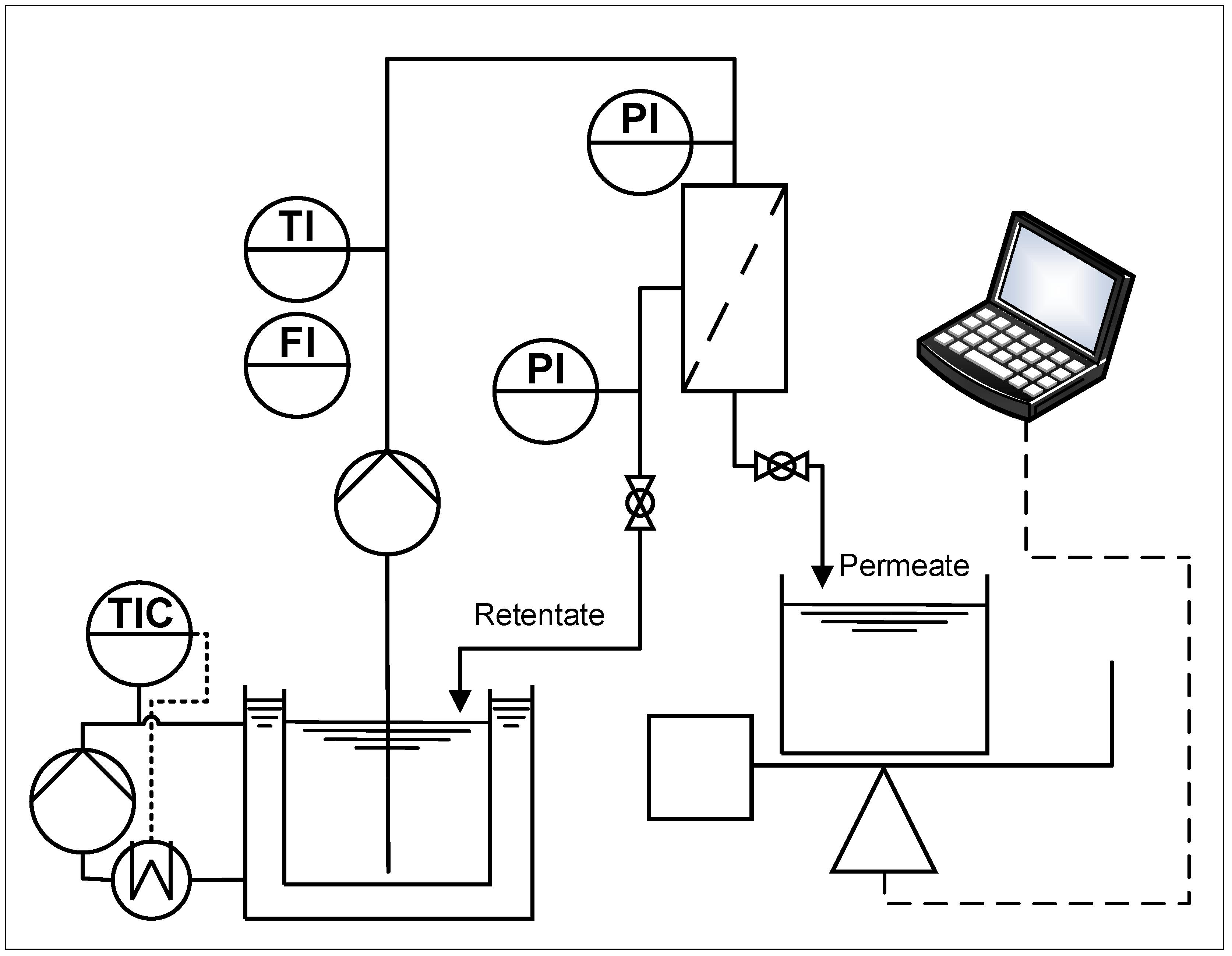
2.3. Filtration Plant
2.4. General Procedure for the Filtration Experiments
2.5. Filtration Experiments to Determine the Compressibility Index
2.6. Chemical Cleaning for Ceramic Membranes
2.7. Data Analysis
3. Results
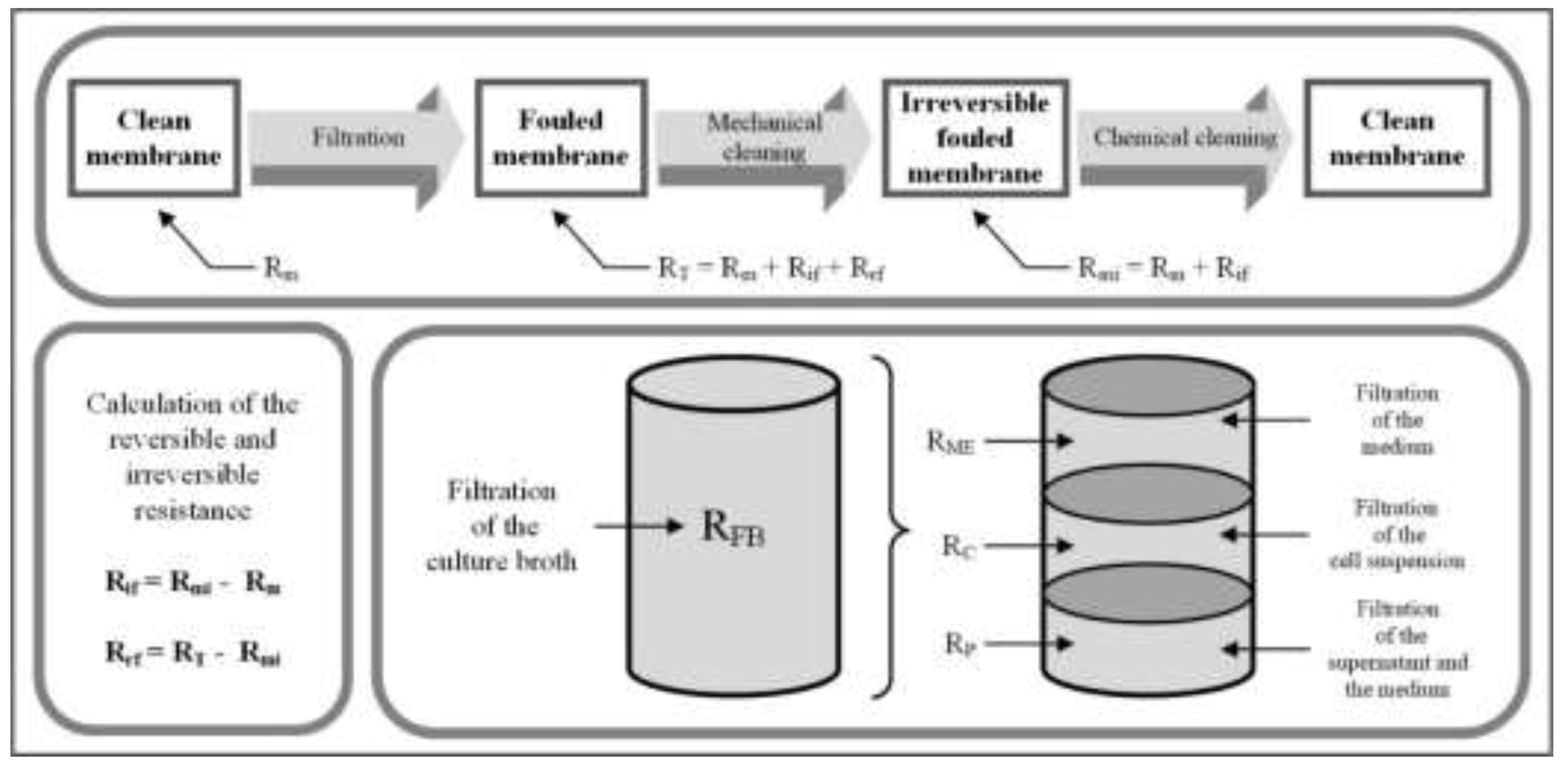
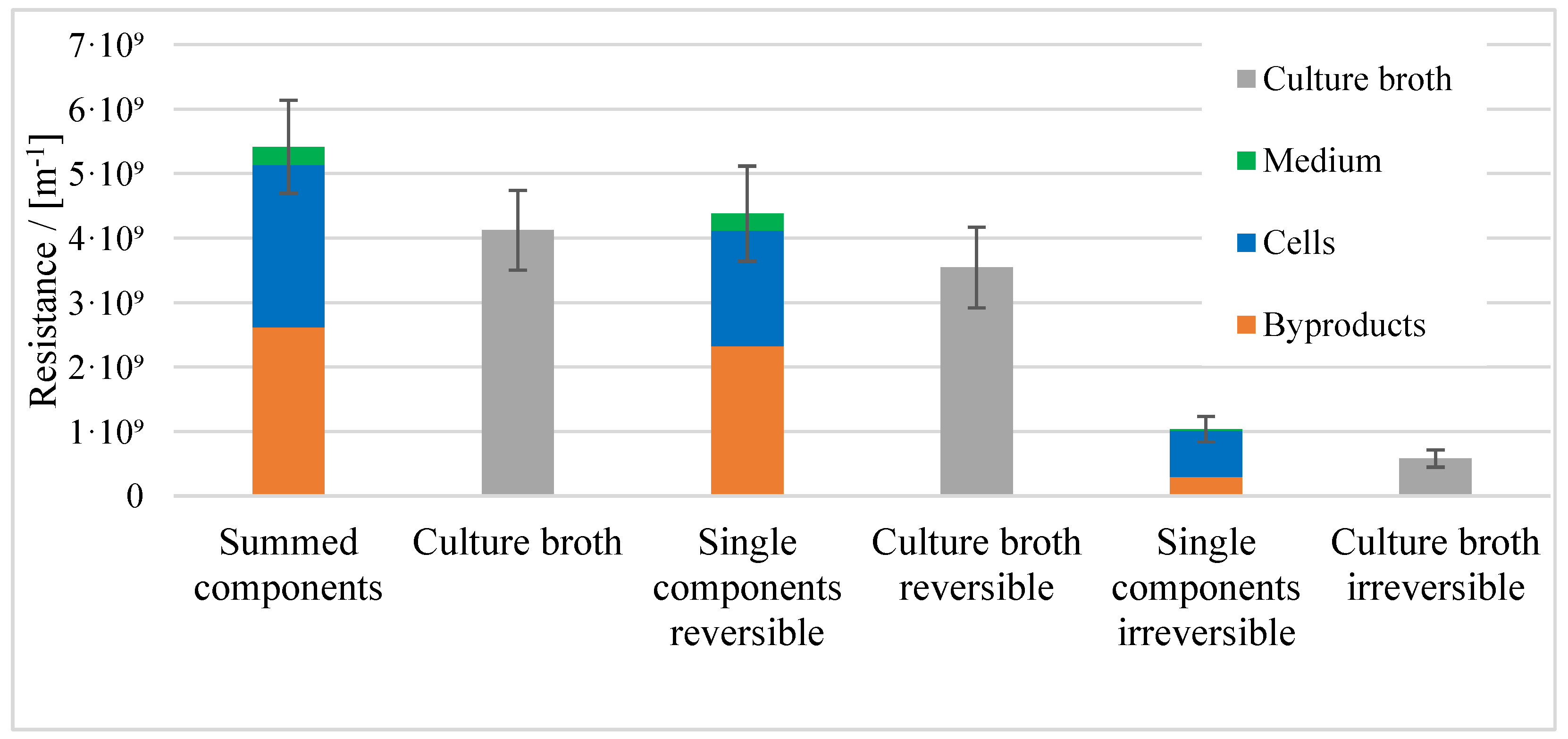
3.1. Resistance in Series Model
3.2. Compressibility Index
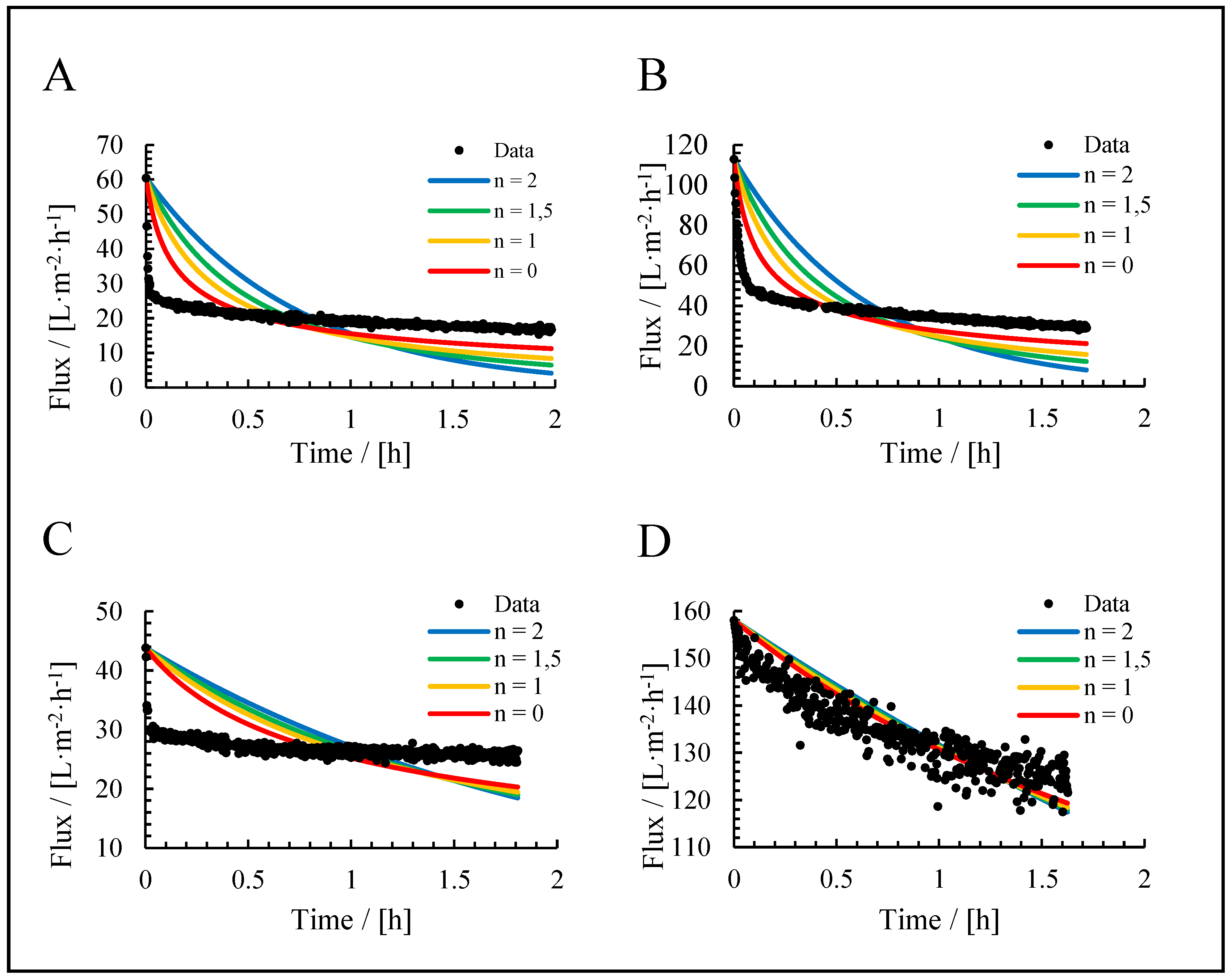
3.3. Application of Hermia’s Laws
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Austin B, Zachary A, Colwell RR. Recognition of Beneckea natriegens (Payne et al.) Baumann et al. as a member of the genus vibrio, as previously proposed by Webb and Payne. Int J Syst Bacteriol 1978;28:315–7. [CrossRef]
- Payne WJ. Studies on bacterial utilization of uronic acids. III. Induction of oxidative enzymes in a marine isolate. J Bacteriol 1958;76:301–7. [CrossRef]
- Eagon RG. Pseudomonas natriegens, a marine bacterium with a generation time of less than 10 minutes. J Bacteriol 1962;83:736–7. [CrossRef]
- Xu J, Dong F, Wu M, Tao R, Yang J, Wu M, et al. Vibrio natriegens as a pET-Compatible Expression Host Complementary to Escherichia coli. Front Microbiol 2021;12:1–11. [CrossRef]
- Maida I, Bosi E, Perrin E, Papaleo MC, Orlandini V, Fondi M, et al. Draft genome sequence of the fast-growing bacterium Vibrio natriegens strain DSMZ 759. Genome Announc 2013;1. [CrossRef]
