Submitted:
30 January 2025
Posted:
03 February 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Primary Human Skeletal Muscle Culture
2.2. Immunofluorescence on Muscle Sections
2.3. Immunofluorescence on Cell Culture
2.4. Wound-Healing (WH) Assay in CD56- Cells
2.5. Confocal Microscopy Analysis and Statistics
3. Results
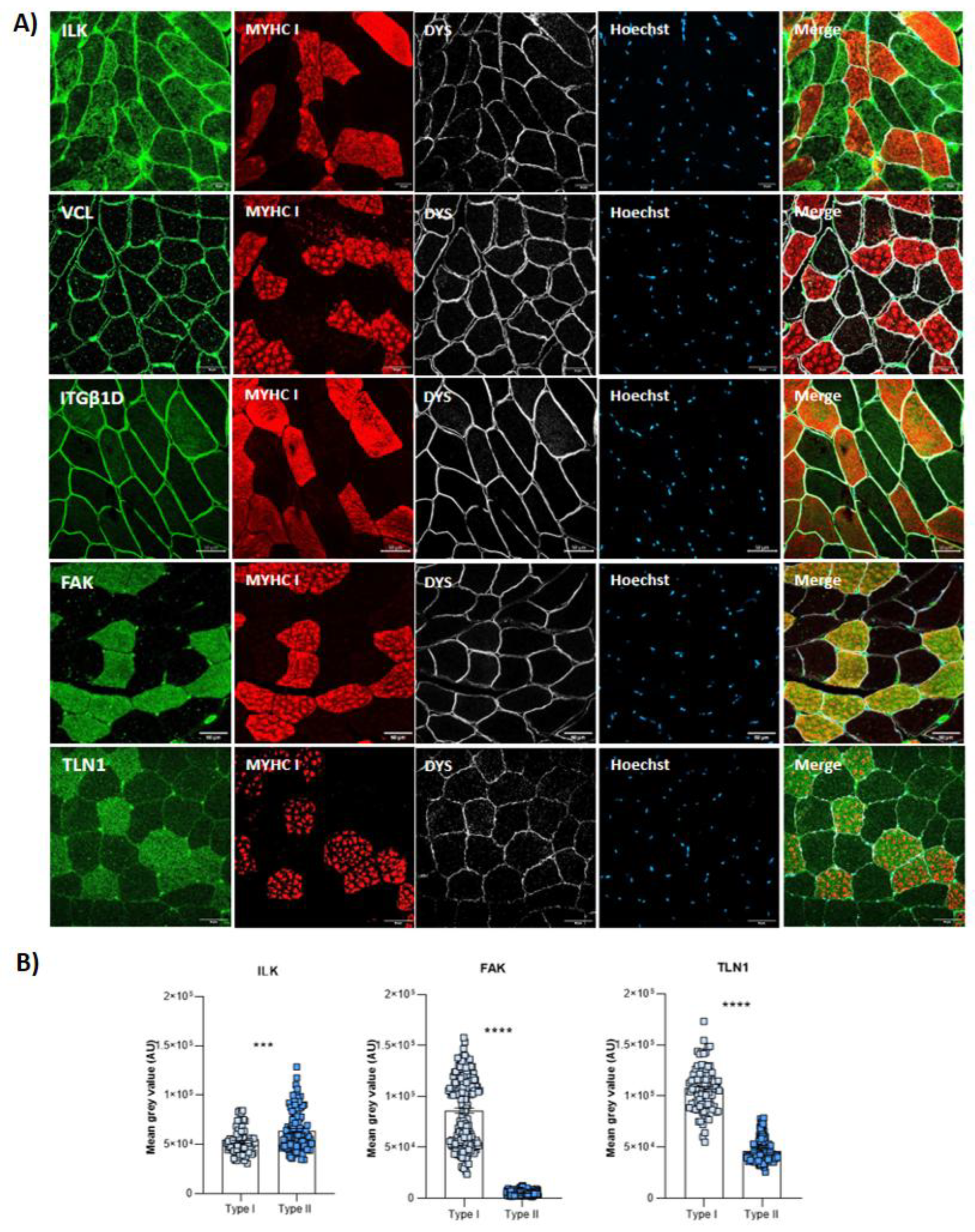
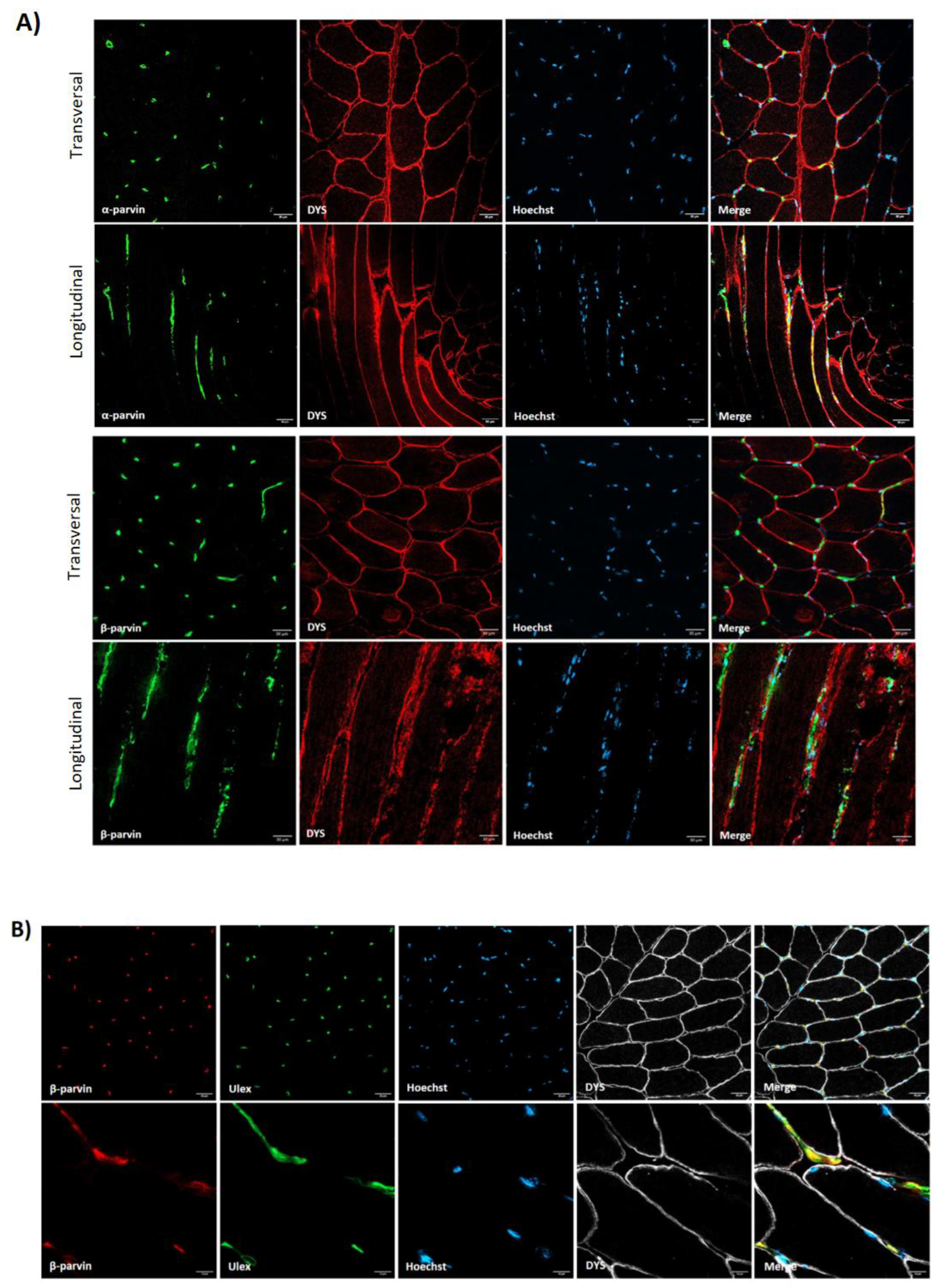
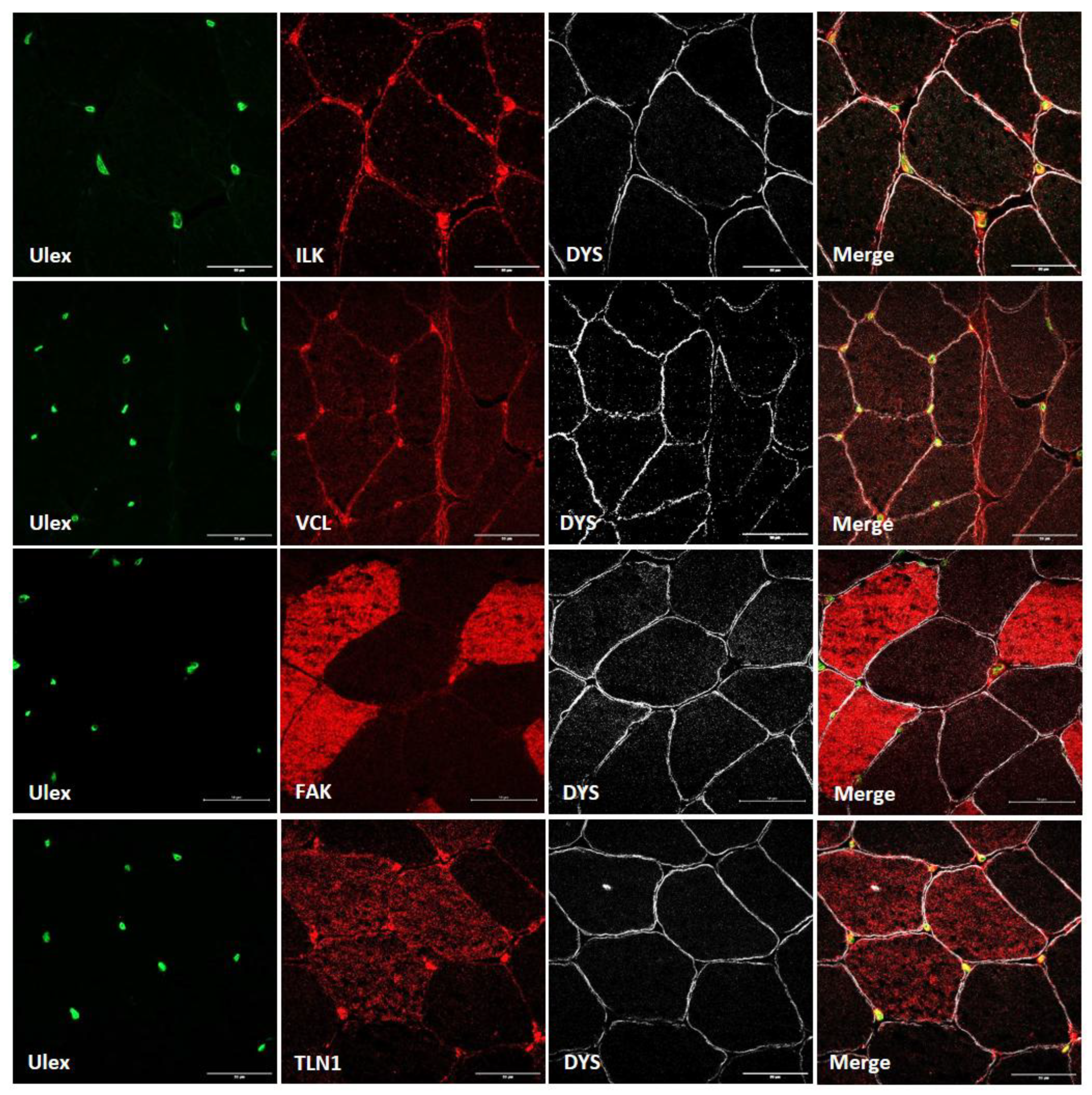
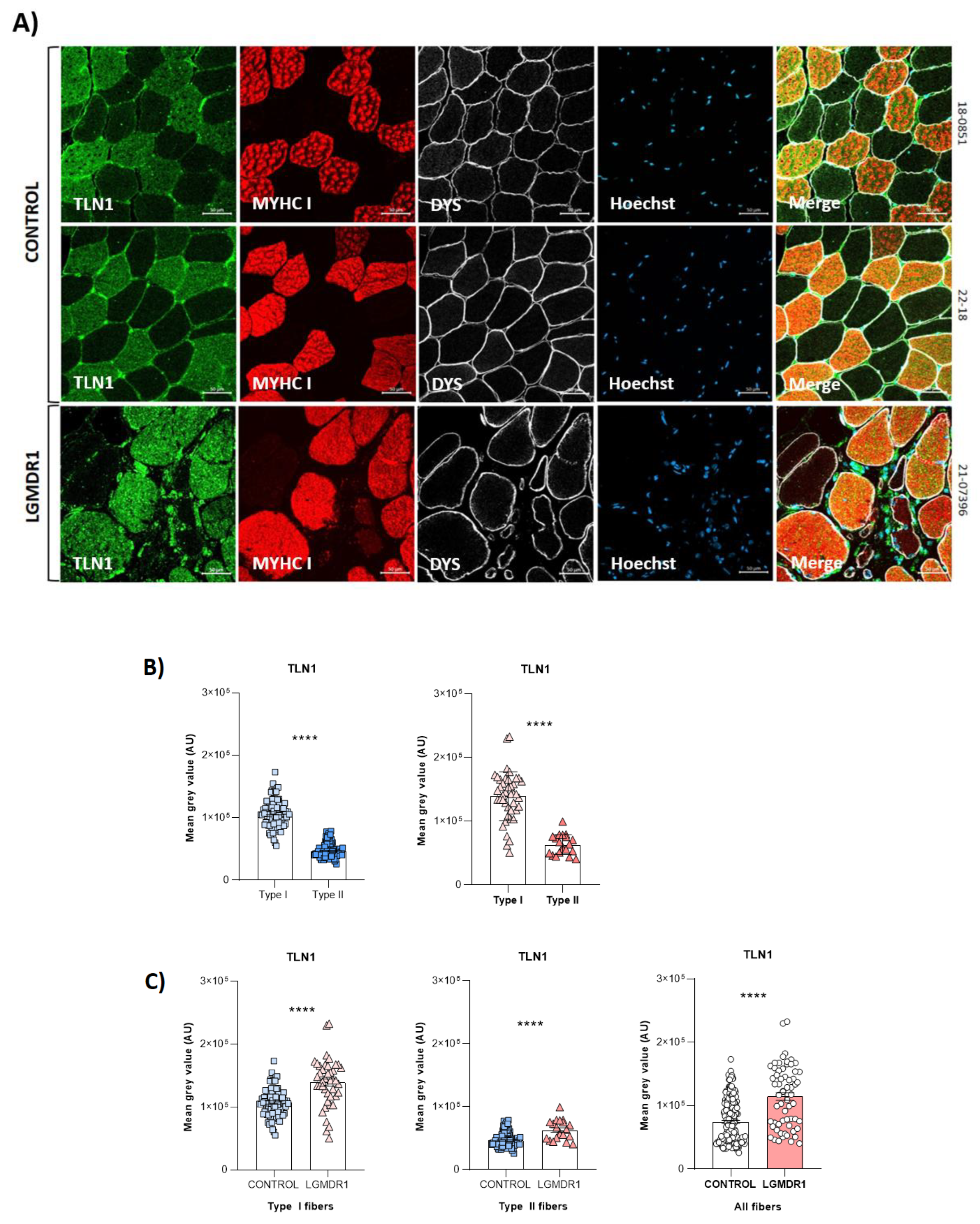
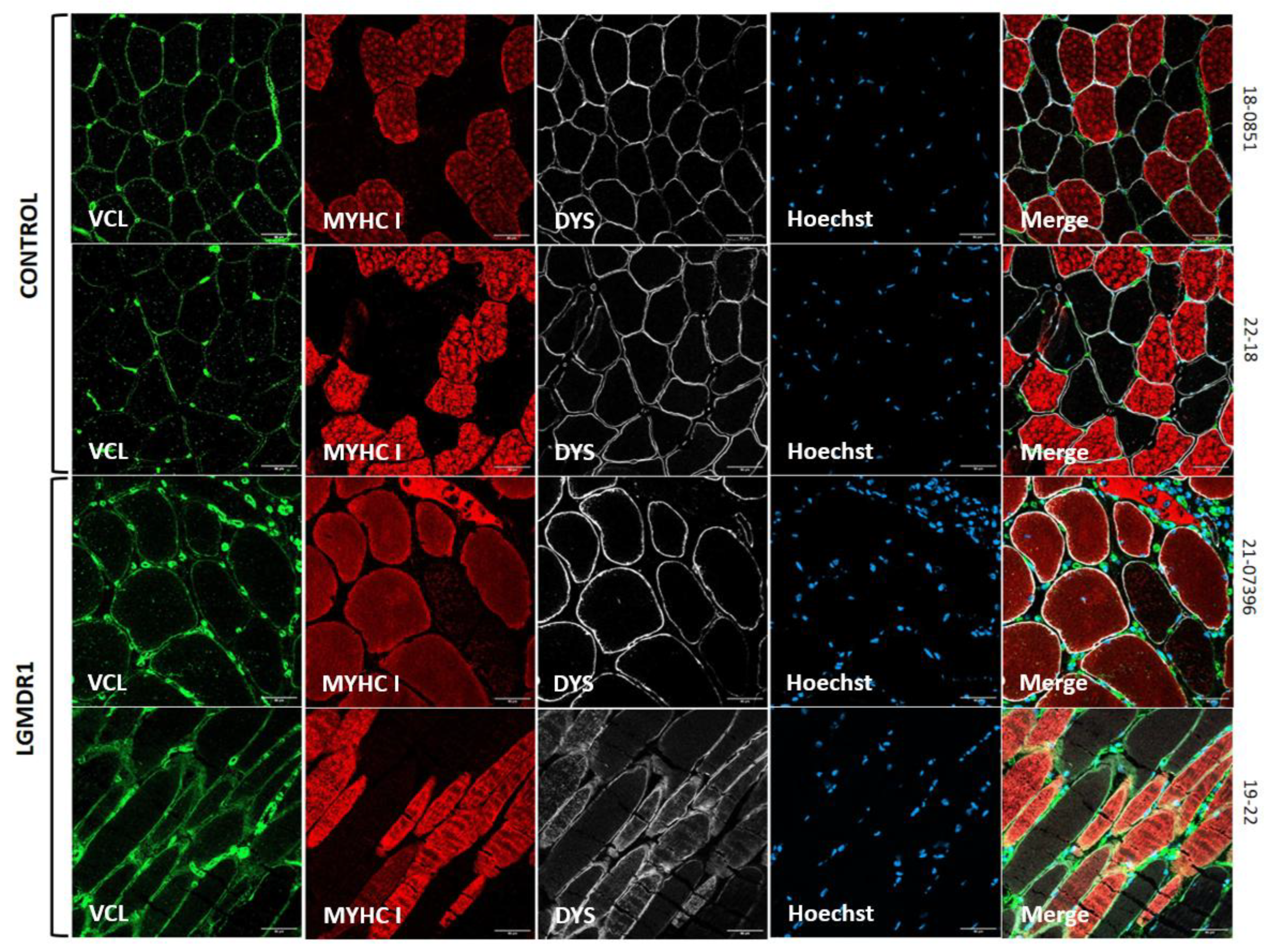
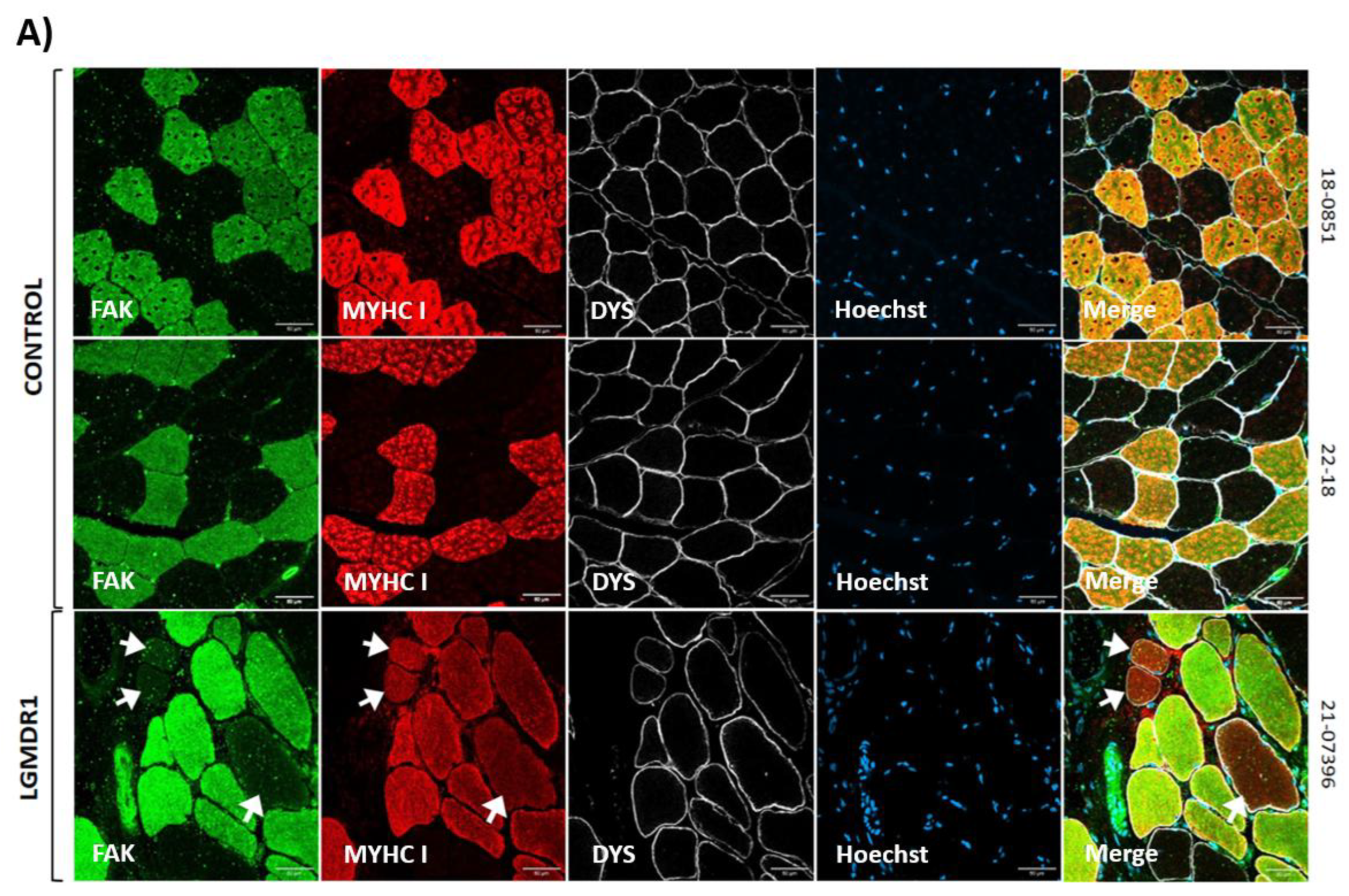
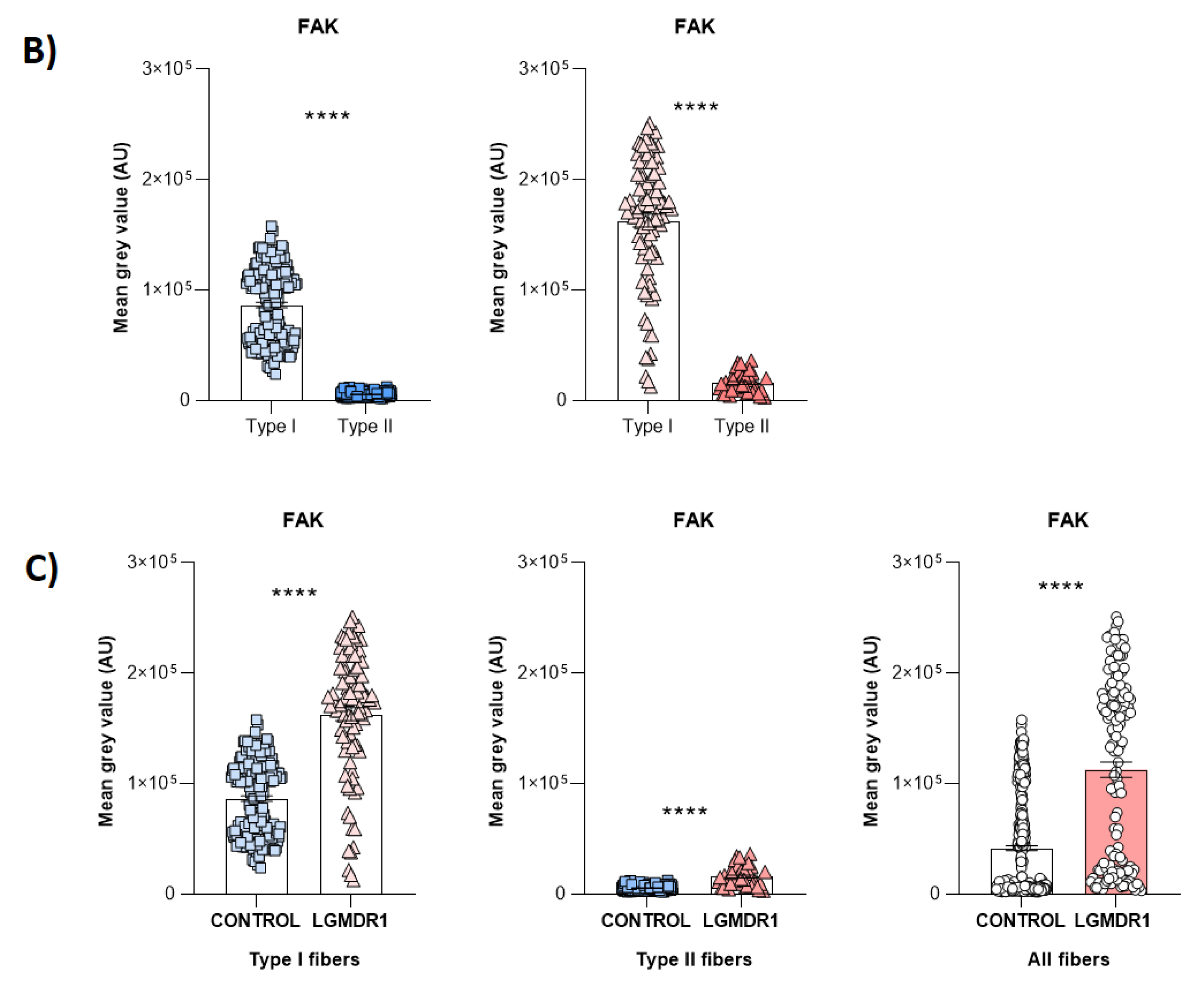
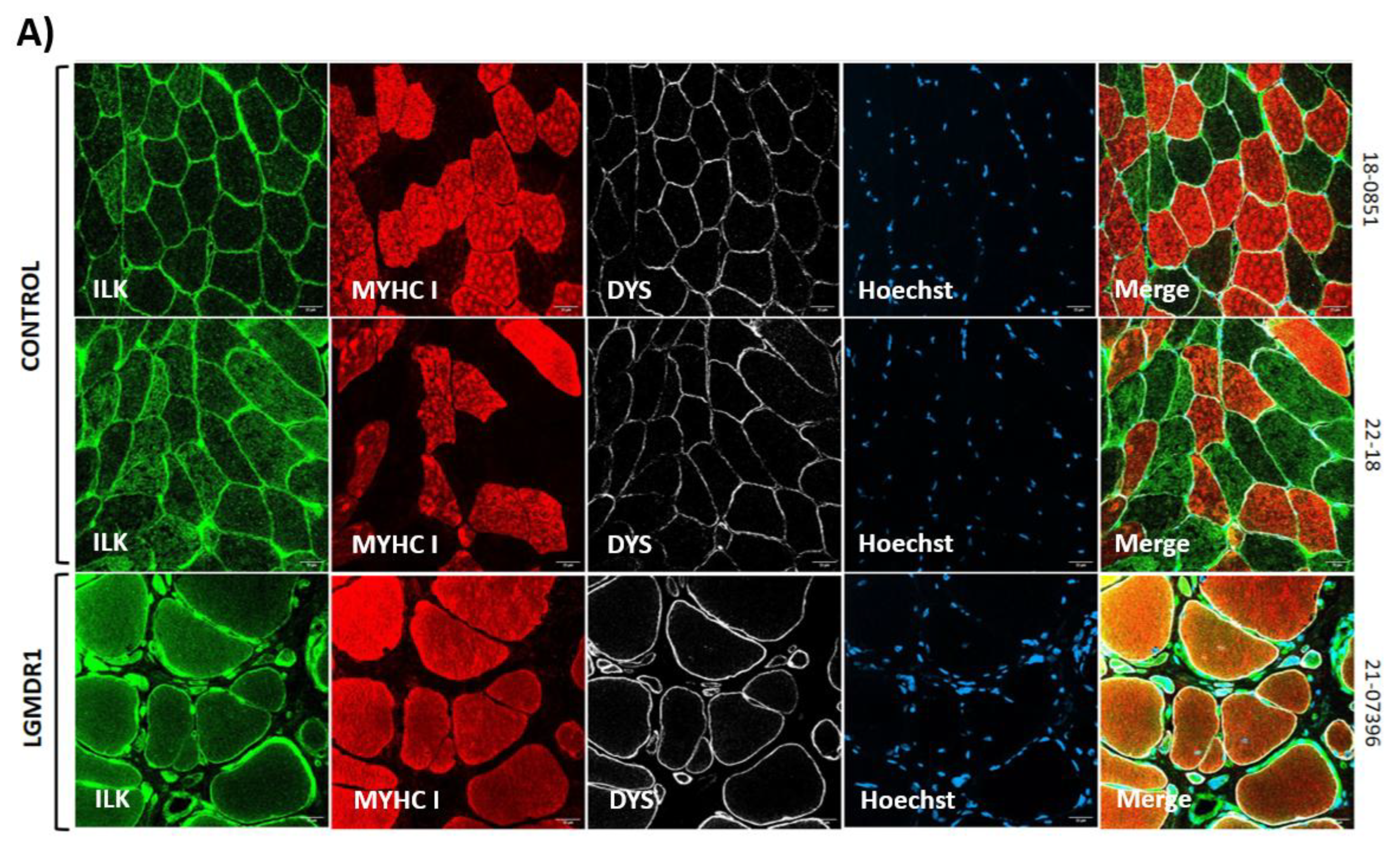
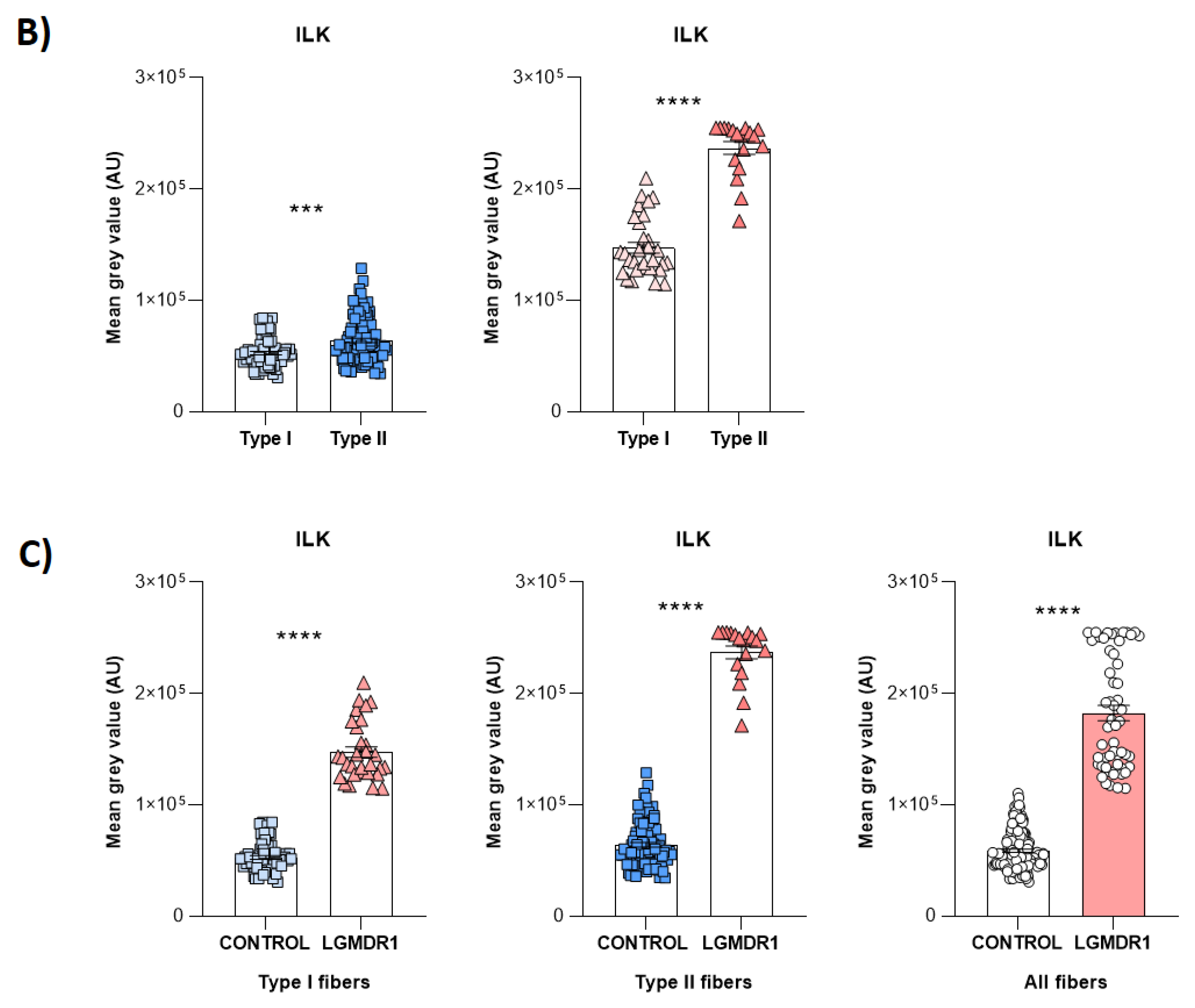
3.1. Costamere Proteins Are Fiber-Type-Dependent and Also Present in the Endothelial Tissue
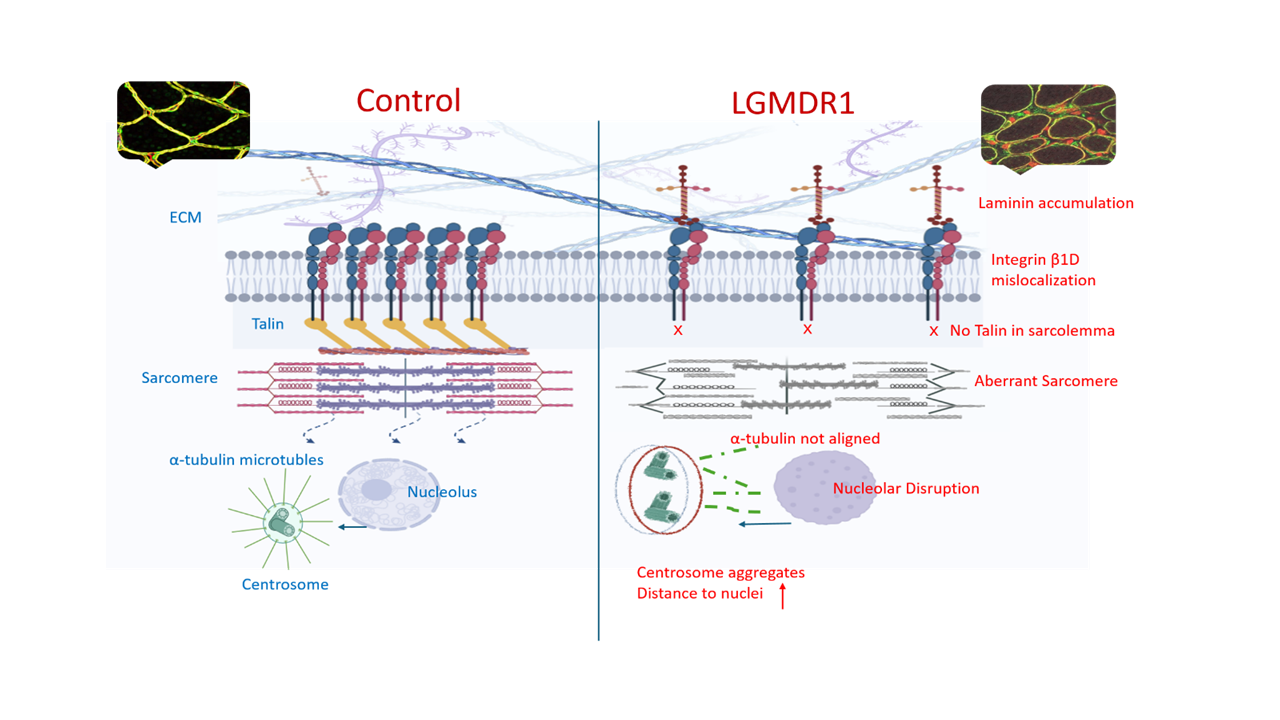
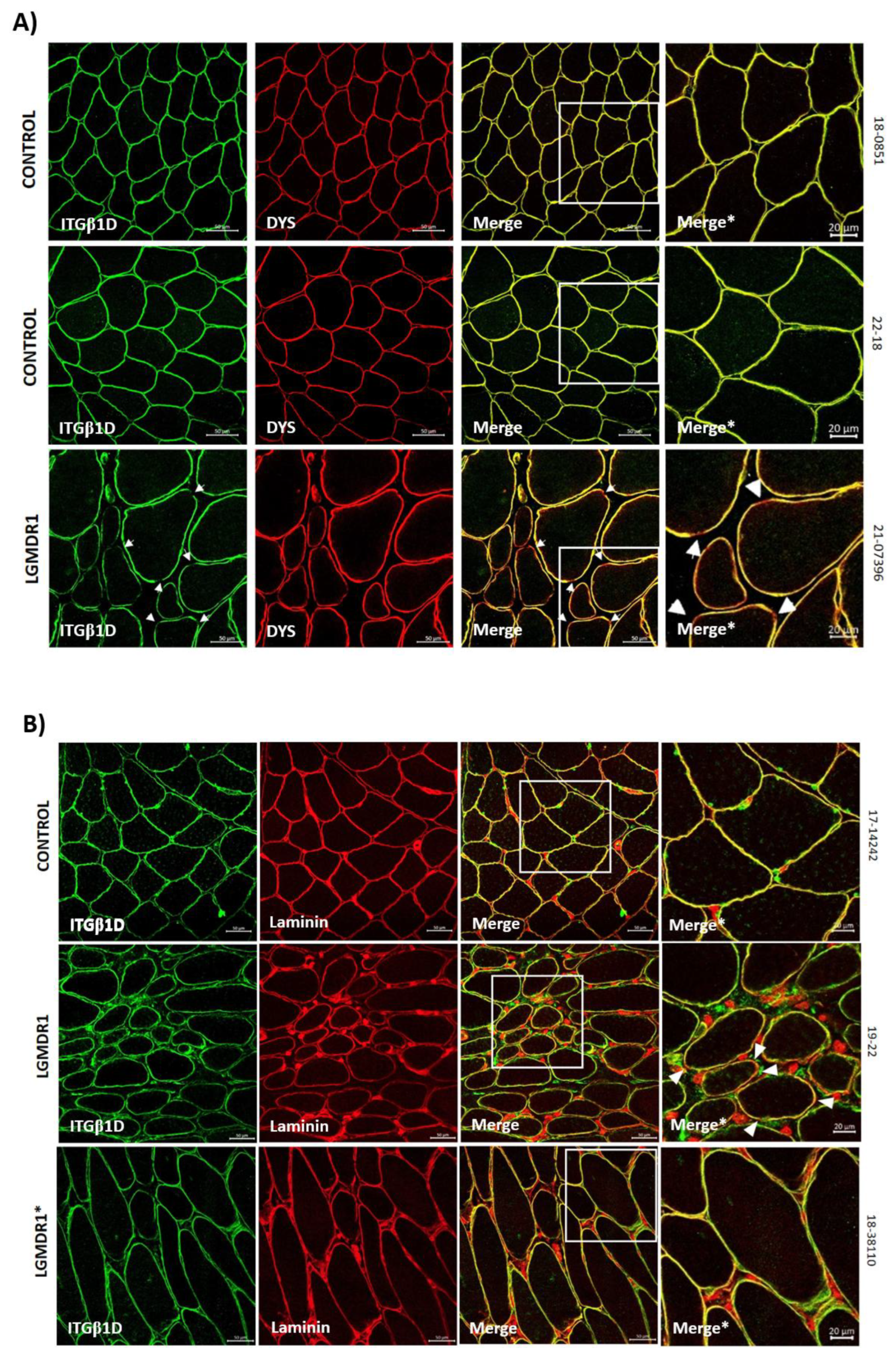
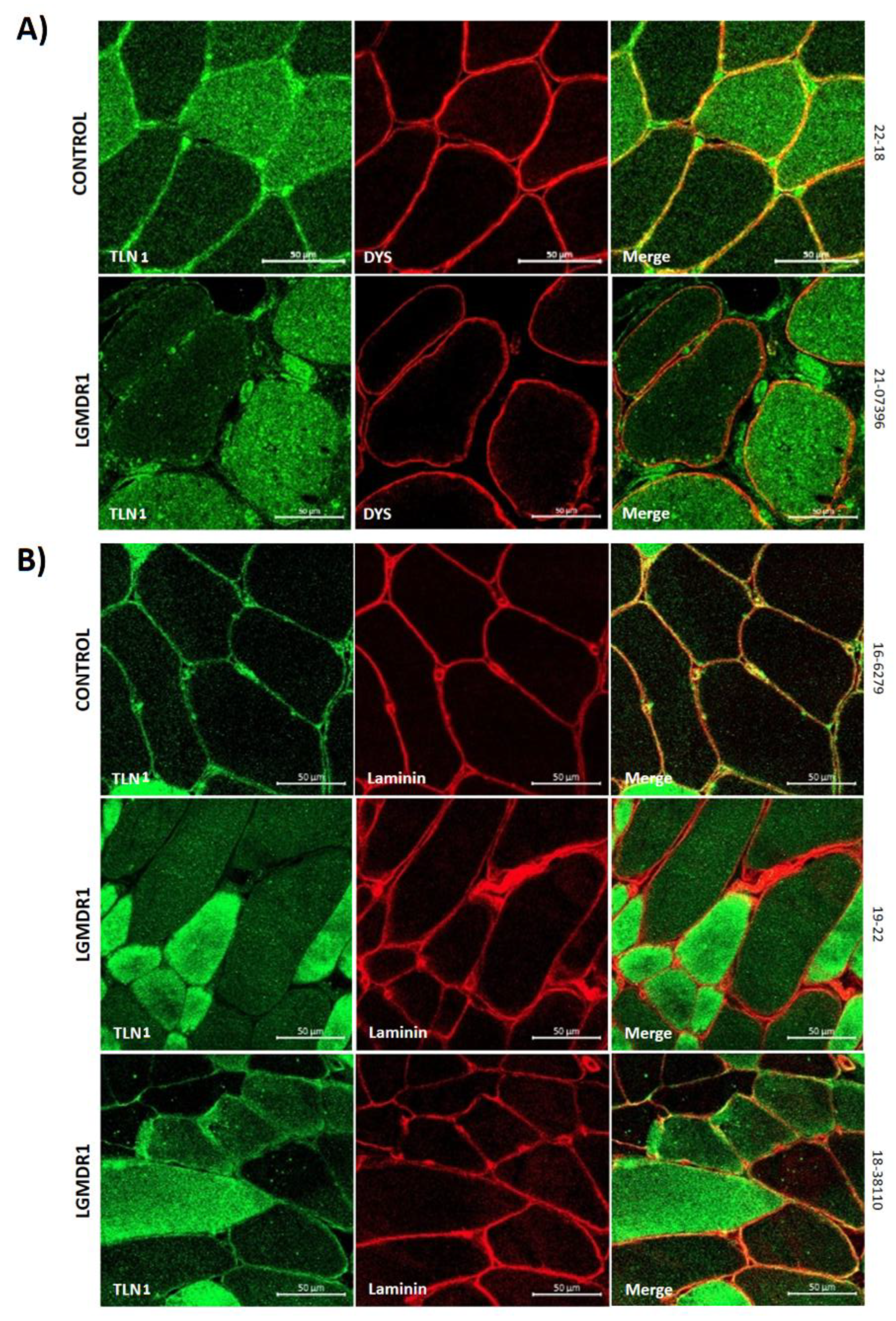
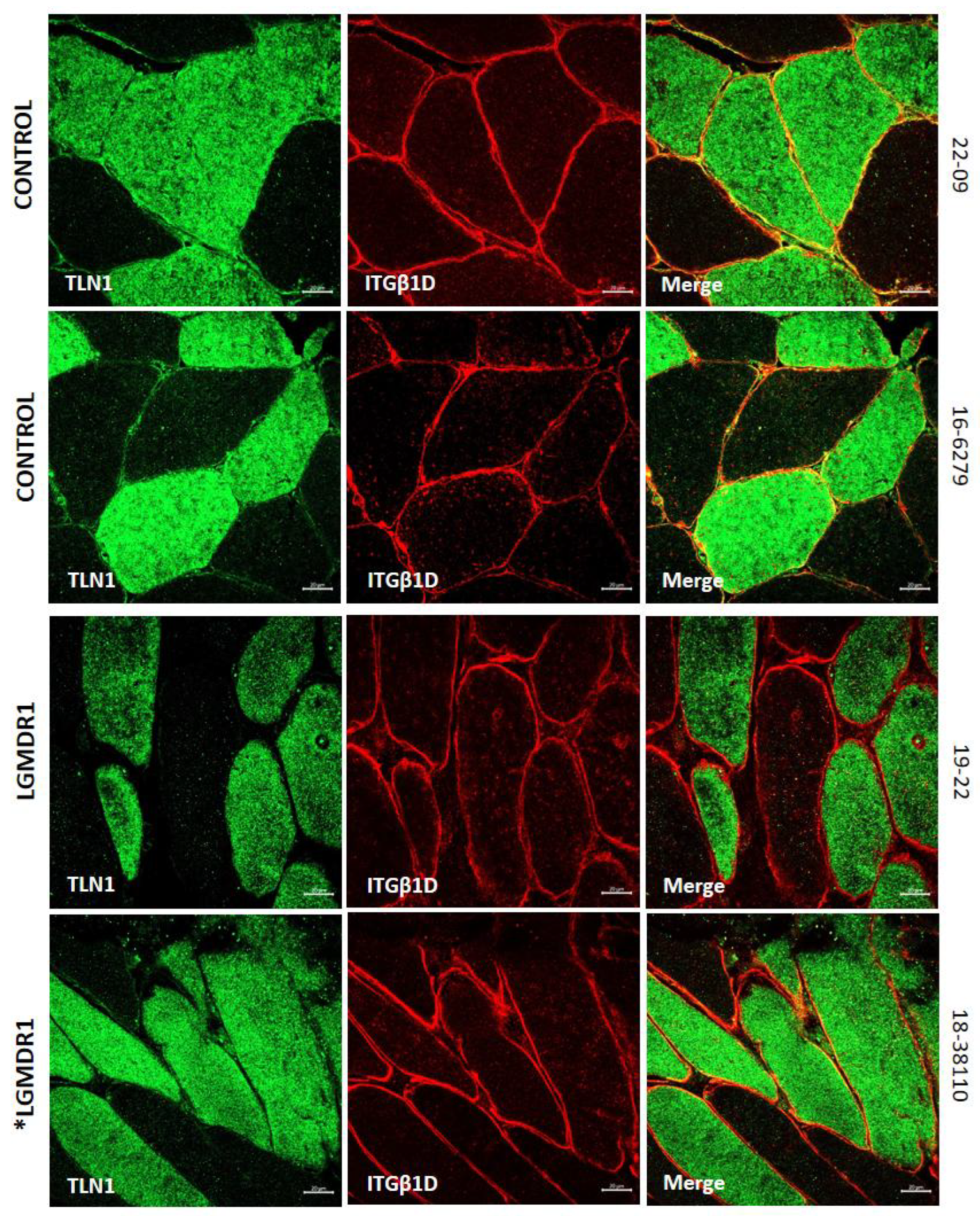
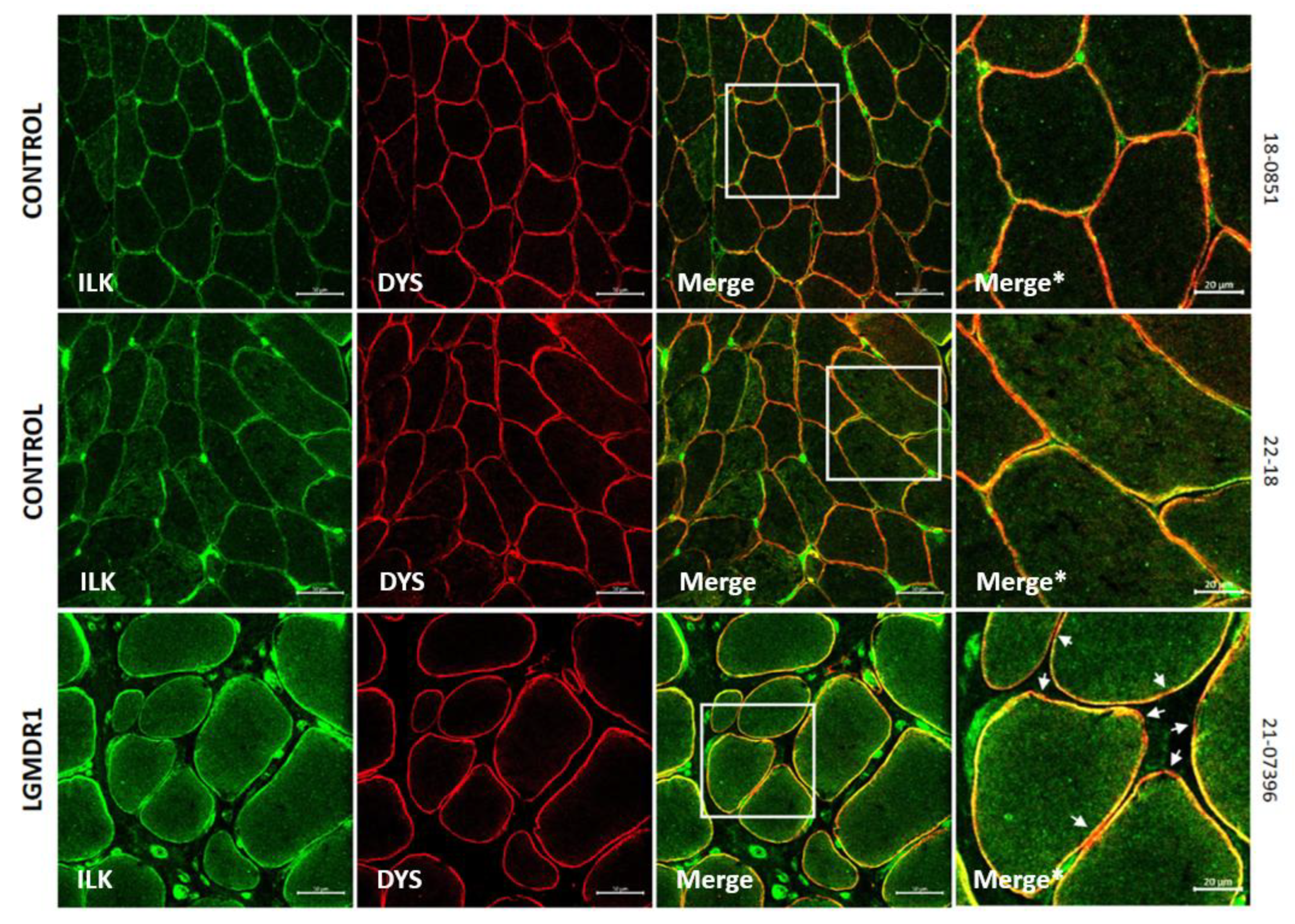
3.2. Abnormal Distribution of ITGβ1D and TLN1 in LGMDR1 Patients’ Muscle
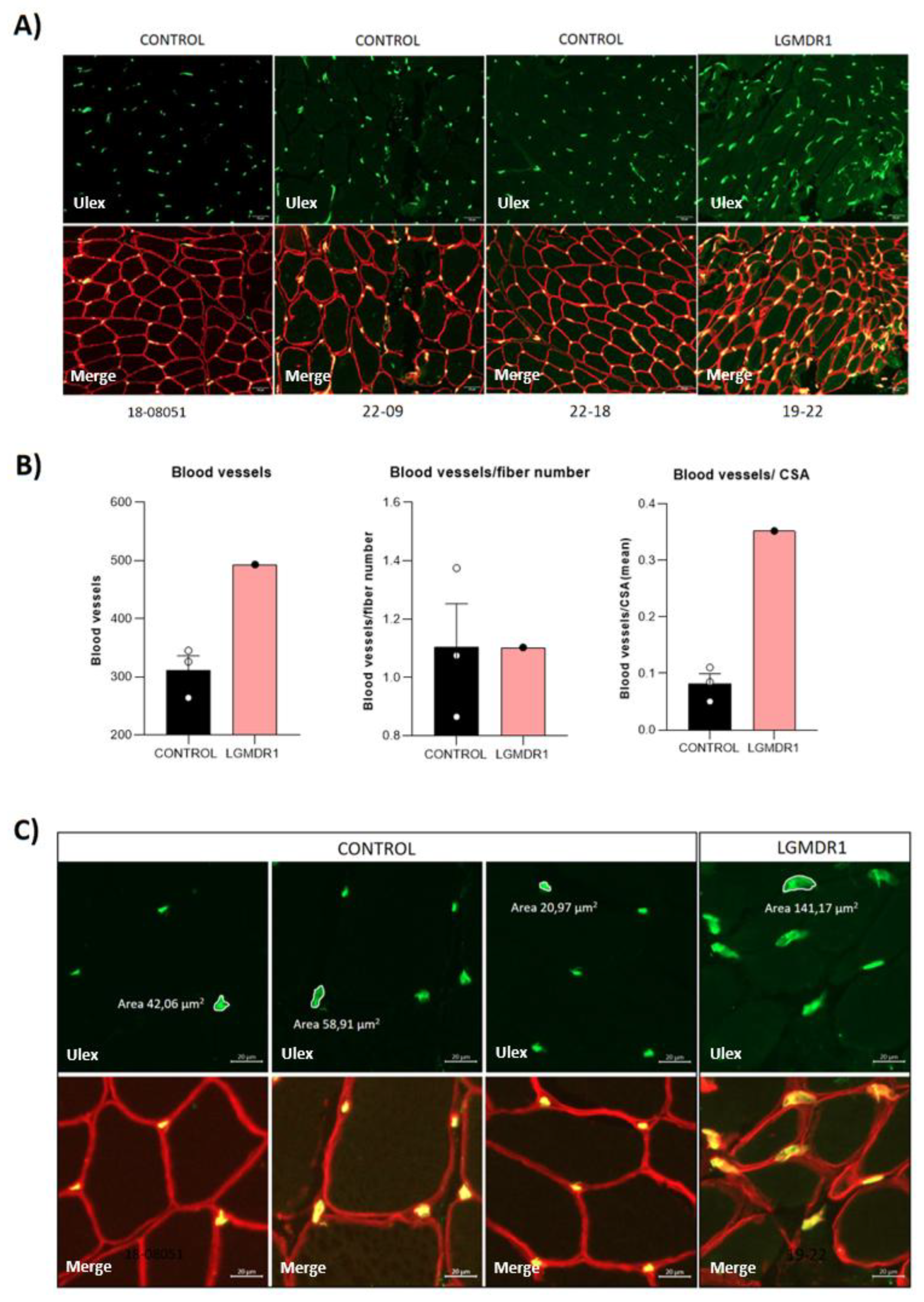
3.3. Abnormal Blood Vessel Morphology in LGMDR1 Patients’ Muscles
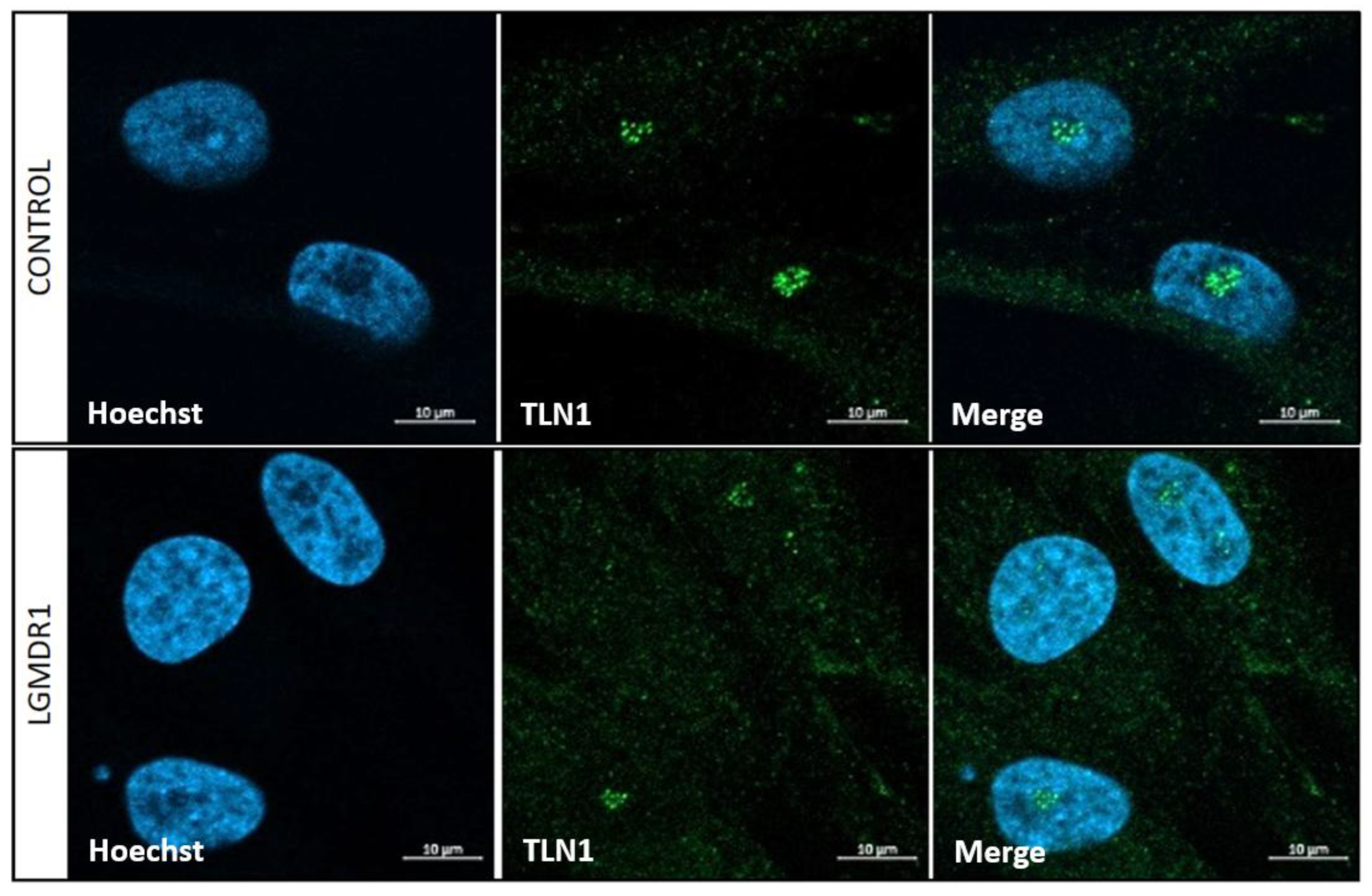
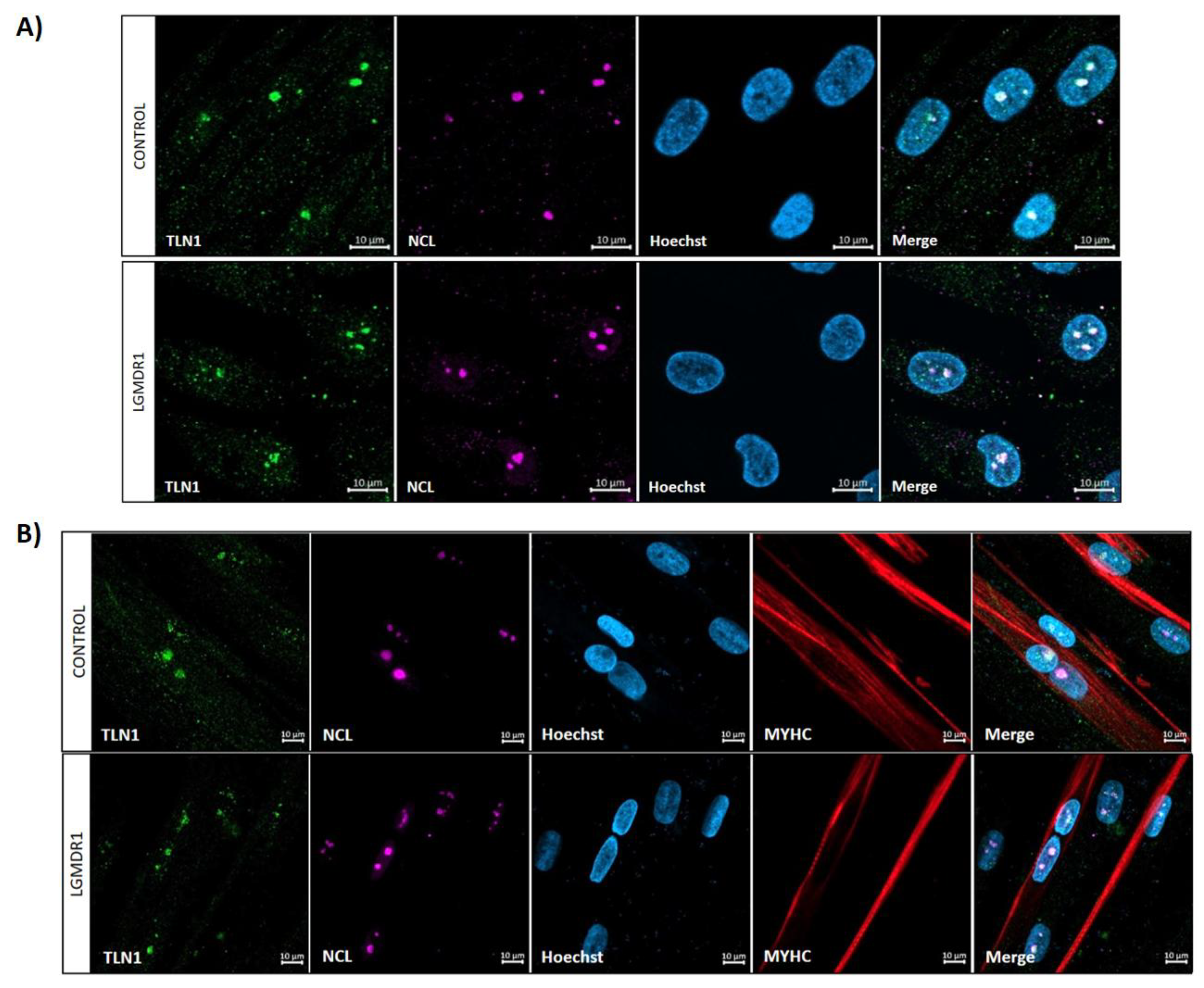
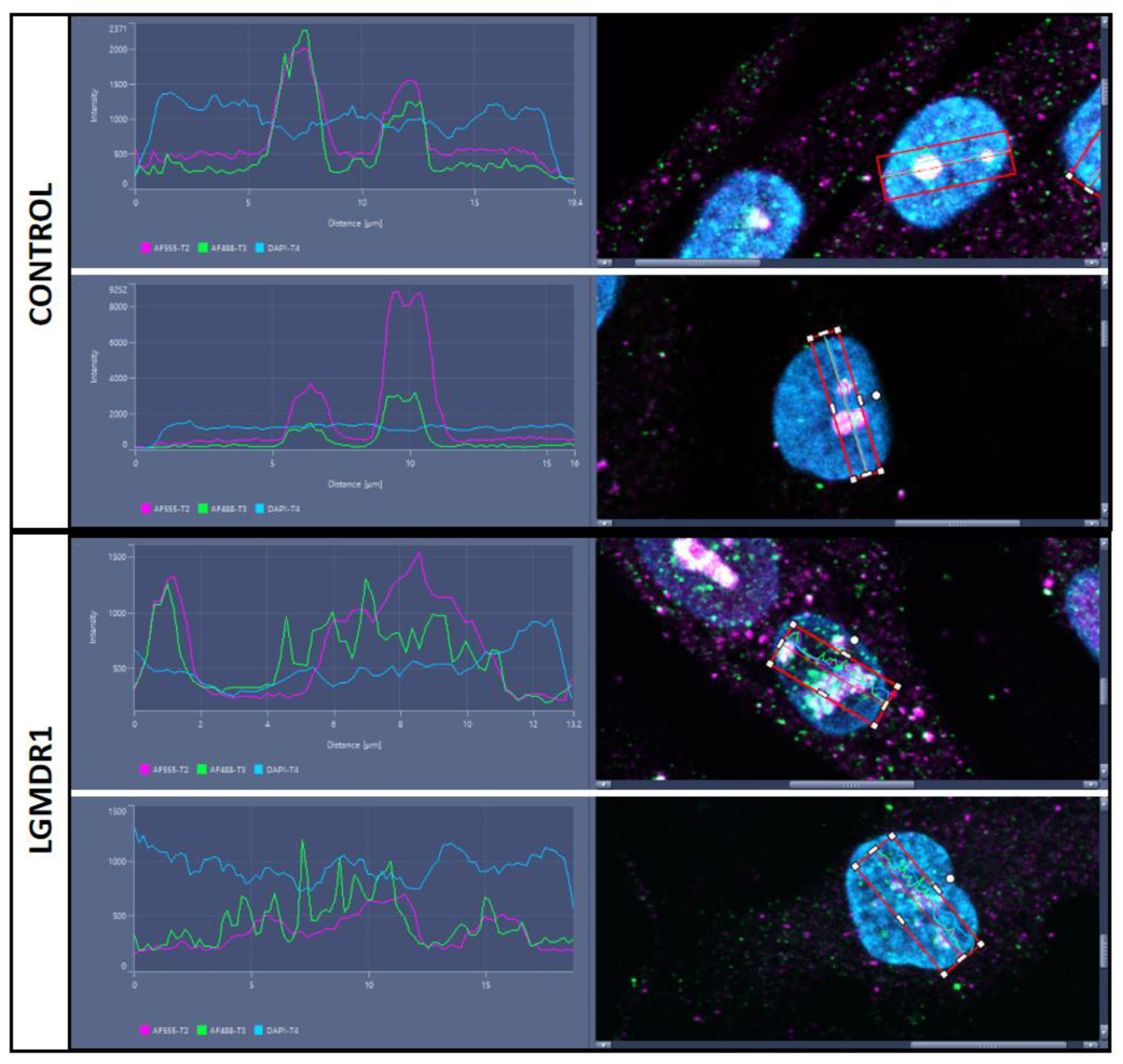
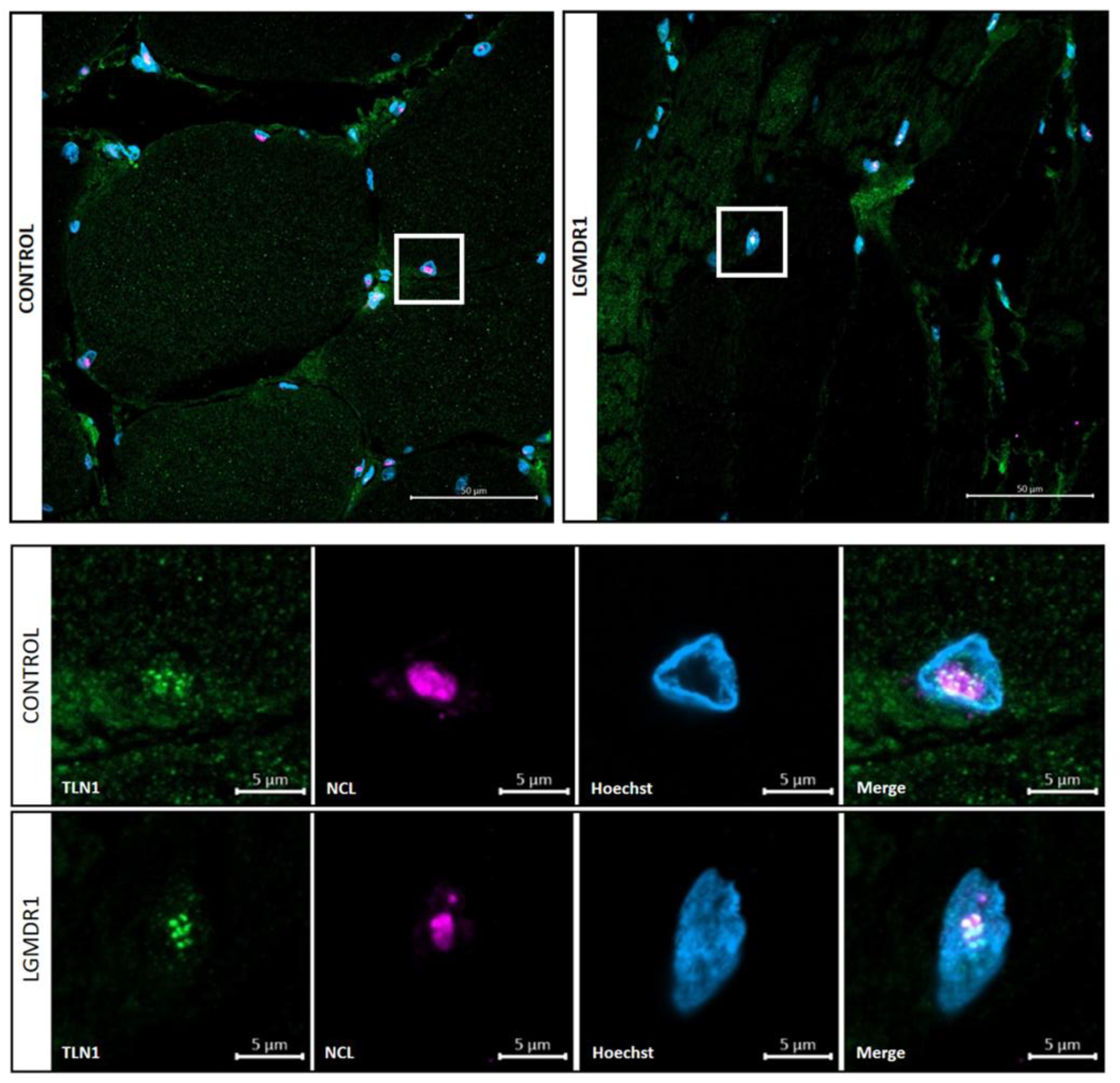
3.4. TLN1 Localizes in the Nucleolus in CD56-, Myoblasts, Myotubes, and Muscle
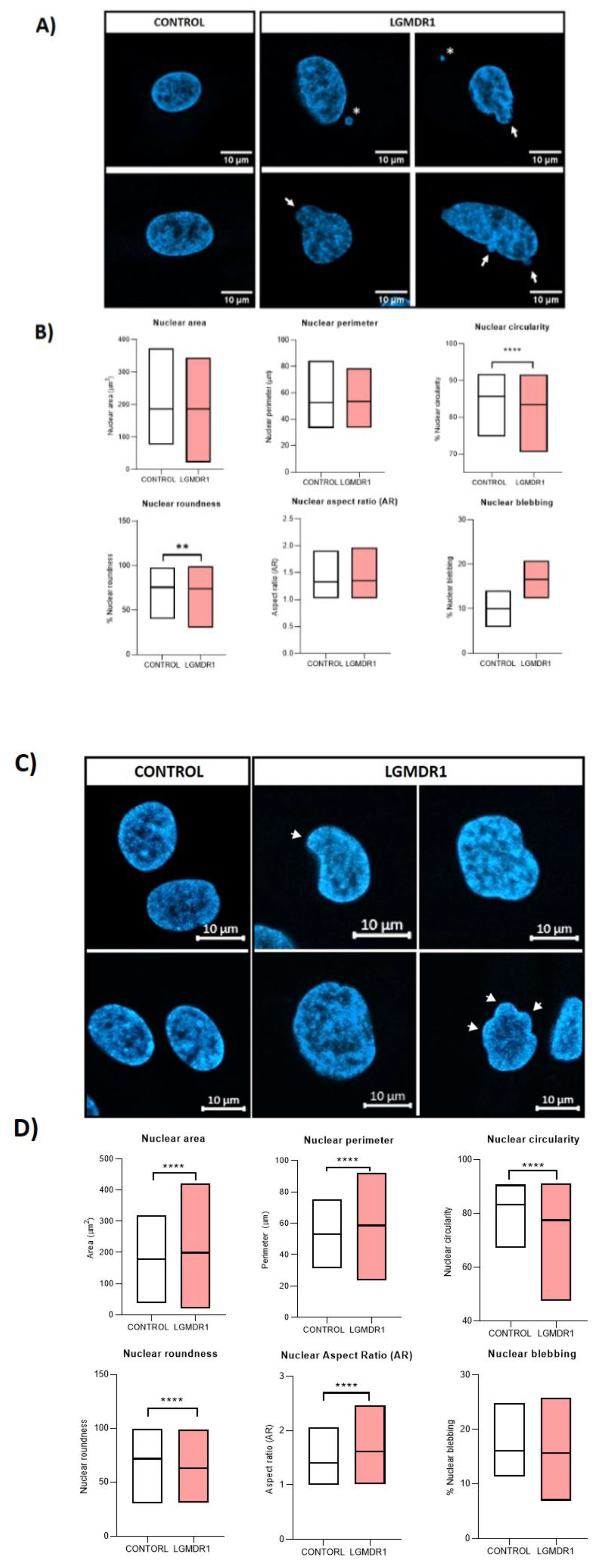
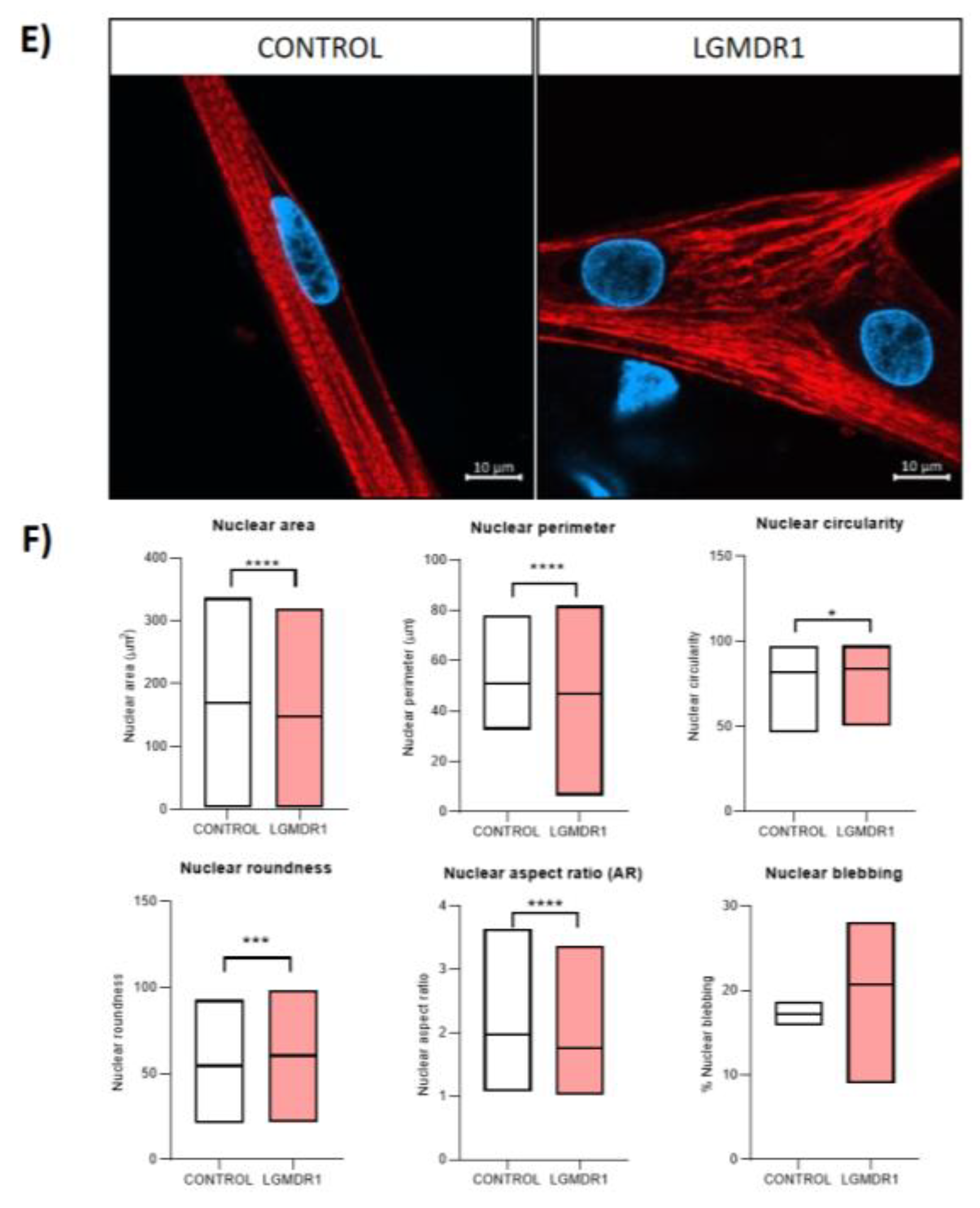
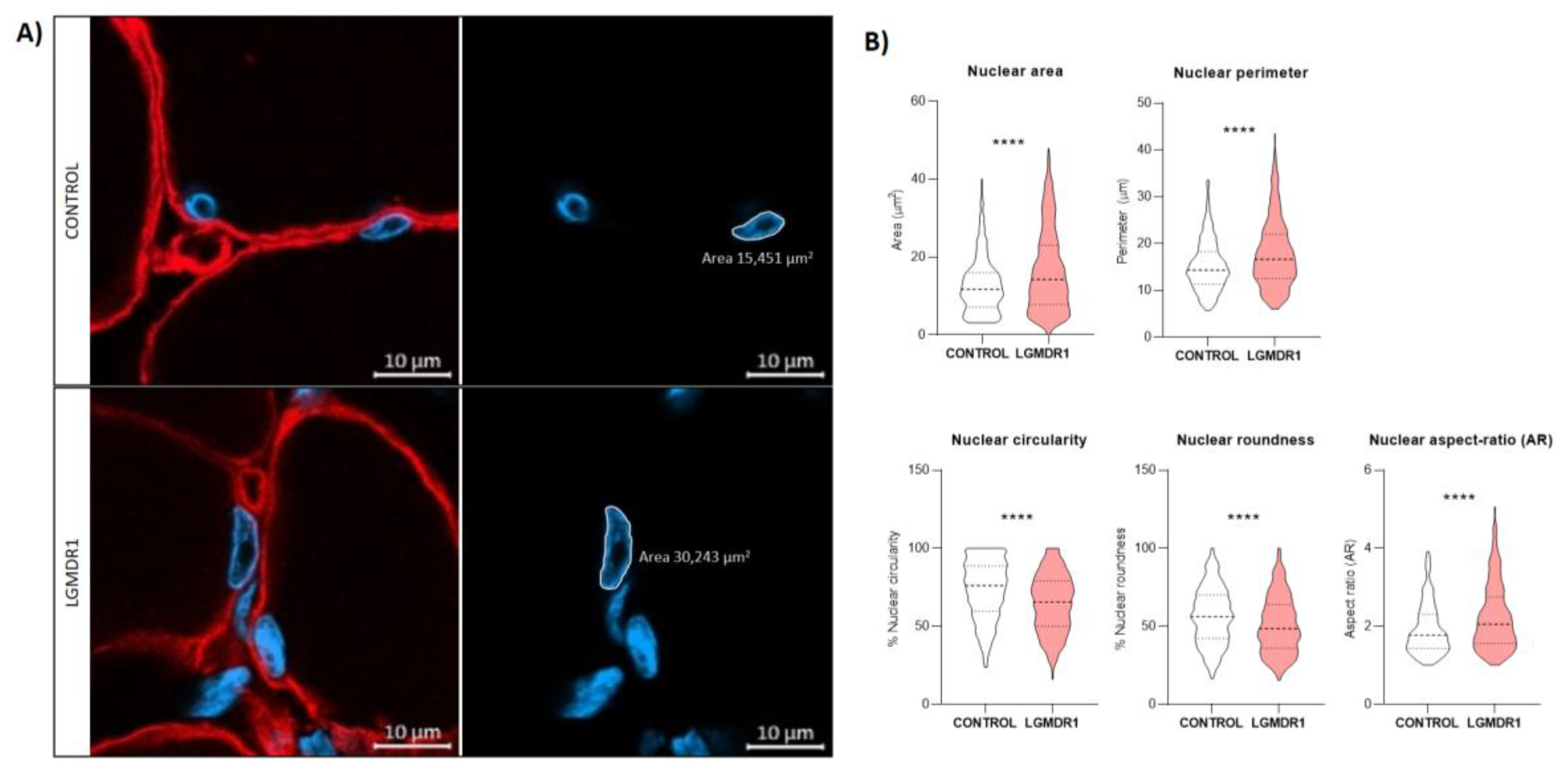
