Materials and methods
198 persons were included in the case-control molecular genetic study. The study was conducted at the clinical bases of Polyclinics No. 3 RSE, Polyclinics No. 5 RSE, Multidisciplinary Hospital No. 1 RSE, Multidisciplinary Hospital No. 2 RSE in Karaganda. The main group included patients with AF (n=75), the control group consisted of 2 subgroups: subgroup 1 (control group 1) included conditionally healthy patients (n=73), subgroup 2 (control group 2) consisted of patients with arterial hypertension (AH) and coronary heart disease (CHD) without diagnosed AF at the time of inclusion in the study (n=50). The scientific study was approved by the Ethics Committee of Karaganda Medical University NC JSC (Protocol No. 14 dated 14.04.2020). All participants were included in the study after the informed consent signing.
At the first stage, the analysis of anamnestic data, medical documentation, results of such clinical research methods as electrocardiography (ECG), Holter ECG monitoring, echocardiography (ECHOCG), selective coronary angiography (SCA), laboratory research methods were carried out. In order to assess the risk factors for AF, the survey of patients included in the study was conducted. The questionnaire was developed based on the recommendations of the European Society of Cardiology (ESC) together with the European Association of Cardiothoracic Surgeons (EACTS) [85] (Appendix A) and included questions to identify clinical risk factors. The copyright for the development of the «Questionnaire for risk factors assessing at atrial fibrillation» is attested in the State Register of Rights to Copyrighted Objects dated 16.11.2020 No. 13249.
The study included patients with AF at the age from 26 to 84 years. The average age of patients in the main group was 66.76±9.13 (CI 64.66-68.86). 57.3% participants of the main group were male and 42.7% – female. The largest group consisted of patients of the age group 61-70 years (n=27 (36%), among whom 39.5% were male patients. Among female , the largest percentage (17%) were patients aged 71-80 years. The average age in the control group 1 was 54.32±8.63, in the control group 2 – 64.96±9.74.
All patients with AF had episodes of this rhythm disorder lasting more than 30 seconds in the anamnesis or at the time of examination. Such episodes were recorded with ECG or Holter ECG monitoring. The types of atrial fibrillation were determined according to the Clinical Guidelines for the Diagnosis and Treatment of Atrial Fibrillation ESC 2016 [
2].
Patients with permanent AF accounted for 63% (n=47). Clinically, AF was manifested in paroxysms form in 20% (n=15) cases and it was persistent in 17% cases (n=13). The study included patients with the average score of 3.05 on the CHA2DS2-VAS scale, most of them had 2 points (n=23 (30.7%). More than 3 points on the scale were noted in 40% (n=30). The study included 50.7% (n=38) patients with chronic heart failure (CHF) with FC ≤ I, 29.3% (n=22) – with FCII, 21.3% (n=16) – with CHF FC III. The decrease in the left ventricular ejection fraction (LVEF) of less than 40% was registered in 21.3% (n=16) patients, LVEF ≥50% – in 50.7% (n=38). According to echocardiography, an increase in the left atrium (LA) of more than 50 mm in diameter was recorded in 56% (n=42) cases. In addition, the increase in the left ventricle of the heart was observed in 20%cases, while in 16% (n=12), normal dimensions of the heart chambers were on ultrasound examination of the heart.
DNA isolation was carried out in Shared Laboratory the Scientific Research Center of Karaganda Medical University NC JSC. The sampling was carried out in accordance with the standards of operational procedures (SOP) in the treatment rooms of medical organizations. 5-6 ml of whole blood was taken from the ulnar vein into vacuum tubes containing EDTA. Until the DNA was isolated, the collected blood was frozen and stored at the temperature of +4-5 0C. DNA was isolated using the salting technique in 2 stages. At the first stage, isolation from blood nuclei was carried out. To do this, 3-6 ml of blood taken from EDTA was added to 9 volumes of well-stirred buffer A in 50 ml test tube. Further, it was centrifuged for 15 minutes at the speed of 2500 rmp at the temperature of 4 0C, after which the supernatant was carefully drained. The core sediment was resuspended in 5 ml of buffer B and transferred to 15 ml polypropylene centrifuge tubes with the volume. Then 500 µl of 10% SDS and 55 µl of proteinase K were added (stock 10 mg/ml). After that the samples were incubated at 37 0C overnight. At the second stage, 1.4 ml of saturated NaCl solution (corresponding to 6 M) was added to each tube and mixed well for 10 minutes. Then the tubes were centrifuged at the rate of 2500 rmp at the temperature of 4 0C on the low-speed centrifuge Beckman. The supernatant was transferred to 15 ml propylene tubes, leaving proteins precipitate. 0.6 volume of chemically pure isopropanol by room temperature were added to each tube. After that the tubes were turned over for several times until the DNA precipitate became visible. DNA was extracted using plastic spatula or tip spout into epindorphs, after which it was left in incubator at 37 0C for 20-30 minutes. Next, DNA was dissolved in 100-200 µl of TE buffer for 2 hours at the temperature of 37 0C. The DNA concentration was quantitatively measured using spectrophotometric method on the NanoDrop1000 Thermoscientific spectrophotometer. After measuring the concentration, the DNA was diluted with H2O to the working concentration of 50 ng/µl.