- Hoffart E, Grenz S, Lange J, Nitschel R, Müller F, Schwentner A, et al. High substrate uptake rates empower Vibrio natriegens as production host for industrial biotechnology. Appl Environ Microbiol 2017;83. [CrossRef]
- Lee HH, Ostrov N, Wong BG, Gold MA, Khalil AS, Church GM. Functional genomics of the rapidly replicating bacterium Vibrio natriegens by CRISPRi. Nat Microbiol 2019;4:1105–13. [CrossRef]
- Weinstock MT, Hesek ED, Wilson CM, Gibson DG. Vibrio natriegens as a fast-growing host for molecular biology. Nat Methods 2016;13:849–51. [CrossRef]
- Becker W, Wimberger F, Zangger K. Vibrio natriegens: An Alternative Expression System for the High-Yield Production of Isotopically Labeled Proteins. Biochemistry 2019;58:2799–803. [CrossRef]
- Eichmann J, Oberpaul M, Weidner T, Gerlach D, Czermak P. Selection of High Producers From Combinatorial Libraries for the Production of Recombinant Proteins in Escherichia coli and Vibrio natriegens. Front Bioeng Biotechnol 2019;7. [CrossRef]
- Schwarz S, Gerlach D, Fan R, Czermak P. GbpA as a secretion and affinity purification tag for an antimicrobial peptide produced in Vibrio natriegens. Electron J Biotechnol 2022;56:75–83. [CrossRef]