3.5. Nuclear-Abnormal Morphology in LGMDR1 Patients
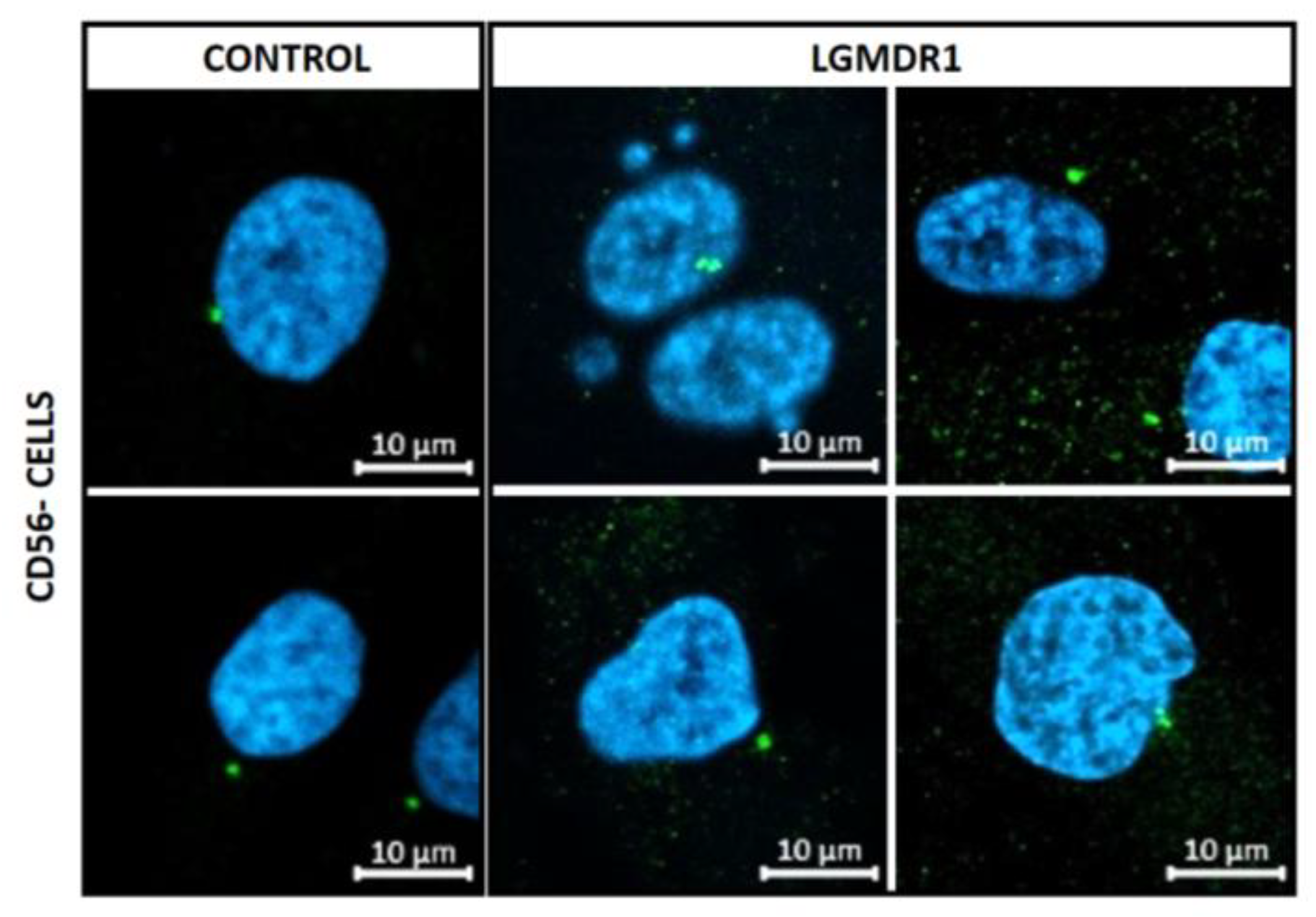
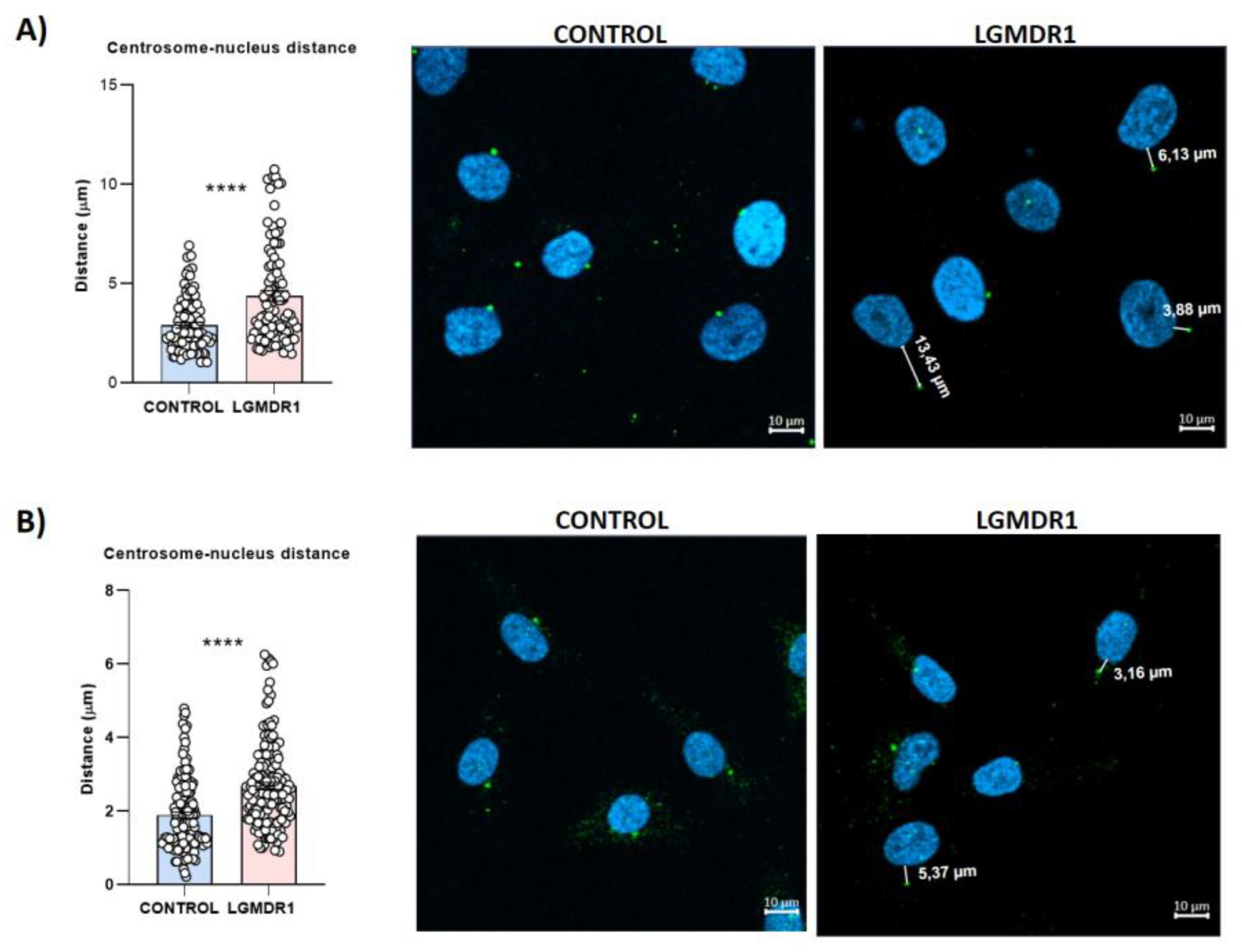
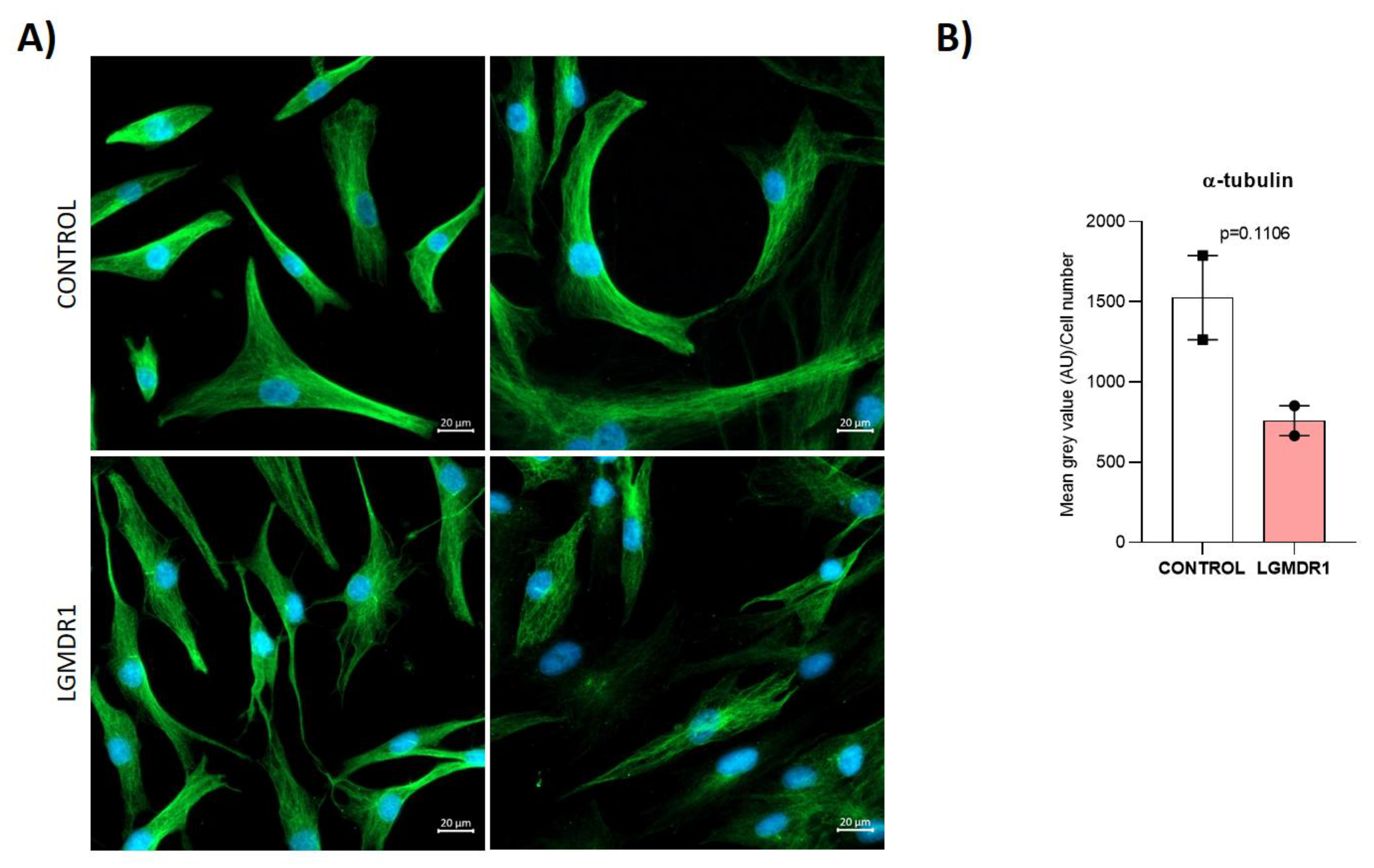
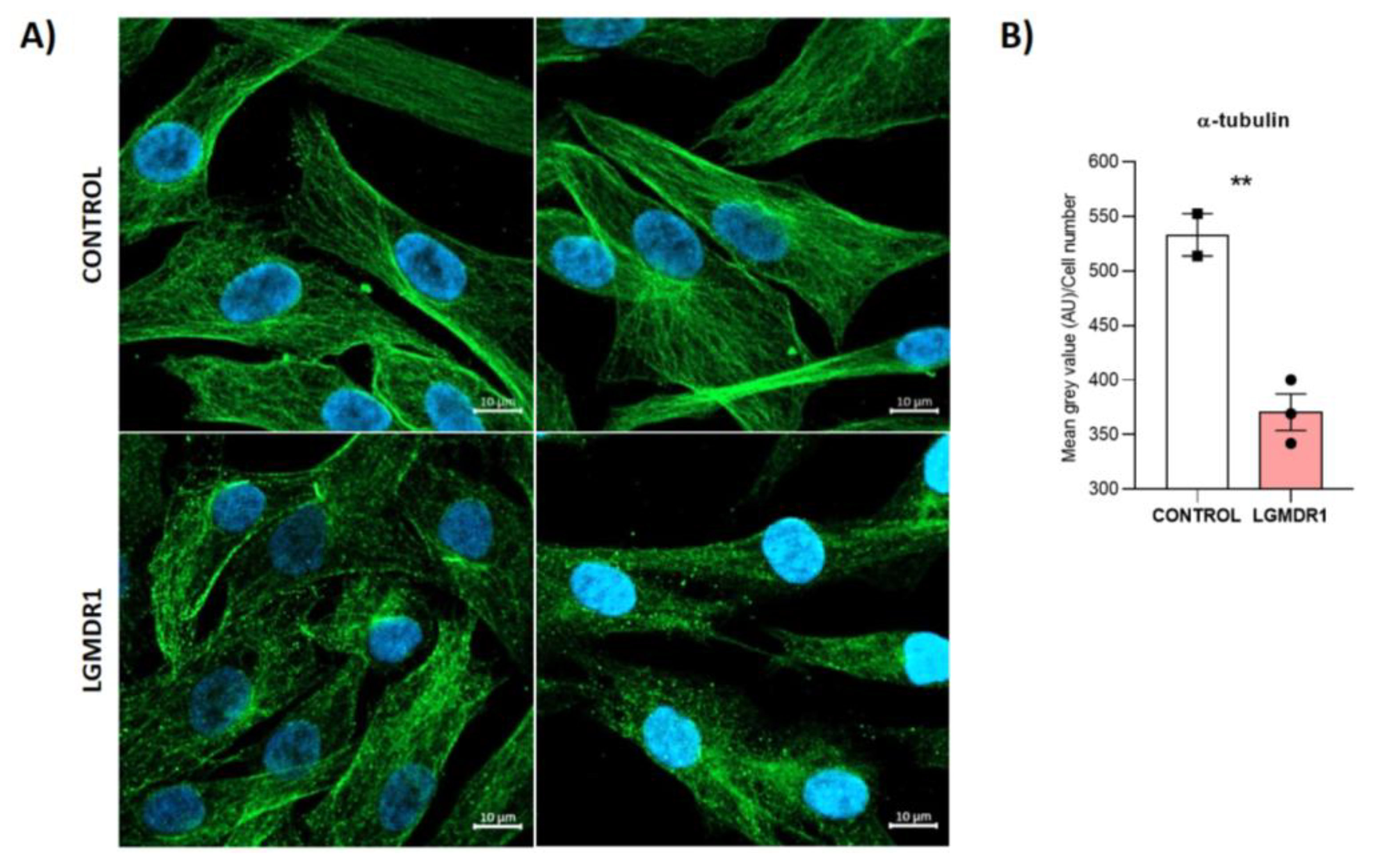
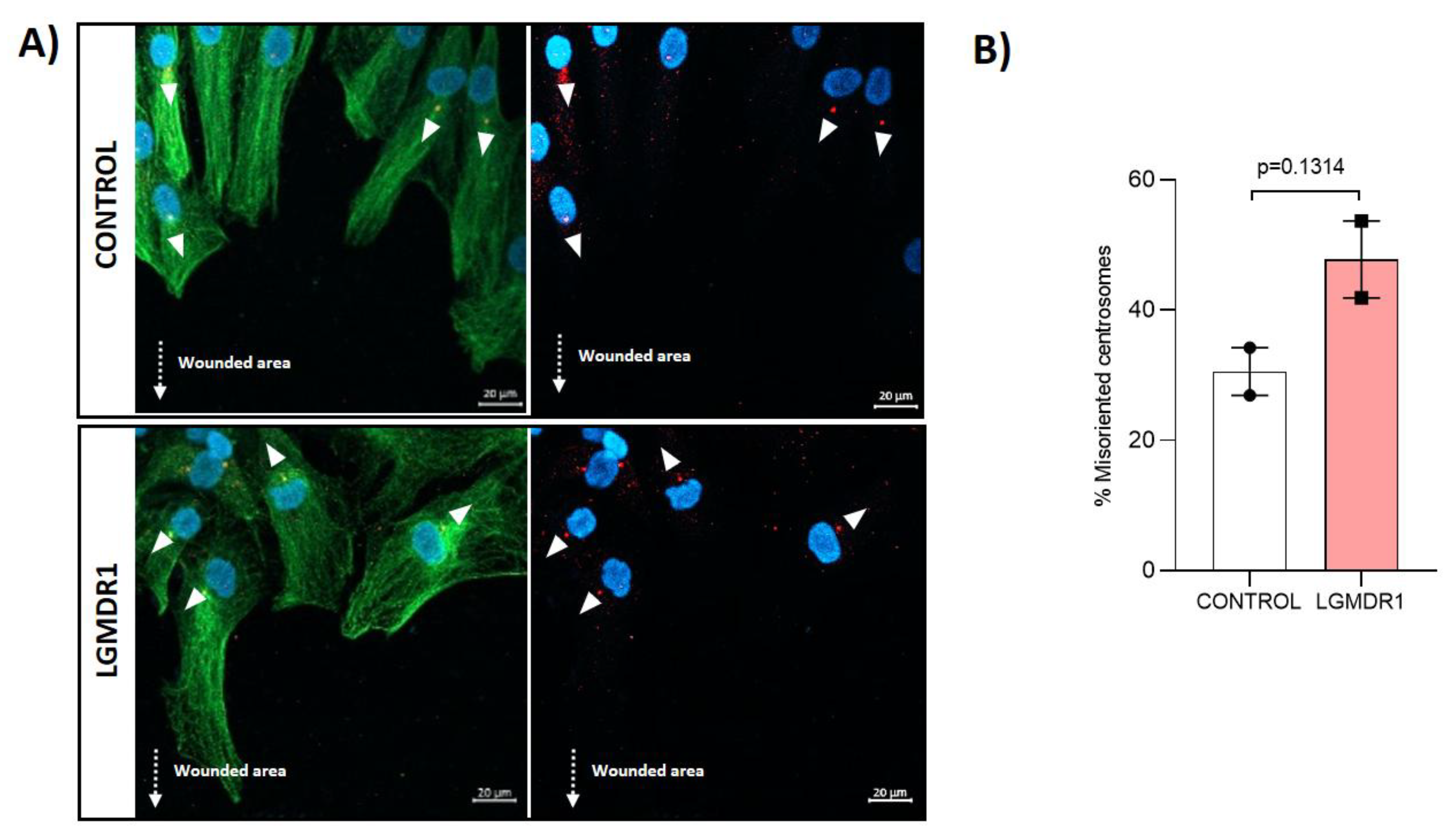
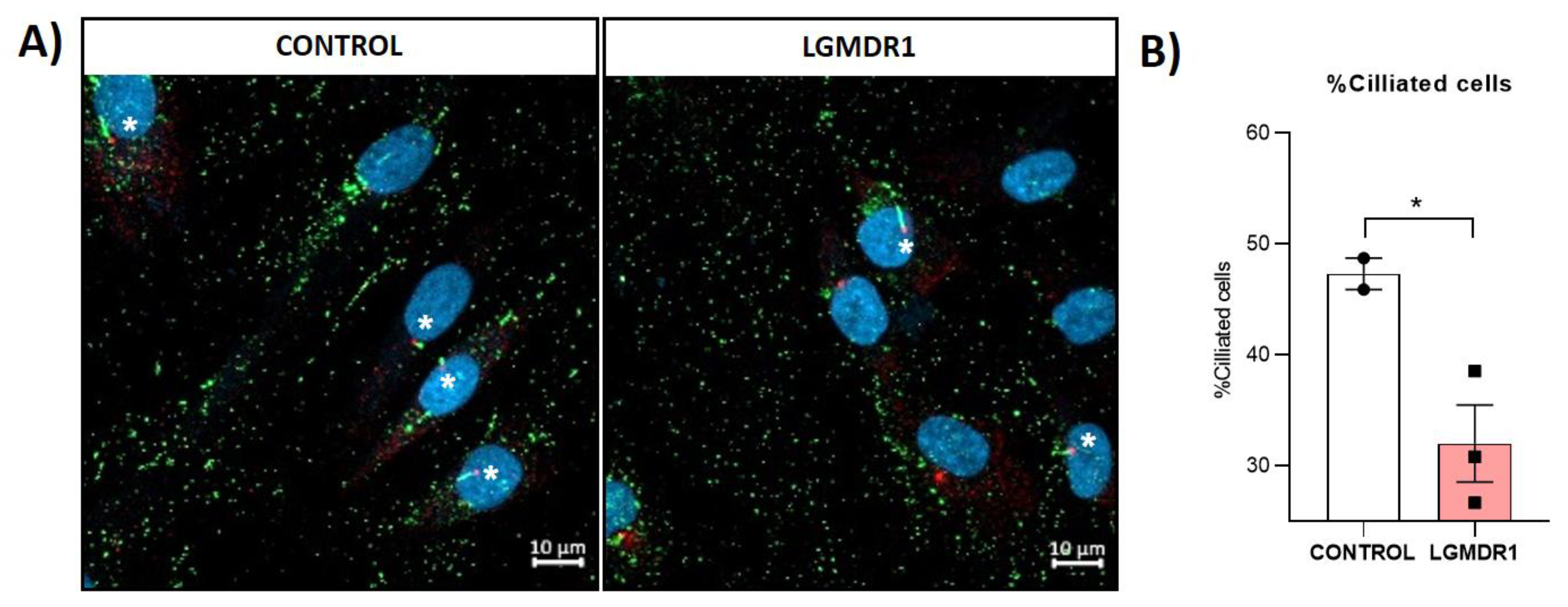
3.6. Centrosome Organization Is Impaired in LGMDR1 Cells
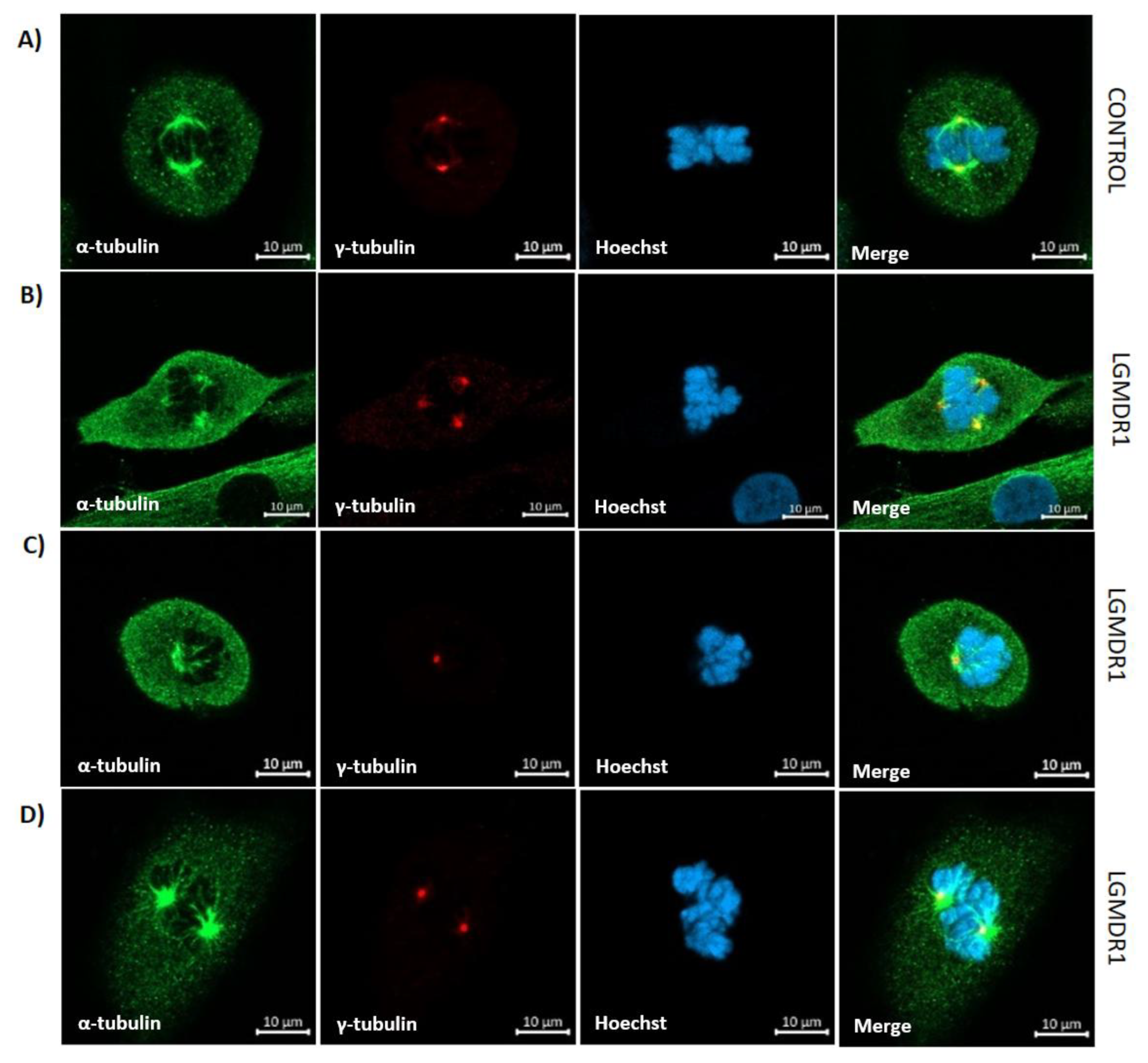
3.7. Aberrant Mitosis in LGMDR1 CD56- Cells
4. Discussion
4.1. Fiber Type Distribution in Healthy Muscle
4.2. Vascular Endothelium Alteration in LGMDR1
4.3. Implication of Altered ITGβ1D Distribution in LGMDR1
Supplementary Materials
Funding
Acknowledgments
References
- Fardeau, M.; Hillaire, D.; Mignard, C.; Feingold, N.; Feingold, J.; Mignard, D.; De Ubeda, B.; Collin, H.; Tomé, F.M.S.; Richard, I.; et al. Juvenile Limb-Girdle Muscular Dystrophy Clinical, Histopathological and Genetic Data from a Small Community Living in the Reunion Island. Brain 1996, 119, 295–308. [CrossRef]
- Urtasun, M.; Sáenz, A.; Roudaut, C.; Poza, J.J.; Urtizberea, J.A.; Cobo, A.M.; Richard, I.; García Bragado, F.; Leturcq, F.; Kaplan, J.C.; et al. Limb-Girdle Muscular Dystrophy in Guipuzcoa (Basque Country, Spain). Brain 1998, 121, 1735–1747. [CrossRef]
- Sorimachi, H.; Imajoh-Ohmi, S.; Emori, Y.; Kawasaki, H.; Ohno, S.; Minami, Y.; Suzuki, K. Molecular Cloning of a Novel Mammalian Calcium-Dependent Protease Distinct from Both m- and μ-Types. Specific Expression of the MRNA in Skeletal Muscle. Journal of Biological Chemistry 1989, 264, 20106–20111. [CrossRef]