The analysis of the rs 3903239 polymorphism of the PRRX1 gene was carried out by genotyping method based on polymerase chain reaction using CFX 96 DNA amplifier (Bio-Rad, USA), detecting in real time. To prepare the samples, the set of TaqMan
™SNP Genotyping Assay primers (Applied Biosystems
™, USA) and the commercial PCR mixture Applied Biosystems
™ TaqMan
™Universal PCR Master Mix were used. The TaqMan
™Universal PCR Master Mix was stored at the temperature of 2-8
0C, and the SNP Genotyping Assay Mix, 20X (the mix of primers and a probe) was stored at -20
0C. The set of reagents for PCR was removed from the freezer 20 minutes before the preparation of the working amplification mixture, the contents were defrosted. Test tubes with the reaction mixture and the set of primers (probe) were first centrifuged on microcentrifuge Vortex to drop droplets from the lid to the bottom, shaken to mix the contents and then centrifuged again. Disposable tips were used to apply reagents, DNA samples and control samples into test tubes. All PCR components were carefully mixed, the drops from the lid were deposited by brief centrifugation for 5 seconds. The components of the reaction mixture were added into the test tube, mixing thoroughly. The protocol of the reaction mixture for real-time PCR per sample is presented in
Table 1.
The mixture was applied into the wells of the tablet, preventing the formation of air bubbles in the mixture. The DNA matrix (1 µl each) was inserted into the specified wells, and the tablet (strip) was sealed with the optical film. To discharge droplets from the walls, the tablet was centrifuged for 5 seconds. The tablet (strip) was installed into the amplifier, noting the location and characteristics of the samples in the program, selecting the working dye (FAM, VIC), and then the PCR program running. The application program is shown in
Table 2.
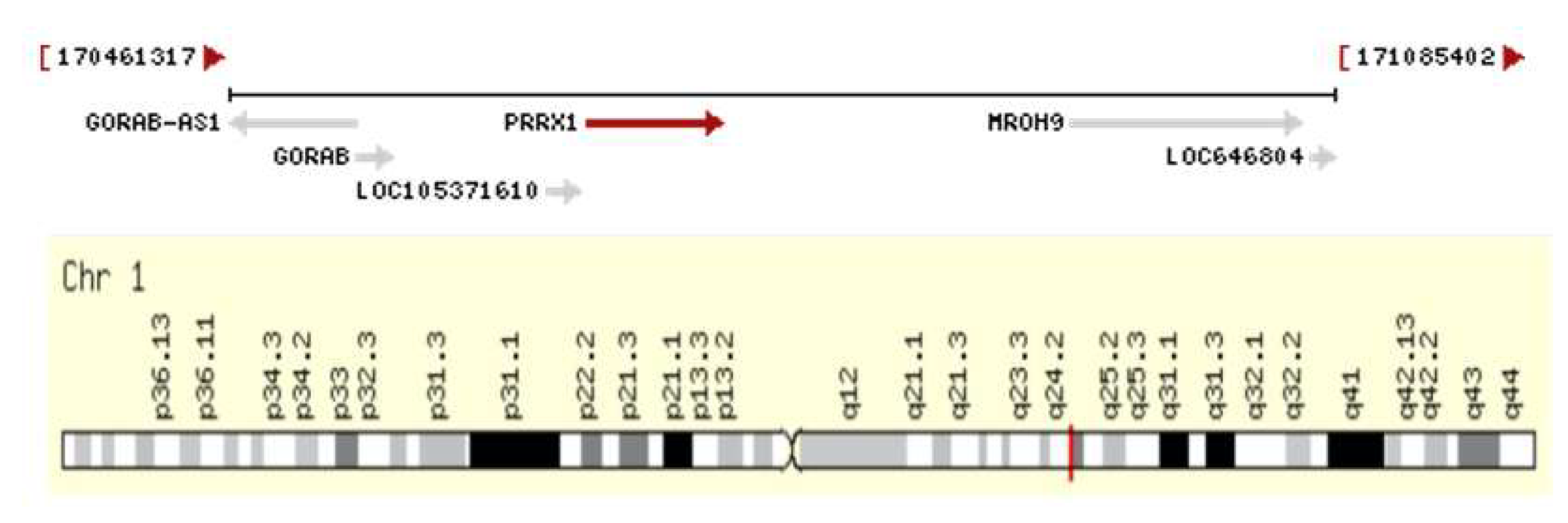
In order to find the association of genes polymorphism with AF in the Kazakh population, we choose the rs 3903239 polymorphism, identified on the 1q24 chromosome of the PRRX gene (
Figure 1). The DNA-associated protein encoded by this gene is a member of the paired family of homeobox localized in the nucleus. The protein acts as a transcription coactivator, enhancing the DNA-binding activity of serum response factor, the protein necessary for gene induction by growth and differentiation factors. The protein regulates creatine kinases in muscles, which indicates its role in the formation of various types of mesodermal muscles. According to the GeneCards.org database, the PRRX1 gene encodes the homeodomain transcription factor highly expressed in the developing heart, especially in connective tissue.
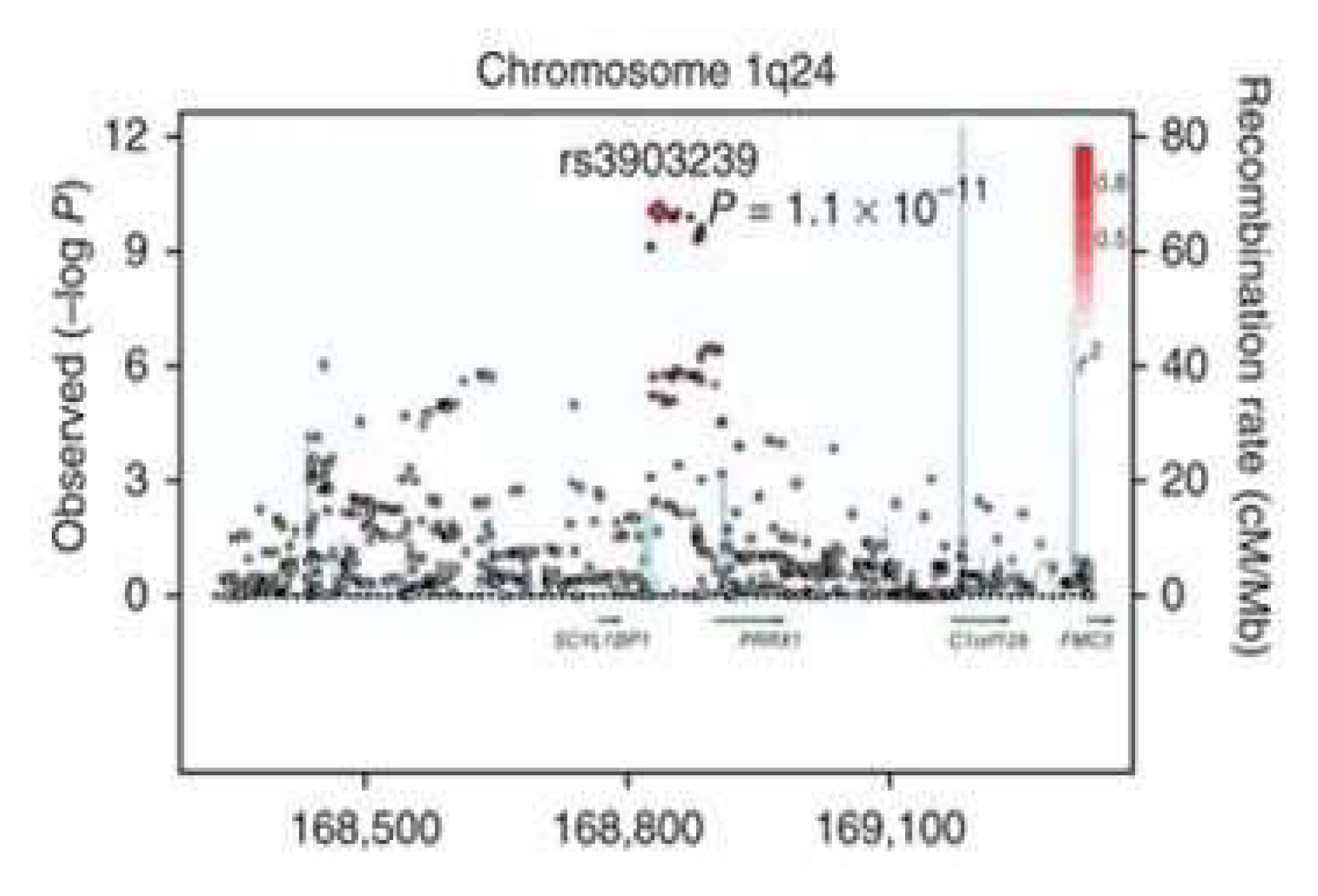
The rs 3903239 polymorphism was identified as a result of GWAS meta-analysis conducted in 2012 [
6]. According to the results of this meta-analysis, this SNP showed the significant association on the 1q24 chromosome (rs3903239; total P = 8.4×10-14) of the PRRX gene (
Figure 2).