- Díaz E, Blázquez ML, Fernández-Llamosas H, Carmona M, Castro L. Speeding up bioproduction of selenium nanoparticles by using Vibrio natriegens as microbial factory. Sci Rep 2017;7:1–9. [CrossRef]
- Schleicher L, Muras V, Claussen B, Pfannstiel J, Blombach B, Dibrov P, et al. Vibrio natriegens as host for expression of multisubunit membrane protein complexes. Front Microbiol 2018;9:1–10. [CrossRef]
- Des Soye BJ, Davidson SR, Weinstock MT, Gibson DG, Jewett MC. Establishing a High-Yielding Cell-Free Protein Synthesis Platform Derived from Vibrio natriegens. ACS Synth Biol 2018;7:2245–55. [CrossRef]
- Stadler KA, Becker W, Darnhofer B, Birner-Gruenberger R, Zangger K. Overexpression of recombinant proteins containing non-canonical amino acids in Vibrio natriegens: p-azido-L-phenylalanine as coupling site for 19F-tags. Amino Acids 2022;54:1041–53. [CrossRef]
- Jørgensen CE, Abrahamsen RK, Rukke EO, Johansen AG, Schüller RB, Skeie SB. Optimization of protein fractionation by skim milk microfiltration: Choice of ceramic membrane pore size and filtration temperature. J Dairy Sci 2016;99:6164–79. [CrossRef]
- Cimini A, Moresi M. Beer Clarification by Novel Ceramic Hollow-Fiber Membranes: Effect of Pore Size on Product Quality. J Food Sci 2016;81:E2521–8. [CrossRef]
- Moreno-García J, García-Martínez T, Mauricio JC, Moreno J. Yeast immobilization systems for alcoholic wine fermentations: Actual trends and future perspectives. Front Microbiol 2018;9. [CrossRef]