- Sorimachi, H.; Toyama-Sorimachi, N.; Saido, T.C.; Kawasaki, H.; Sugita, H.; Miyasaka, M.; Arahata, K.; Ishiura, S.; Suzuki, K. Muscle-Specific Calpain, P94, Is Degraded by Autolysis Immediately after Translation, Resulting in Disappearance from Muscle. J Biol Chem 1993, 268, 10593–10605.
- Sorimachi, H.; Kinbara, K.; Kimura, S.; Takahashi, M.; Ishiura, S.; Sasagawa, N.; Sorimachi, N.; Shimada, H.; Tagawa, K.; Maruyama, K.; et al. Muscle-Specific Calpain, P94, Responsible for Limb Girdle Muscular Dystrophy Type 2A, Associates with Connectin through IS2, a P94-Specific Sequence. Journal of Biological Chemistry 1995, 270, 31158–31162. [CrossRef]
- Kramerova, I.; Kudryashova, E.; Tidball, J.G.; Spencer, M.J. Null Mutation of Calpain 3 (P94) in Mice Causes Abnormal Sarcomere Formation in Vivo and in Vitro. Hum Mol Genet 2004, 13, 1373–1388. [CrossRef]
- Sáenz, A.; Azpitarte, M.; Armañanzas, R.; Leturcq, F.; Alzualde, A.; Inza, I.; García-Bragado, F.; De la Herran, G.; Corcuera, J.; Cabello, A.; et al. Gene Expression Profiling in Limb-Girdle Muscular Dystrophy 2A. PLoS One 2008, 3. [CrossRef]
- Pardo, J. V; Siliciano, J.D.; Craig, S.W. A Vinculin-Containing Cortical Lattice in Skeletal Muscle: Transverse Lattice Elements (“costameres”) Mark Sites of Attachment between Myofibrils and Sarcolemma. Proc Natl Acad Sci U S A 1983, 80, 1008–1012. [CrossRef]
- Danowski, B.A.; Imanaka-Yoshida, K.; Sanger, J.M.; Sanger, J.W. Costameres Are Sites of Force Transmission to the Substratum in Adult Rat Cardiomyocytes. J Cell Biol 1992, 118, 1411–1420. [CrossRef]