The analysis of the results was carried out using the Bio-Rad CFX Manager 2 software (
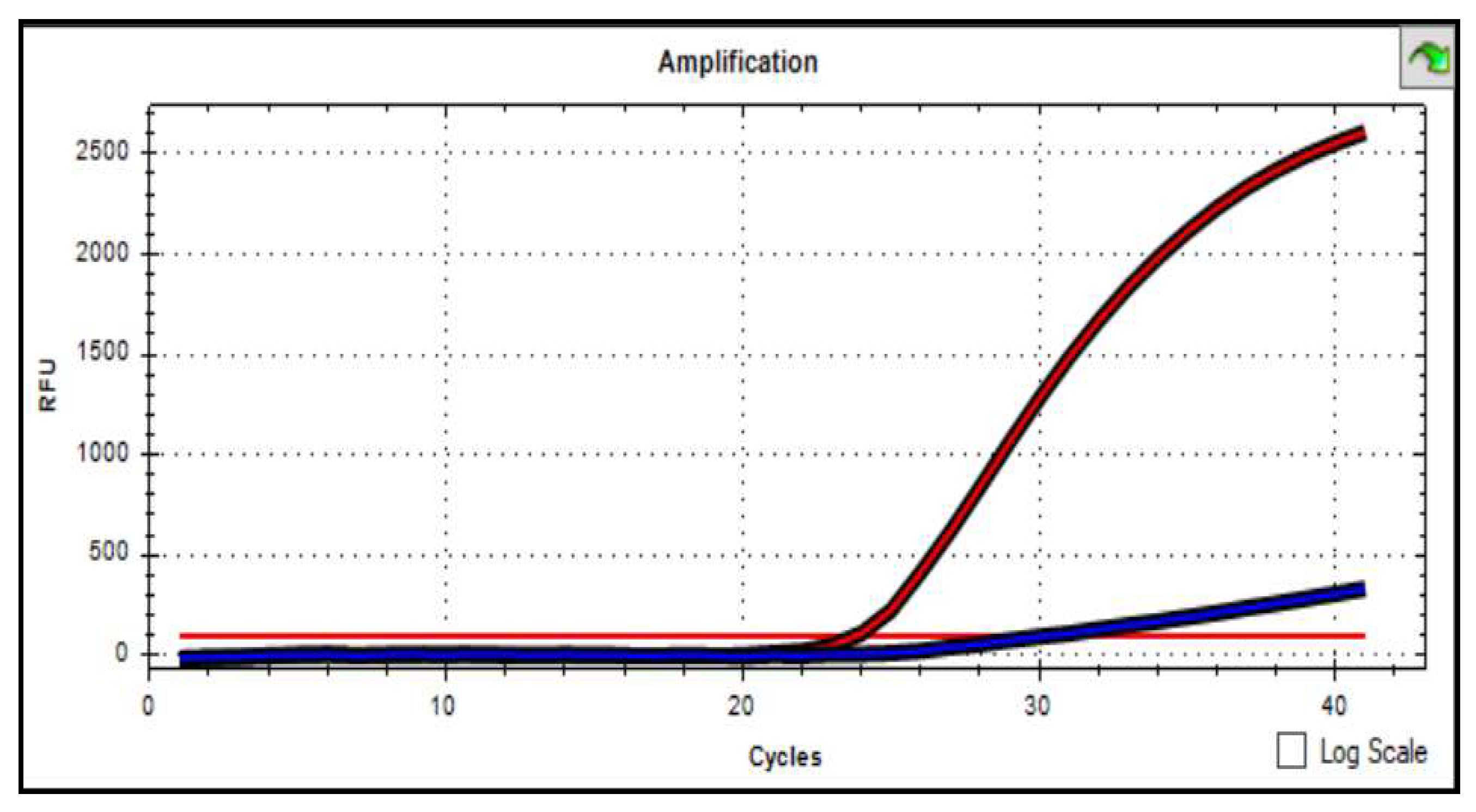
Figure 3). Based on the automatic detection of the accumulation level of PCR products by the device, accumulation curves were constructed along two corresponding channels of fluorophores. As a result, curves of different colors were built for one sample, the FAM fluorescence level curve was displayed in blue, and the VIC curve was displayed in