- Oppermann T. Mannheimia haemolytica – Bioprocess improvement of leukotoxin production using a fed-batch process and a ceramic membrane based downstream strategy. Peter Czermak, 2018.
- Ebrahimi M, Busse N, Kerker S, Schmitz O, Hilpert M, Czermak P. Treatment of the bleaching effluent from sulfite pulp production by ceramic membrane filtration. Membranes (Basel) 2015;6. [CrossRef]
- Tomczak W, Gryta M. Clarification of 1,3-propanediol fermentation broths by using a ceramic fine UF membrane. Membranes (Basel) 2020;10:1–21. [CrossRef]
- Persson A, Jönsson AS, Zacchi G. Separation of lactic acid-producing bacteria from fermentation broth using a ceramic microfiltration membrane with constant permeate flow. Biotechnol Bioeng 2001;72:269–77. [CrossRef]
- Ebrahimi M, Kerker S, Daume S, Geile M, Ehlen F, Unger I, et al. Innovative ceramic hollow fiber membranes for recycling/reuse of oilfield produced water. Desalin Water Treat 2015;55:3554–67. [CrossRef]
- Hoffmann D, Leber J, Loewe D, Lothert K, Oppermann T, Zitzmann J, et al. Purification of new biologicals using membrane-based processes. Elsevier Inc.; 2018. [CrossRef]
- Influence of Ceramic Membrane Surface Characteristics on the Flux Behavior of a Complex Fermentation Broth 2021.