- Trimarchi, F.; Favaloro, A.; Fulle, S.; Magaudda, L.; Puglielli, C.; Di Mauro, D. Culture of Human Skeletal Muscle Myoblasts: Timing Appearance and Localization of Dystrophin-Glycoprotein Complex and Vinculin-Talin-Integrin Complex. Cells Tissues Organs 2006, 183, 87–98. [CrossRef]
- Jaka, O.; Casas-Fraile, L.; Azpitarte, M.; Aiastui, A.; López de Munain, A.; Sáenz, A. FRZB and Melusin, Overexpressed in LGMD2A, Regulate Integrin Β1D Isoform Replacement Altering Myoblast Fusion and the Integrin-Signalling Pathway. Expert Rev Mol Med 2017, 19. [CrossRef]
- Pickett-Heaps, J.D. Preprophase Microtubule Bands in Some Abnormal Mitotic Cells of Wheat. J Cell Sci 1969, 4, 397–420. [CrossRef]
- Bornens, M. Centrosome Composition and Microtubule Anchoring Mechanisms. Curr Opin Cell Biol 2002, 14, 25–34. [CrossRef]
- O’Toole, E.; Greenan, G.; Lange, K.I.; Srayko, M.; Müller-Reichert, T. The Role of γ-Tubulin in Centrosomal Microtubule Organization. PLoS One 2012, 7, e29795. [CrossRef]
- Tassin, A.M.; Maro, B.; Bornens, M. Fate of Microtubule-Organizing Centers during Myogenesis in Vitro. J Cell Biol 1985, 100, 35–46. [CrossRef]
- Bugnard, E.; Zaal, K.J.M.; Ralston, E. Reorganization of Microtubule Nucleation during Muscle Differentiation. Cell Motil Cytoskeleton 2005, 60, 1–13. [CrossRef]
- de Andrade Rosa, I.; Corrêa, S.; Costa, M.L.; Mermelstein, C. The Scaffolding Protein Calpain-3 Has Multiple Distributions in Embryonic Chick Muscle Cells and It Is Essential for the Formation of Muscle Fibers. Tissue Cell 2020, 67, 101436. [CrossRef]