red. For polymorphism in the PRXX1 gene (rs 3903229 (A>G) homozygous genotype of the «wild» type, the VIC fluorescence level increased in the graph, but there was no increase in FAM (
Graph 1). Conversely, if the FAM fluorescence level increased, but VIC did not increase, then this genotype was regarded as homozygous for the «mutant» type (
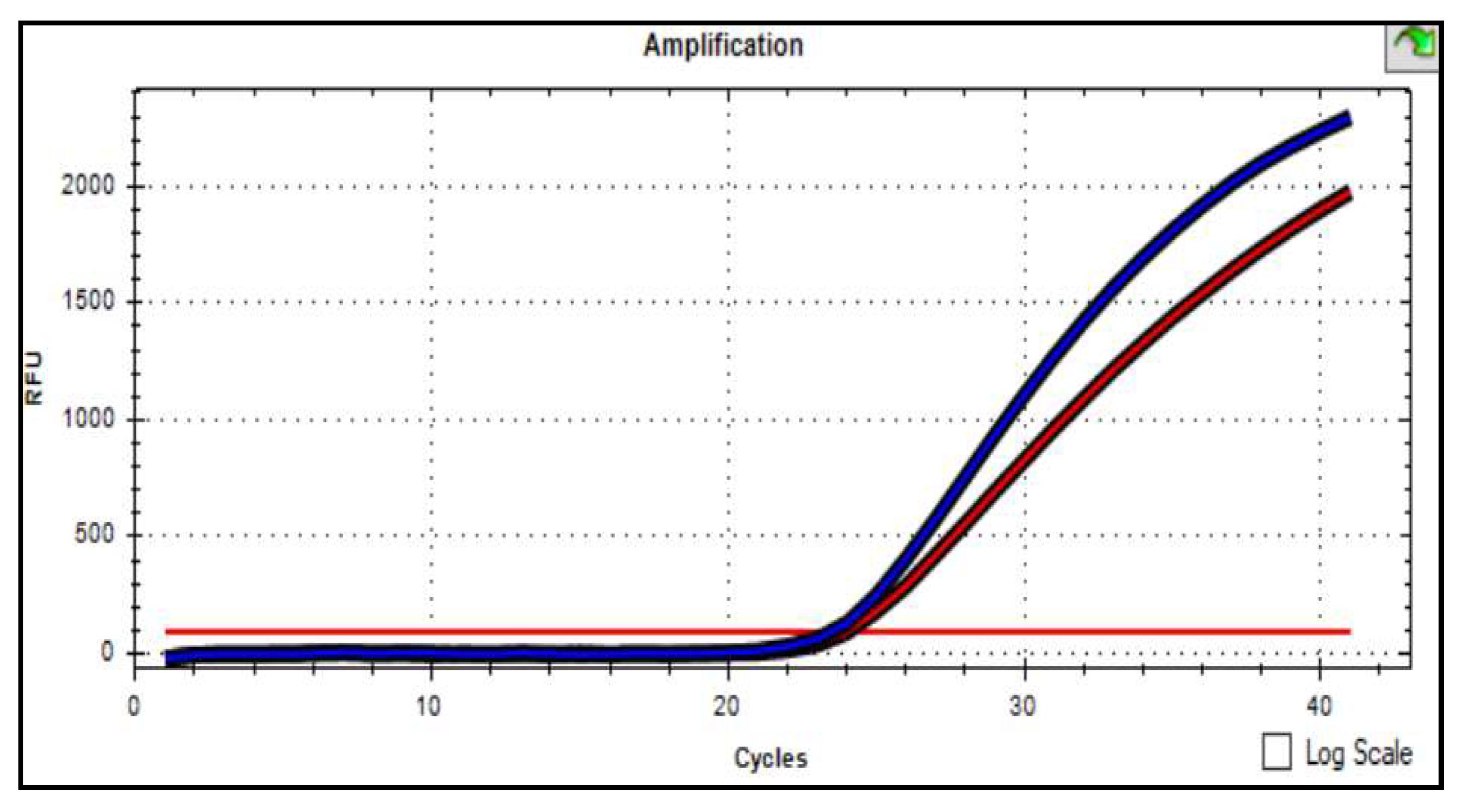
Graph 2). If both fluorescence curves grew, then this genotype was heterozygous (
Graph 3).