- McCarthy AA, Walsh PK, Foley G. Experimental techniques for quantifying the cake mass, the cake and membrane resistances and the specific cake resistance during crossflow filtration of microbial suspensions. J Memb Sci 2002;201:31–45. [CrossRef]
- Di Bella G, Di Trapani D. A brief review on the resistance-in-series model in membrane bioreactors (MBRs). Membranes (Basel) 2019;9. [CrossRef]
- Mota M, Teixeira JA, Yelshin A. Influence of cell-shape on the cake resistance in dead-end and cross-flow filtrations. Sep Purif Technol 2002;27:137–44. [CrossRef]
- Riesmeier B, Kroner KH, Kula MR. Studies on secondary layer formation and its characterization during cross-flow filtration of microbial cells. J Memb Sci 1987;34:245–66. [CrossRef]
- Tanaka T, Tsuneyoshi SI, Kitazawa W, Nakanishi K. Characteristics in Crossflow Filtration Using Different Yeast Suspensions. Sep Sci Technol 1997;32:1885–98. [CrossRef]
- Vyas HK, Mawson AJ, Bennett RJ, Marshall AD. A new method for estimating cake height and porosity during crossflow filtration of particulate suspensions. J Memb Sci 2000;176:113–9. [CrossRef]
- Diez V, Ezquerra D, Cabezas JL, García A, Ramos C. A modified method for evaluation of critical flux, fouling rate and in situ determination of resistance and compressibility in MBR under different fouling conditions. J Memb Sci 2014;453:1–11. [CrossRef]
- Gul A, Hruza J, Yalcinkaya F. Fouling and chemical cleaning of microfiltration membranes: A mini-review. Polymers (Basel) 2021;13. [CrossRef]