- Winter, L.; Kustermann, M.; Ernhofer, B.; Hoger, H.; Bittner, R.E.; Schmidt, W.M. Proteins Implicated in Muscular Dystrophy and Cancer Are Functional Constituents of the Centrosome. Life Sci Alliance 2022, 5, 1–16. [CrossRef]
- Peng, H.; Ong, Y.M.; Shah, W.A.; Holland, P.C.; Carbonetto, S. Integrins Regulate Centrosome Integrity and Astrocyte Polarization Following a Wound. Dev Neurobiol 2013, 73, 333–353. [CrossRef]
- Barker, A.R.; McIntosh, K. V; Dawe, H.R. Centrosome Positioning in Non-Dividing Cells. Protoplasma 2016, 253, 1007–1021. [CrossRef]
- Satir, P. Landmarks in Cilia Research from Leeuwenhoek to US. Cell Motil 1995, 32, 90–94. [CrossRef]
- Satir, P.; Christensen, S.T. Overview of Structure and Function of Mammalian Cilia. Annu Rev Physiol 2007, 69, 377–400. [CrossRef]
- Yuan, S.; Li, J.; Diener, D.R.; Choma, M.A.; Rosenbaum, J.L.; Sun, Z. Target-of-Rapamycin Complex 1 (Torc1) Signaling Modulates Cilia Size and Function through Protein Synthesis Regulation. Proceedings of the National Academy of Sciences 2012, 109, 2021–2026. [CrossRef]
- Rosengren, T.; Larsen, L.J.; Pedersen, L.B.; Christensen, S.T.; Møller, L.B. TSC1 and TSC2 Regulate Cilia Length and Canonical Hedgehog Signaling via Different Mechanisms. Cellular and Molecular Life Sciences 2018, 75, 2663–2680. [CrossRef]
- Anvarian, Z.; Mykytyn, K.; Mukhopadhyay, S.; Pedersen, L.B.; Christensen, S.T. Cellular Signalling by Primary Cilia in Development, Organ Function and Disease. Nat Rev Nephrol 2019, 15, 199–219. [CrossRef]