Statistical processing of the obtained data was carried out using the Statistica 6.0 software package. The assessment of the normality of the numerical variables’ distribution was realized using the Kolmogorov – Smirnov criteria. The data was analyzed at a significance level of α=0.05. The description of quantitative data was carried out on the basis of median and quartiles. For qualitative data, the proportion of persons with the characteristics of interest and a 95% confidence interval of the proportion determined by the Pearson χ2 method were calculated.
The frequency of the rs 3903239 polymorphism genotypes of the PRRX1 gene in the main group and in the control groups was in the Hardy – Weinberg equilibrium. The frequency of the minor G allele occurrence in the Kazakh population corresponded to the frequency of the allele in the general population in accordance with the base 1000genome G=0.3393 (1699/5008, 1000G).
The strength of the associations of the analyzed features was determined using the magnitude of the odds ratio. The odds ratio (OR) was considered statistically significant at the value >1.
The calculation of the odds ratio was used to identify the association between gene polymorphism and AF. The Pearson χ2 criterion was used to check the statistical significance of the differences between the «cases» and «controls» groups.
In order to study the relationship of the rs 3903239 polymorphism of the PRRX1 gene with the development of AF, 198 persons were examined, among whom 75 patients had AF. The control group consisted of 2 subgroups: subgroup 1 (control group 1) included conditionally healthy patients (n=3), subgroup 2 (control group 2) consisted of patients with arterial hypertension (AH) and coronary heart disease (CHD) without diagnosed AF at the time of inclusion in the study (n=50). The initial characteristics of the study participants are presented in
Table 3.
As
Table 4 demonstrates, in the main group of patients with AF, the homozygous AA genotype for the common allele was detected in 33.3% of cases, the heterozygous AG genotype – in 52% cases, and the homozygous GG genotype for the rare allele – in 14.7% cases. In the group of conditionally healthy patients, the frequency of genotypes occurrence was distributed as follows: the homozygous AA genotype – in 45.2%, the heterozygous AG genotype – in 42.5%, the homozygous GG genotype – in 12.3%. In the group of patients with AH and CHD, the frequency of genotypes occurrence was as follows: the homozygous AA genotype – in 34%, the heterozygous AG genotype – in 48%, the homozygous GG genotype – in 18%. The frequency distribution of the minor rare G allele and the common A allele of the rs 3903239 polymorphism of the PRRX1 gene in the main and the control groups was distributed as follows: the frequency of the rare G allele in the main group of patients with AF was 40.7%, the frequency of the common A allele was 59.3%.
In the group of healthy patients, the frequency of the rare G allele was 33.6%, and the frequency of the common A allele was 66.4%. In the control group 2, the frequency of the rare G allele was 42%, and the frequency of the common A allele was 58% (
Table 5).
Comparative analysis of the data of the main group and the group of conditionally healthy patients showed that the frequency of occurrence of the rare G allele in the main group of patients with AF prevailed in comparison with the group of healthy patients (40.7% versus 33.6%, OR 1.357; 95% CI 0.845-2.178) (
Table 6).
We analyzed the distribution of genotypes of the rs 3903239 polymorphism of the PRRX1 gene in the group of patients with AF and in healthy patients, depending on the inheritance model (
Table 7).