- Shi X, Tal G, Hankins NP, Gitis V. Fouling and cleaning of ultrafiltration membranes: A review. J Water Process Eng 2014;1:121–38. [CrossRef]
- Slimane FZ, Ellouze F, Amar N Ben. Fouling mechanism and screening of backwash parameters: Seawater ultrafiltration case. Environ Eng Res 2019;24:298–308. [CrossRef]
- Liang J, Yu L, Wu J. The dynamic change of specific cake resistance in membrane bioreactor due to periodical cake relaxation. J Environ Chem Eng 2020;8:103837. [CrossRef]
- Ghaffour N, Qamar A. Membrane fouling quantification by specific cake resistance and flux enhancement using helical cleaners. Sep Purif Technol 2020;239. [CrossRef]
- Fane AG, Fell CJD, Hodgson PH, Leslie G, Marshall K. Microfiltration of biomass and biofluids: Effects of membrane morphology and operating conditions. Filtr Sep 1991;28:332–40. [CrossRef]
- Nakanish K, Tadokoro T, Matsuno R. On the Specific Resistance of Cakes of Microorganisms. Chem Eng Commun 1987;62:187–201. [CrossRef]
- Tanaka T, Abe KI, Asakawa H, Yoshida H, Nakanishi K. Filtration characteristics and structure of cake in crossflow filtration of bacterial suspension. J Ferment Bioeng 1994;78:455–61. [CrossRef]
- Loginov M, Doudiès F, Hengl N, Pignon F, Gésan-Guiziou G. Influence of membrane resistance on swelling and removal of colloidal filter cake after filtration pressure release. J Memb Sci 2020;595. [CrossRef]
- Sørensen BL, Sørensen PB. Structure Compression in Cake Filtration. J Environ Eng 1997;123:345–53. [CrossRef]