- Jin, X.; Mohieldin, A.M.; Muntean, B.S.; Green, J.A.; Shah, J. V.; Mykytyn, K.; Nauli, S.M. Cilioplasm Is a Cellular Compartment for Calcium Signaling in Response to Mechanical and Chemical Stimuli. Cellular and Molecular Life Sciences 2014, 71, 2165–2178. [CrossRef]
- Rico, A.; Guembelzu, G.; Palomo, V.; Martínez, A.; Aiastui, A.; Casas-fraile, L.; Valls, A.; de Munain, A.L.; Sáenz, A. Allosteric Modulation of GSK-3β as a New Therapeutic Approach in Limb Girdle Muscular Dystrophy R1 Calpain 3-related. Int J Mol Sci 2021, 22. [CrossRef]
- Ding, F.; Huang, D.; Wang, M.; Peng, J. An 86 Amino Acids Motif in CAPN3 Is Essential for Formation of the Nucleolus-Localized Def-CAPN3 Complex. Biochem Biophys Res Commun 2022, 623, 66–73. [CrossRef]
- Schmidt, W.M.; Uddin, M.H.; Dysek, S.; Moser-Thier, K.; Pirker, C.; Höger, H.; Ambros, I.M.; Ambros, P.F.; Berger, W.; Bittner, R.E. DNA Damage, Somatic Aneuploidy, and Malignant Sarcoma Susceptibility in Muscular Dystrophies. PLoS Genet 2011, 7, e1002042. [CrossRef]
- Teixidó-Travesa, N.; Roig, J.; Lüders, J. The Where, When and How of Microtubule Nucleation – One Ring to Rule Them All. J Cell Sci 2012. [CrossRef]
- Tovey, C.A.; Conduit, P.T. Microtubule Nucleation by γ-Tubulin Complexes and Beyond. Essays Biochem 2018, 62, 765–780. [CrossRef]
- Caspary, T.; Larkins, C.E.; Anderson, K. V The Graded Response to Sonic Hedgehog Depends on Cilia Architecture. Dev Cell 2007, 12, 767–778. [CrossRef]
- Mansour, H.; de Tombe, P.P.; Samarel, A.M.; Russell, B. Restoration of Resting Sarcomere Length After Uniaxial Static Strain Is Regulated by Protein Kinase Cε and Focal Adhesion Kinase. Circ Res 2004, 94, 642–649. [CrossRef]