As can be seen from
Table 7, in the dominant inheritance model, the frequency of genotypes with rare G (AG+GG) allele in patients with AF prevailed when compared with the control group (66.7% versus 54.8%, OR 1.650; 95% CI 0.848-3.210). In the recessive inheritance model, the frequency of the homozygous GG genotype in patients with AF was 14.7%, in the group of conditionally healthy patients – 12.3%. In the overdominant inheritance model, the frequency of heterozygous AG genotype in the main group was 52%, in the control group 1 – 42.5%.
Discussion
According to the results of the largest GWAS meta-analysis conducted in 2012, the rs 3903239 polymorphism of the PRRX1 gene was the most significant association on the 1q24 chromosome of the PRRX gene [
8]. In 2018, the largest multiethnic study of the association of the genome with the left atrium in more than 500,000 participants, which included 84.2% European, 12.5% Japanese, 2% African American and 1.3% Brazilian and Hispanic populations, confirmed the role of the PRRX1 gene in the occurrence of AF [
9]. In this regard, the purpose of our study was to identify the association of the rs 3903239 polymorphism of the PRRX1 gene with the development of AF in the Kazakh population. We conducted the genetic study of 198 patients of Kazakh nationality, of which 75 patients with AF were included in the main group. For comparison, the control group was formed in the number of 73 conditionally healthy individuals who do not have diseases of the cardiovascular system and 50 patients with clinical and anamnestic data of AH and CHD disease without AF.
The distribution of genotypes and alleles of the rs 3903239 polymorphism of the PRRX1 gene was analyzed, and the obtained results were compared with data from patients belonging to the group of conditionally healthy individuals without cardiovascular pathology (control group 1). According to the results, in patients with AF, the homozygous AA genotype for the common allele was detected in 33.3% of cases, the heterozygous AG genotype – in 52% cases, and the homozygous GG genotype for the rare allele – in 14.7% cases. In the group of conditionally healthy patients, the frequency of genotypes occurrence was distributed as follows: the homozygous AA genotype – in 45.2%, the heterozygous AG genotype – in 42.5%, the homozygous GG genotype – in 12.3%. The frequency distribution of the minor rare G allele and the common A allele of the rs 3903239 polymorphism of the PRRX1 gene in the main and the control groups was distributed as follows: the frequency of the rare G allele in the main group of patients with AF was 40.7%, the frequency of the common A allele was 59.3%. In the group of healthy patients, the frequency of the rare G allele was 33.6%, and the frequency of the common A allele was 66.4%. Comparative analysis of data from patients with AF with data from conditionally healthy patients showed that the homozygous AA genotype was less common in the group of patients with AF compared with the control group 1 (33.3% versus 45.2%, p<0.05). The homozygous GG genotype for the rare allele in the group of patients with AF was more common compared with the control group (14.7% versus 12.3%, p<0.05). The frequency of occurrence of the rare G allele in the main group of patients with AF prevailed in comparison with the group of healthy patients (40.7% versus 33.6%, p<0.05, OR 1.357; 95% CI 0.845-2.178). We analyzed the distribution of genotypes of the rs 3903239 polymorphism of the PRRX1 gene in the group of patients with AF and in healthy patients, depending on the inheritance model. In the dominant inheritance model, the frequency of occurrence of genotypes with the rare G (AG+GG) allele in patients with AF prevailed when compared with the group of conditionally healthy patients (66.7% relative to 54.8%, p<0.05, OR 1.650; 95% CI 0.848-3.210). Thus, the homozygous GG genotype for the rare allele and the G allele of the rs 3903239 polymorphism of the PRRX1 gene can be considered as predictors of AF in the Kazakh population in the dominant inheritance model: the frequency of occurrence of genotypes with the rare G (AG+GG) allele in patients with AF prevailed when compared with a group of conditionally healthy patients (66.7% relative to 54.8%, p<0.05, OR 1.650; 95% CI 0.848-3.210). Similar results, associations of the rs 3903239 polymorphism of the PRRX1 gene with AF were obtained as a result of the studies in the European, Latin American and Korean cohort of patients [
6,
10,
11]. However, as the result of the study conducted in the Greek and Japanese cohort of patients, this polymorphism did not show the significant association with AF development [
12,
13]. The limitation of our study may be the small sample of patients with AF, so the obtained results, taking into account the CI/OR, are statistically insignificant. The significant association of the rs 3903239 polymorphism of the PRRX1 gene has been identified in numerous genome-wide population studies. Therefore, the obtained results need to be tested on a larger population of patients of Kazakh nationality with AF.