- Kujundzic E, Greenberg AR, Fong R, Moore B, Kujundzic D, Hernandez M. Biofouling potential of industrial fermentation broth components during microfiltration. J Memb Sci 2010;349:44–55. [CrossRef]
- Fonseca AC, Summers RS, Greenberg AR, Hernandez MT. Extra-cellular polysaccharides, soluble microbial products, and natural organic matter impact on nanofiltration membranes flux decline. Environ Sci Technol 2007;41:2491–7. [CrossRef]
- Rushton A, editor. Mathematical Models and Design Methods in Solid-Liquid Separation. Dordrecht: Springer Netherlands; 1985. [CrossRef]
- Bowen WR, Calvo JI, Hernández A. Steps of membrane blocking in flux decline during protein microfiltration. J Memb Sci 1995;101:153–65. [CrossRef]
- Burghardt JP, Coletta LA, van der Bolt R, Ebrahimi M, Gerlach D, Czermak P. Development and characterization of an enzyme membrane reactor for fructo-oligosaccharide production. Membranes (Basel) 2019;9. [CrossRef]
- Field RW, Wu D, Howell JA, Gupta BB. Critical flux concept for microfiltration fouling. J Memb Sci 1995;100:259–72. [CrossRef]
- Gunde-Cimerman N, Plemenitaš A, Oren A. Strategies of adaptation of microorganisms of the three domains of life to high salt concentrations. FEMS Microbiol Rev 2018;42:353–75. [CrossRef]
- Gregory GJ, Boyd EF. Stressed out: Bacterial response to high salinity using compatible solute biosynthesis and uptake systems, lessons from Vibrionaceae. Comput Struct Biotechnol J 2021;19:1014–27. [CrossRef]
- Magpusao J, Oey I, Kebede B. Opportunities and Challenges of Algal Protein Extraction and Production. Innov Food Process Technol A Compr Rev 2020:216–33. [CrossRef]