- Ervasti, J.M. Costameres: The Achilles’ Heel of Herculean Muscle. Journal of Biological Chemistry 2003, 278, 13591–13594. [CrossRef]
- Hodges, B.L.; Kaufman, S.J. Developmental Regulation and Functional Significance of Alternative Splicing of NCAM and 7 1 Integrin in Skeletal Muscle. BAM-PADOVA 1996, 6, 437–448.
- Pardo, J. V; Siliciano, J.D.; Craig, S.W. Vinculin Is a Component of an Extensive Network of Myofibril-Sarcolemma Attachment Regions in Cardiac Muscle Fibers. J Cell Biol 1983, 97, 1081–1088. [CrossRef]
- Anastasi, G.; Cutroneo, G.; Rizzo, G.; Arco, A.; Santoro, G.; Bramanti, P.; Vitetta, A.G.; Pisani, A.; Trimarchi, F.; Favaloro, A. Sarcoglycan and Integrin Localization in Normal Human Skeletal Muscle: A Confocal Laser Scanning Microscope Study. European Journal of Histochemistry 2004, 48, 245–252.
- Pardo, J. V; D’Angelo Siliciano, J.; Craig, S.W. A Vinculin-Containing Cortical Lattice in Skeletal Muscle: Transverse Lattice Elements ('costameres’) Mark Sites of Attachment between Myofibrils and Sarcolemma. Proc Natl Acad Sci U S A 1983, 80, 1008–1012. [CrossRef]
- Korthuis, R.J. Skeletal Muscle Circulation. Colloquium Series on Integrated Systems Physiology: From Molecule to Function 2011, 3, 1–144. [CrossRef]
- Di Tomaso, M.V.; Liddle, P.; Lafon-Hughes, L.; Laura, A.; Folle, G. Chromatin Damage Patterns Shift According to Eu/ Heterochromatin Replication. In The Mechanisms of DNA Replication; InTech, 2013.