- Liu D, Wu C, Wu R, Huang J, Liao B, Lei M, et al. Comprehensive analysis of the phylogeny and extracellular proteases in genus Vibrio strain. Microb Pathog 2019;131:1–8. [CrossRef]
- Thoma F, Blombach B. Metabolic engineering of Vibrio natriegens. Essays Biochem 2021;65:381–92. [CrossRef]
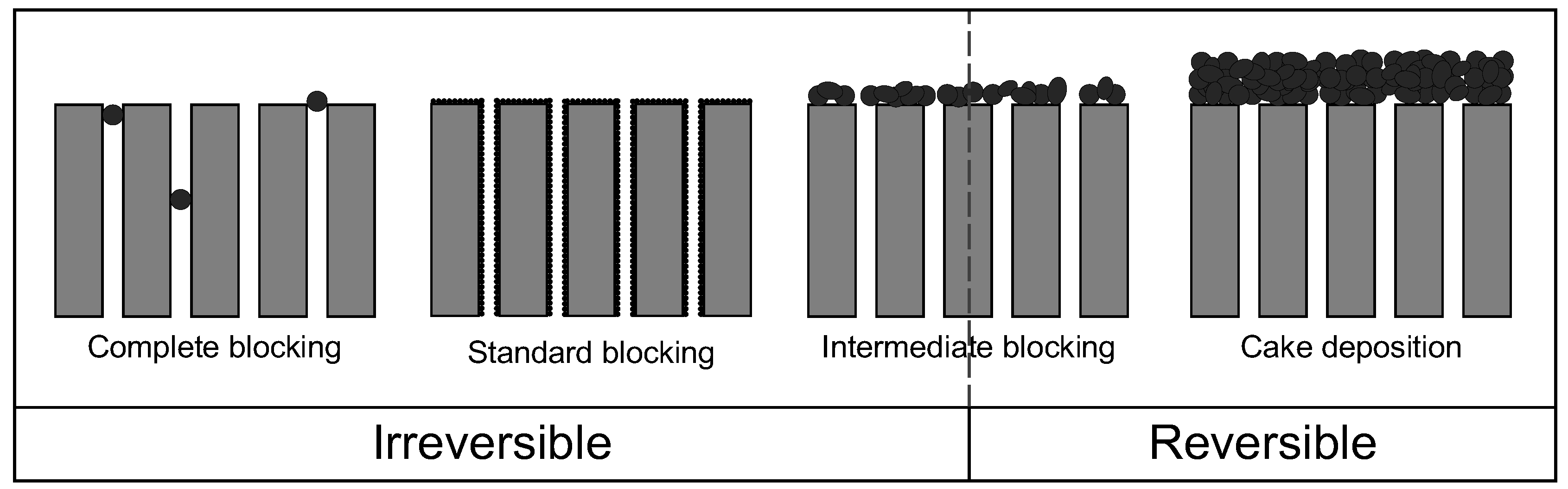
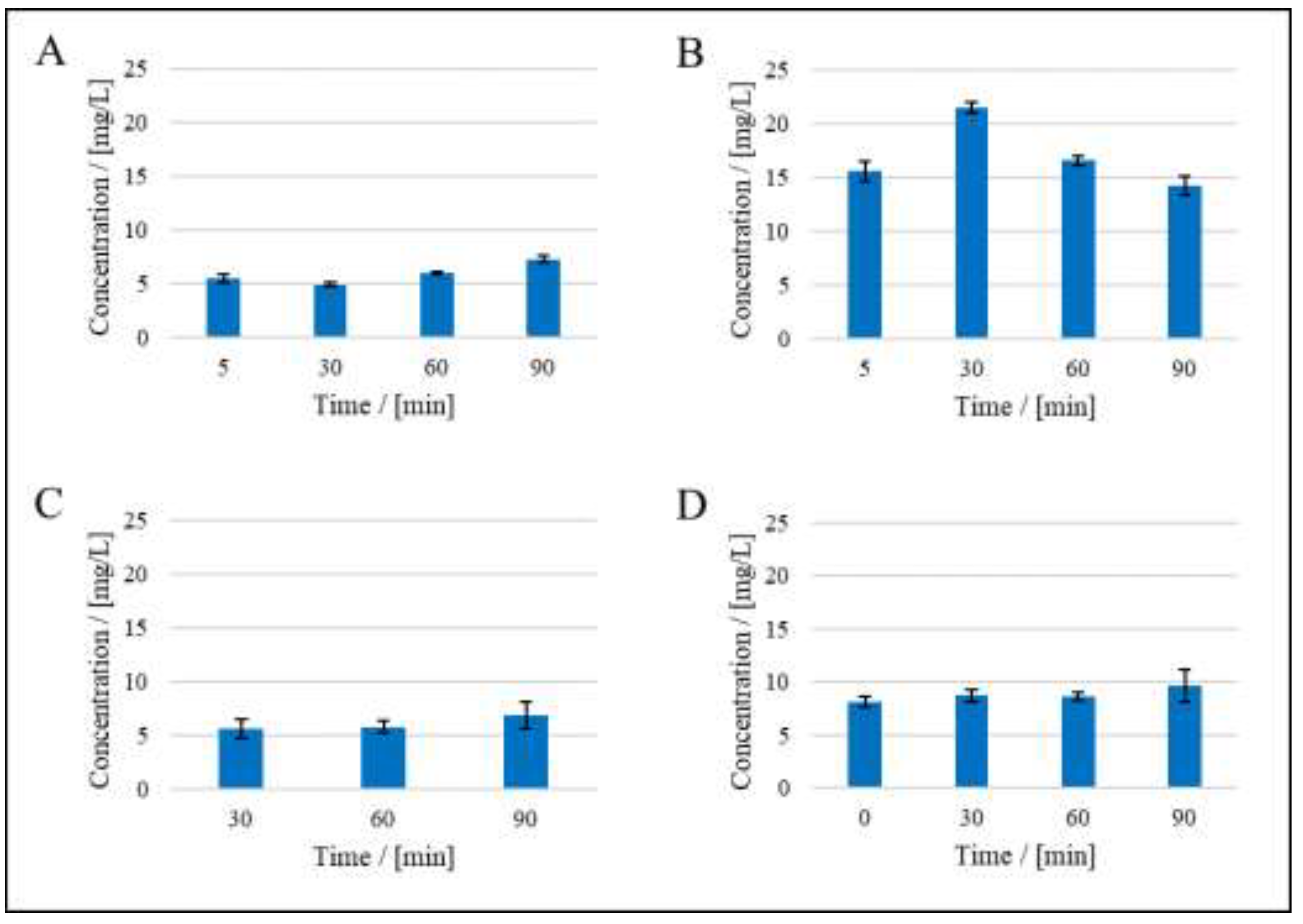
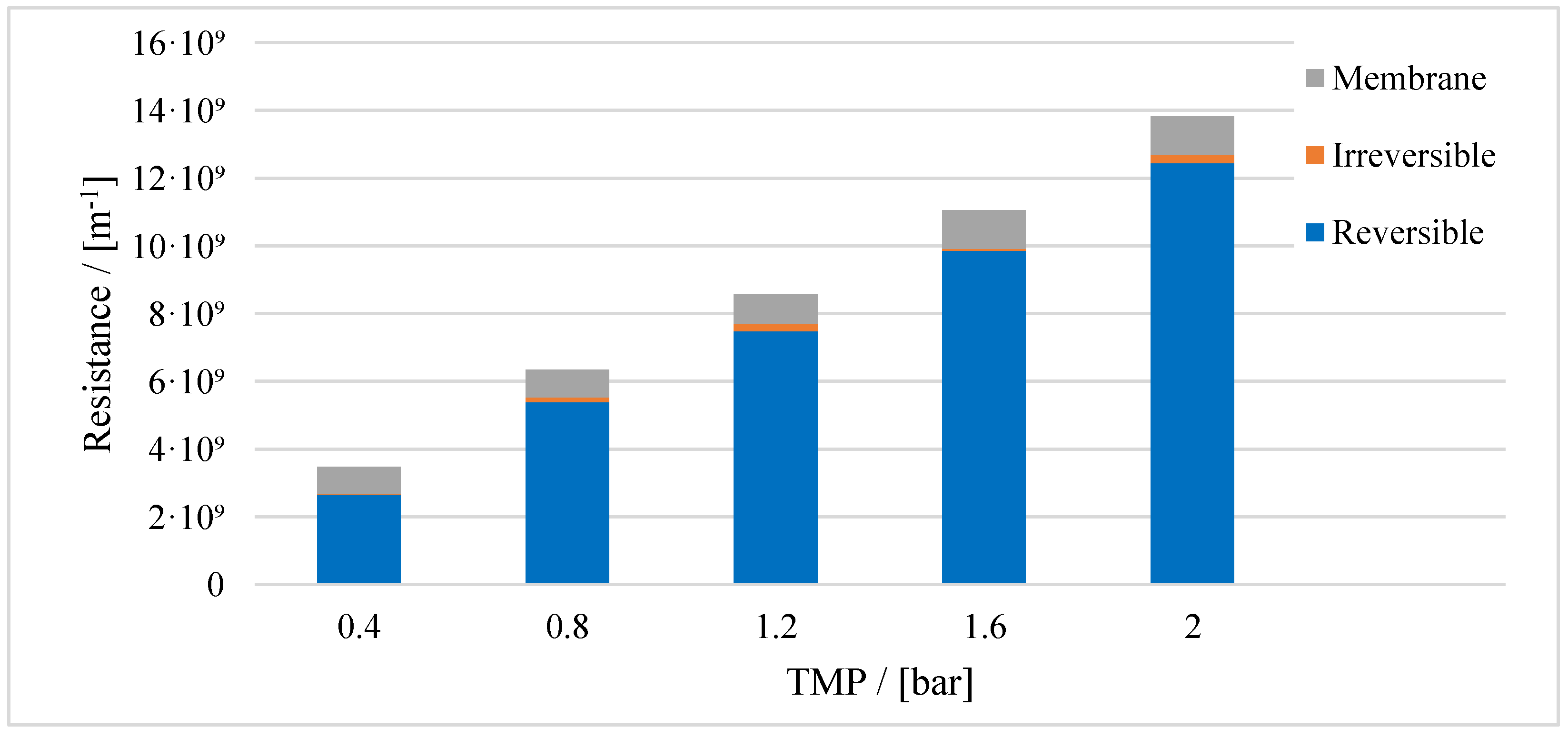
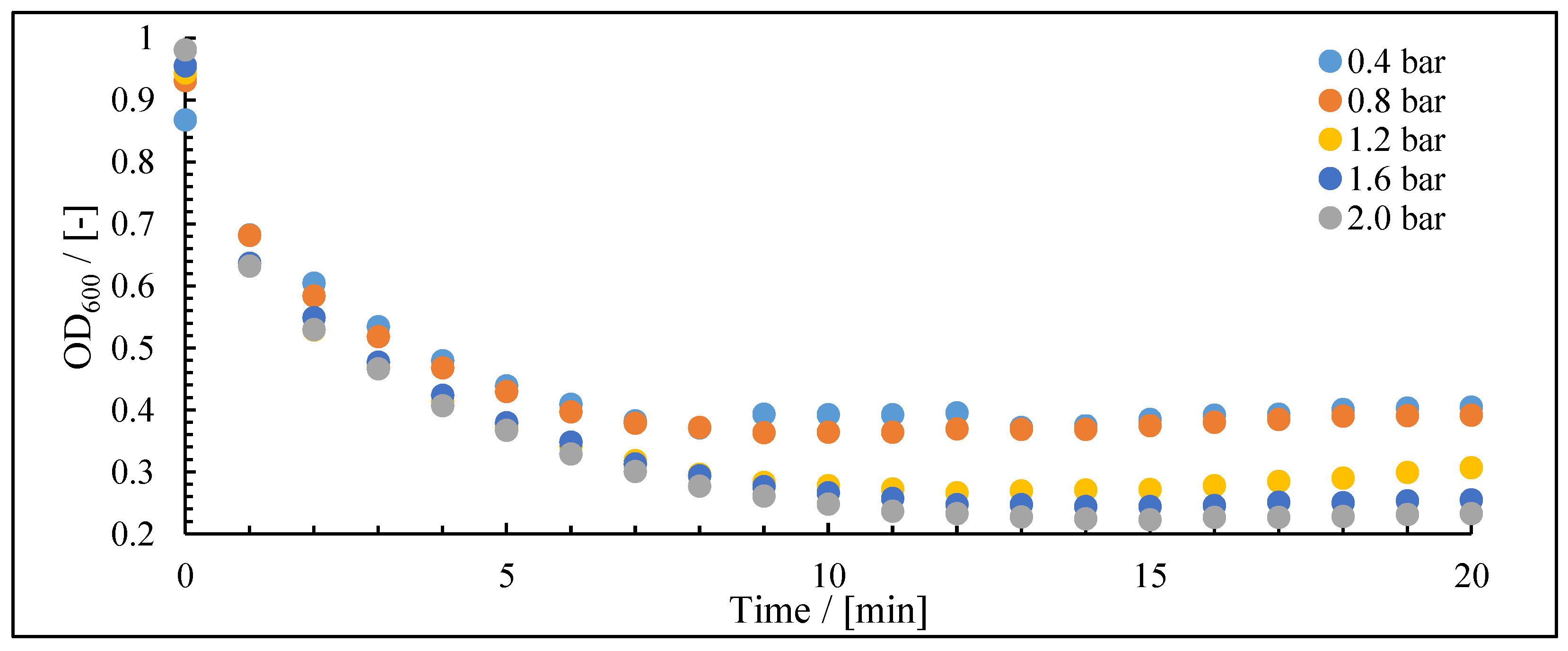

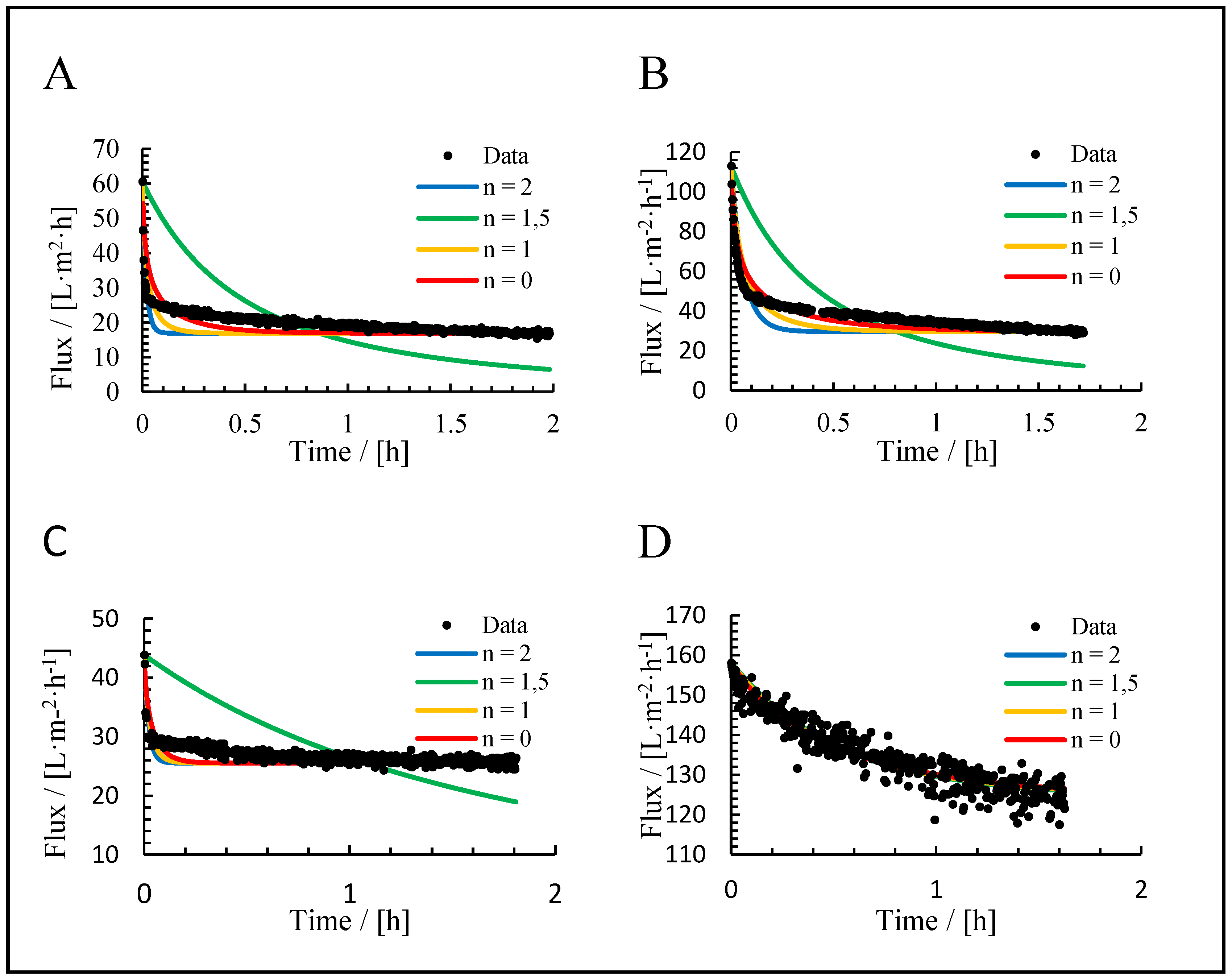
| Mechanism | n | Equation | |
| Complete blocking | 2 | 12 | |
| Standard blocking | 1.5 | 13 | |
| Intermediate blocking | 1 | 14 | |
| Cake deposition | 0 | 15 |
| Mechanism | n | Equation | |
| Complete blocking | 2 | 17 | |
| Standard blocking | 1.5 | 18 | |
| Intermediate blocking | 1 | 19 | |
| Cake deposition | 0 | 20 |
| Fit model | R2 of the fit | |||
| Culture broth | Cells | Supernatant | Medium | |
| Complete blocking | 0.53 | 0.81 | 0.50 | 0.88 |
| Standard blocking | 0.74 | 0.74 | 0.58 | 0.88 |
| Intermediate blocking | 0.61 | 0.88 | 0.53 | 0.88 |
| Cake filtration | 0.72 | 0.92 | 0.51 | 0.88 |
| Fit model | R2 of the fit | |||
| Culture broth | Cells | Supernatant | Medium | |
| Complete blocking | 0.71 | 0.69 | 0.56 | 0.86 |
| Standard blocking | 0.74 | 0.74 | 0.58 | 0.87 |
|
Intermediate blocking |
0.31 | 0.14 | 0.30 | 0.87 |
| Cake filtration | 0.88 | 0.88 | 0.71 | 0.88 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