- Reverte, C.G.; Benware, A.; Jones, C.W.; LaFlamme, S.E. Perturbing Integrin Function Inhibits Microtubule Growth from Centrosomes, Spindle Assembly, and Cytokinesis. Journal of Cell Biology 2006, 174, 491–497. [CrossRef]
- Ng, D.C.H.; Ho, U.Y.; Grounds, M.D. Cilia, Centrosomes and Skeletal Muscle. Int J Mol Sci 2021, 22, 1–16. [CrossRef]
- Park, R.S.; Légier G., M.F.; Cartwright, J.; Goldstein, M.A. Perinuclear Microtubules in Postnatal Rat Heart. J Morphol 1984, 179, 13–19. [CrossRef]
- Espigat-Georger, A.; Dyachuk, V.; Chemin, C.; Emorine, L.; Merdes, A. Nuclear Alignment in Myotubes Requires Centrosome Proteins Recruited by Nesprin-1. J Cell Sci 2016, 129, 4227–4237. [CrossRef]
- Roman, W.; Gomes, E.R. Nuclear Positioning in Skeletal Muscle. Semin Cell Dev Biol 2018, 82, 51–56. [CrossRef]
- Khodjakov, A.; Cole, R.W.; Oakley, B.R.; Rieder, C.L. Centrosome-Independent Mitotic Spindle Formation in Vertebrates. Current Biology 2000, 10, 59–67. [CrossRef]
- Mathes, S.; Vanmunster, M.; Bloch, W.; Suhr, F. Evidence for Skeletal Muscle Fiber Type-Specific Expressions of Mechanosensors. Cellular and Molecular Life Sciences 2019, 76, 2987–3004. [CrossRef]
- Andresen, B.; de Marees, M.; Schiffer, T.; Bloch, W.; Suhr, F. Skeletal Muscle Fiber Type-Specific Expressions of Mechanosensors Integrin-Linked Kinase, Talin, and Vinculin and Their Modulation by Loading and Environmental Conditions in Humans. FASEB Journal 2022, 36, 1–19. [CrossRef]
- Graham, Z.A.; Gallagher, P.M.; Cardozo, C.P. Focal Adhesion Kinase and Its Role in Skeletal Muscle. J Muscle Res Cell Motil 2015, 36, 305–315. [CrossRef]
- Turner, C.E.; Kramarcy, N.; Sealock, R.; Burridge, K. Localization of Paxillin, a Focal Adhesion Protein, to Smooth Muscle Dense Plaques, and the Myotendinous and Neuromuscular Junctions of Skeletal Muscle. Exp Cell Res 1991, 192, 651–655. [CrossRef]
- Draeger, A.; Amos, W.B.; Ikebe, M.; Small, J. V The Cytoskeletal and Contractile Apparatus of Smooth Muscle: Contraction Bands and Segmentation of the Contractile Elements. Journal of Cell Biology 1990, 111, 2463–2473. [CrossRef]
- Javerzat, S.; Franco, M.; Herbert, J.; Platonova, N.; Peille, A.-L.; Pantesco, V.; De Vos, J.; Assou, S.; Bicknell, R.; Bikfalvi, A.; et al. Correlating Global Gene Regulation to Angiogenesis in the Developing Chick Extra-Embryonic Vascular System. PLoS One 2009, 4, e7856. [CrossRef]
- Sit, B.; Gutmann, D.; Iskratsch, T. Costameres, Dense Plaques and Podosomes: The Cell Matrix Adhesions in Cardiovascular Mechanosensing. J Muscle Res Cell Motil 2019, 40, 197–209. [CrossRef]
- Yamaji, S.; Suzuki, A.; Sugiyama, Y.; Koide, Y.I.; Yoshida, M.; Kanamori, H.; Mohri, H.; Ohno, S.; Ishigatsubo, Y. A Novel Integrin-Linked Kinase-Binding Protein, Affixin, Is Involved in the Early Stage of Cell-Substrate Interaction. Journal of Cell Biology 2001, 153, 1251–1264. [CrossRef]
- Matsuda, C.; Kameyama, K.; Tagawa, K.; Ogawa, M.; Suzuki, A.; Yamaji, S.; Okamoto, H.; Nishino, I.; Hayashi, Y.K. Dysferlin Interacts with Affixin (β-Parvin) at the Sarcolemma. J Neuropathol Exp Neurol 2005, 64, 334–340. [CrossRef]
- Sepulveda, J.L.; Gkretsi, V.; Wu, C. Assembly and Signaling of Adhesion Complexes. In; 2005; pp. 183–225.
- Budatha, M.; Zhang, J.; Schwartz, M.A. Fibronectin-Mediated Inflammatory Signaling Through Integrin A5 in Vascular Remodeling. J Am Heart Assoc 2021, 10. [CrossRef]
- Krahn, M.; Goicoechea, M.; Hanisch, F.; Groen, E.; Bartoli, M.; Pécheux, C.; Garcia-Bragado, F.; Leturcq, F.; Jeannet, P.Y.; Lobrinus, J.A.; et al. Eosinophilic Infiltration Related to CAPN3 Mutations: A Pathophysiological Component of Primary Calpainopathy? Clin Genet 2011, 80, 398–402. [CrossRef]
- Pitter, B.; Werner, A.-C.; Montanez, E. Parvins Are Required for Endothelial Cell–Cell Junctions and Cell Polarity During Embryonic Blood Vessel Formation. Arterioscler Thromb Vasc Biol 2018, 38, 1147–1158. [CrossRef]
- Dalakas, M.C. Immunopathogenesis of Inflammatory Myopathies. Ann Neurol 1995, 37, 74–86. [CrossRef]
- Thompson, C.; Piguet, V.; Choy, E. The Pathogenesis of Dermatomyositis. British Journal of Dermatology 2018, 179, 1256–1262. [CrossRef]
- Palecek, S.P.; Schmidt, C.E.; Lauffenburger, D.A.; Horwitz, A.F. Integrin Dynamics on the Tail Region of Migrating Fibroblasts. J Cell Sci 1996, 109, 941–952. [CrossRef]
- Schwander, M.; Leu, M.; Stumm, M.; Dorchies, O.M.; Ruegg, U.T.; Schittny, J.; Müller, U. Β1 Integrins Regulate Myoblast Fusion and Sarcomere Assembly. Dev Cell 2003, 4, 673–685. [CrossRef]
- Maniotis, A.J.; Chen, C.S.; Ingber, D.E. Demonstration of Mechanical Connections between Integrins, Cytoskeletal Filaments, and Nucleoplasm That Stabilize Nuclear Structure. Proc Natl Acad Sci U S A 1997, 94, 849–854. [CrossRef]
- Thomas, C.H.; Collier, J.H.; Sfeir, C.S.; Healy, K.E. Engineering Gene Expression and Protein Synthesis by Modulation of Nuclear Shape. Proc Natl Acad Sci U S A 2002, 99, 1972–1977. [CrossRef]
- Zhang, X.; Cook, P.C.; Zindy, E.; Williams, C.J.; Jowitt, T.A.; Streuli, C.H.; MacDonald, A.S.; Redondo-Muñoz, J. Integrin A4β1 Controls G9a Activity That Regulates Epigenetic Changes and Nuclear Properties Required for Lymphocyte Migration. Nucleic Acids Res 2015, 44, 3031–3044. [CrossRef]
- Zwerger, M.; Ho, C.Y.; Lammerding, J. Nuclear Mechanics in Disease. Annu Rev Biomed Eng 2011, 13, 397–428. [CrossRef]
- Belkin, A.M.; Zhidkova, N.I.; Balzac, F.; Altruda, F.; Tomatis, D.; Maier, A.; Tarone, G.; Koteliansky, V.E.; Burridge, K. Β1D Integrin Displaces the Β1A Isoform in Striated Muscles: Localization at Junctional Structures and Signaling Potential in Nonmuscle Cells. Journal of Cell Biology 1996, 132, 211–226. [CrossRef]
- Kramerova, I.; Kudryashova, E.; Wu, B.; Spencer, M.J. Regulation of the M-Cadherin-β-Catenin Complex by Calpain 3 during Terminal Stages of Myogenic Differentiation. Mol Cell Biol 2006, 26, 8437–8447. [CrossRef]
- Papageorgiou, A.P.; Heymans, S. Interactions between the Extracellular Matrix and Inflammation during Viral Myocarditis. Immunobiology 2012, 217, 503–510. [CrossRef]
- Gaudet, A.D.; Popovich, P.G. Extracellular Matrix Regulation of Inflammation in the Healthy and Injured Spinal Cord. Exp Neurol 2014, 258, 24–34. [CrossRef]
- Bollyky, P.L.; Bogdani, M.; Bollyky, J.B.; Hull, R.L.; Wight, T.N. The Role of Hyaluronan and the Extracellular Matrix in Islet Inflammation and Immune Regulation. Curr Diab Rep 2012, 12, 471–480. [CrossRef]
- Yun, S.; Budatha, M.; Dahlman, J.E.; Coon, B.G.; Ryan, T.; Langer, R.; Anderson, D.G.; Baillie, G.; Martin, A. Inflammatory Signalling. 2017, 18, 1043–1053.
- Yurdagul, A.; Green, J.; Albert, P.; McInnis, M.C.; Mazar, A.P.; Orr, A.W. A5Β1 Integrin Signaling Mediates Oxidized Low-Density Lipoprotein-Induced Inflammation and Early Atherosclerosis. Arterioscler Thromb Vasc Biol 2014, 34, 1362–1373. [CrossRef]
- Orr, A.W.; Ginsberg, M.H.; Shattil, S.J.; Deckmyn, H.; Schwartz, M.A. Matrix-Specific Suppression of Integrin Activation in Shear Stress Signaling. Mol Biol Cell 2006, 17, 4686–4697. [CrossRef]
- Tejeda-Muñoz, N.; Morselli, M.; Moriyama, Y.; Sheladiya, P.; Pellegrini, M.; De Robertis, E.M. Canonical Wnt Signaling Induces Focal Adhesion and Integrin Beta-1 Endocytosis. iScience 2022, 25, 104123. [CrossRef]
- Hynes, R.O. Integrins. Cell 2002, 110, 673–687. [CrossRef]
- Calderwood, D.A.; Zent, R.; Grant, R.; Rees, D.J.G.; Hynes, R.O.; Ginsberg, M.H. The Talin Head Domain Binds to Integrin β Subunit Cytoplasmic Tails and Regulates Integrin Activation. Journal of Biological Chemistry 1999, 274, 28071–28074. [CrossRef]
- Tadokoro, S.; Shattil, S.J.; Eto, K.; Tai, V.; Liddington, R.C.; De Pereda, J.M.; Ginsberg, M.H.; Calderwood, D.A. Talin Binding to Integrin β Tails: A Final Common Step in Integrin Activation. Science (1979) 2003, 302, 103–106. [CrossRef]
- Hu, Y.L.; Lu, S.; Szeto, K.W.; Sun, J.; Wang, Y.; Lasheras, J.C.; Chien, S. FAK and Paxillin Dynamics at Focal Adhesions in the Protrusions of Migrating Cells. Sci Rep 2014, 4, 1–7. [CrossRef]
- Bellis, S.L.; Miller, J.T.; Turner, C.E. Characterization of Tyrosine Phosphorylation of Paxillin in Vitro by Focal Adhesion Kinase. Journal of Biological Chemistry 1995, 270, 17437–17441. [CrossRef]
- Boppart, M.D.; Mahmassani, Z.S. Integrin Signaling: Linking Mechanical Stimulation to Skeletal Muscle Hypertrophy. Am J Physiol Cell Physiol 2019, 317, C629–C641. [CrossRef]
- Hour, T.C.; Lan Nhi, N.T.; Lai, I.J.; Chuu, C.P.; Lin, P.C.; Chang, H.W.; Su, Y.F.; Chen, C.H.; Chen, Y.K. Kaempferol-Enhanced Migration and Differentiation of C2C12 Myoblasts via ITG1B/FAK/Paxillin and IGF1R/AKT/MTOR Signaling Pathways. Mol Nutr Food Res 2024, 68, 1–14. [CrossRef]
- Zhu, W.; Wu, C.; Liu, Z.; Zhao, S.; Cheng, X.; Huang, J. SULF1 Regulates Malignant Progression of Colorectal Cancer by Modulating ARSH via FAK/PI3K/AKT/MTOR Signaling. Cancer Cell Int 2024, 24, 1–19. [CrossRef]
- Guilak, F.; Tedrow, J.R.; Burgkart, R. Viscoelastic Properties of the Cell Nucleus. Biochem Biophys Res Commun 2000, 269, 781–786. [CrossRef]
- Rowat, A.C.; Lammerding, J.; Herrmann, H.; Aebi, U. Towards an Integrated Understanding of the Structure and Mechanics of the Cell Nucleus. BioEssays 2008, 30, 226–236. [CrossRef]
- Silva, A.J. Da; Hästbacka, H.S.E.; Puustinen, M.C.; Pessa, J.C.; Goult, B.T.; Jacquemet, G.; Henriksson, E.; Sistonen, L. A Subpopulation of Talin 1 Resides in the Nucleus and Regulates Gene Expression. bioRxiv preprint 2022, 2022.03.15.484419.
- Rubbi, C.P.; Milner, J. Disruption of the Nuclear Organisation of P53 in Response to DNA Stress. Embo 2003, 22, 6068–6077.
- Yang, K.; Yang, J.; Yi, J. Nucleolar Stress: Hallmarks, Sensing Mechanism and Diseases. Cell Stress 2018, 2, 125–140. [CrossRef]
- Goudarzi, K.M.; Nistér, M.; Lindström, M.S. MTOR Inhibitors Blunt the P53 Response to Nucleolar Stress by Regulating RPL11 and MDM2 Levels. Cancer Biol Ther 2014, 15, 1499–1514. [CrossRef]
- Rico, A.; Valls, A.; Guembelzu, G.; Azpitarte, M.; Aiastui, A.; Zufiria, M.; Jaka, O.; López de Munain, A.; Sáenz, A. Altered Expression of Proteins Involved in Metabolism in LGMDR1 Muscle Is Lost in Cell Culture Conditions. Orphanet J Rare Dis 2023, 18, 315. [CrossRef]
- LaFlamme, S.E.; Mathew-Steiner, S.; Singh, N.; Colello-Borges, D.; Nieves, B. Integrin and Microtubule Crosstalk in the Regulation of Cellular Processes. Cellular and Molecular Life Sciences 2018, 75, 4177–4185. [CrossRef]
- Taranum, S.; Vaylann, E.; Meinke, P.; Abraham, S.; Yang, L.; Neumann, S.; Karakesisoglou, I.; Wehnert, M.; Noegel, A.A. LINC Complex Alterations in DMD and EDMD/CMT Fibroblasts. Eur J Cell Biol 2012, 91, 614–628. [CrossRef]
- Oakley, B.R. An Abundance of Tubulins. Trends Cell Biol 2000, 10, 537–542. [CrossRef]
- Joseph, R.; Robinson, M.L.; Lambert, L.; Srivastava, O.P. Lens-Specific ΒA3/A1-Conditional Knockout Mice: Phenotypic Characteristics and Calpain Activation Causing Protein Degradation and Insolubilization. PLoS One 2023, 18, 1–24. [CrossRef]
- Piemonte, K.M.; Anstine, L.J.; Keri, R.A. Centrosome Aberrations as Drivers of Chromosomal Instability in Breast Cancer. Endocrinology (United States) 2021, 162, 1–14. [CrossRef]
| Biopsy Number | Gender | Status | Mutation 1 | Mutation 2 | Muscle | Age # | Age at onset | Functional Status |
Clinical information |
|---|---|---|---|---|---|---|---|---|---|
| 22-09 | Male | Control | - | - | Biceps | 47 | - | - | - |
| 22-18 | Male | Control | - | - | Semitendinosus | 24 | - | - | - |
| 17-08051 | Female | Control | - | - | Quadriceps | 46 | - | - | - |
| 23-10 | Male | Control | - | - | Deltoid | 75 | - | - | - |
| 16-6279 | Male | Control | - | - | Quadriceps | 35 | - | - | - |
| 19-22 | Female | LGMDR1 | p.(Arg788Serfs*14) | Complete deletion of CANP3 gene | Biceps | 23 | 23 | Ambulant | Benign |
| 21-07396 | Male | LGMDR1 | p.(Arg490Trp) | p.(Arg490Trp) | Tibialis anterior | 46 | 33 | Ambulant | Mild facial weakness. Proximal weakness. Bilateral scapular winging. Bilateral atrophy of biceps and pectoral muscles. |
| B09-83 | Female |
LGMDR1 |
DelEx2-6 | DelEx2-6 | Quadriceps | 12 | 12 | Ambulant | Proximal weakness. Not able to climb stairs. |
| 18-38110 | Female | LGMDR1 | p.(Arg489Gln) | c.1116-2A>C | Biceps | 47 | Fifth decade | Ambulant | First symptoms were myalgia and fatigue, with persistently elevated CK, X9 (1800 UI/l). No muscle weakness at 52 year old. Unspecific slight changes in the biopsy. |
|
97-168 |
Male | LGMDR1 | p.(Ser479Gly) | c.1992+1G>T | N.A. | 41 | 20 | Ambulant | Muscle weakness of the pelvic and scapular girdles. |
| BIOPSY N. | STATUS | MUSCLE | AGE | MUTATION 1 | MUTATION 2 |
|---|---|---|---|---|---|
| 15-12 | CONTROL | Deltoid | 36 | - | - |
| 13-07 | CONTROL | Deltoid | 36 | - | - |
| 13-09 | CONTROL | Vastus lateralis | 37 | - | - |
| 22-18 | CONTROL | Semitendinosus | 24 | - | - |
| 23-10 | CONTROL | Deltoid | 75 | - | - |
| 10-39 | LGMDR1 | Deltoid | 29 | p.(Lys254del) | p.(X822Leuext62X) |
| 09-21 | LGMDR1 | Biceps | 19 | p.(His690Argfs*9) | p.(His690Argfs*9) |
| 09-25 | LGMDR1 | Deltoid | 28 | p.(Lys254Glu) | p.(Pro637HisfsX25) |
| Exp05 | LGMDR1 | Deltoid | 13 | p.(Arg788SersX14) | p.(Arg788SersX14) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





