Submitted:
14 January 2025
Posted:
14 January 2025
You are already at the latest version
Abstract
The genomes from complex eukaryotes are enriched in non-coding genes whose transcription products (non-coding RNAs) are involved in the regulation of genomic output at different levels. Non-coding RNA action is predominantly driven by sequence and structural motifs that interact with specific functional partners. Despite the exponential growth in primary RNA sequence data facilitated by next-generation sequencing studies, the availability of tridimensional RNA data is comparatively more limited. The subjacent reasons for this relative lack of information regarding RNA structure are related to the specific chemical nature of RNA molecules and the limitations of the current available methods for structural characterization of biomolecules. In this review, we describe and analyze the different structural motifs involved in non-coding RNA function, and the wet-lab and computational methods used to characterize their structure-function relationships, highlighting the current need for detailed structural studies to explore the molecular determinants of non-coding RNA function.
Keywords:
1. Prevalence of Non-Coding Genes in the Genomes of Complex Eukaryotes
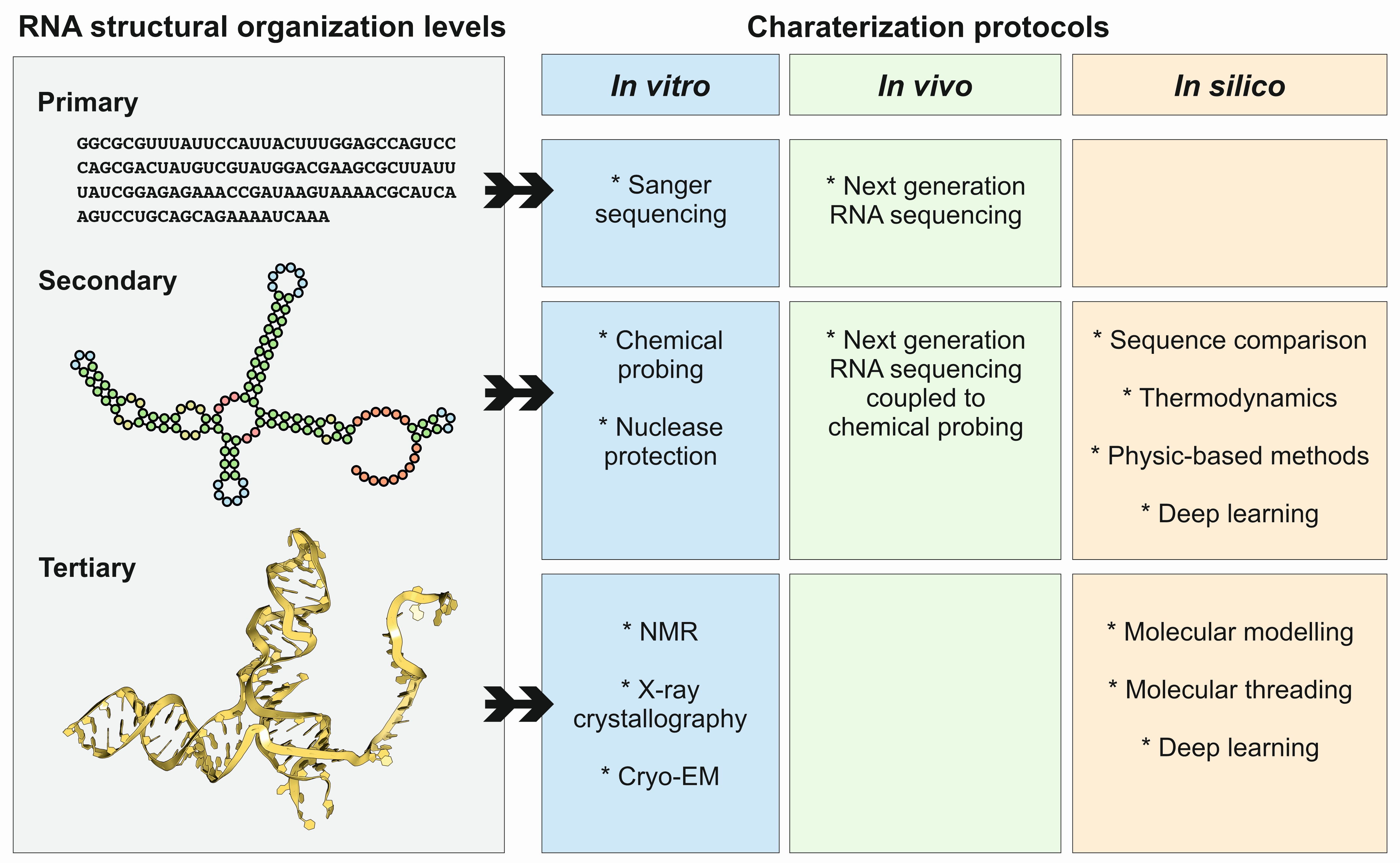
2. General Principles and Rules Governing RNA Structure
3. Functional Relevance of Structural Elements in Non-Coding RNAs
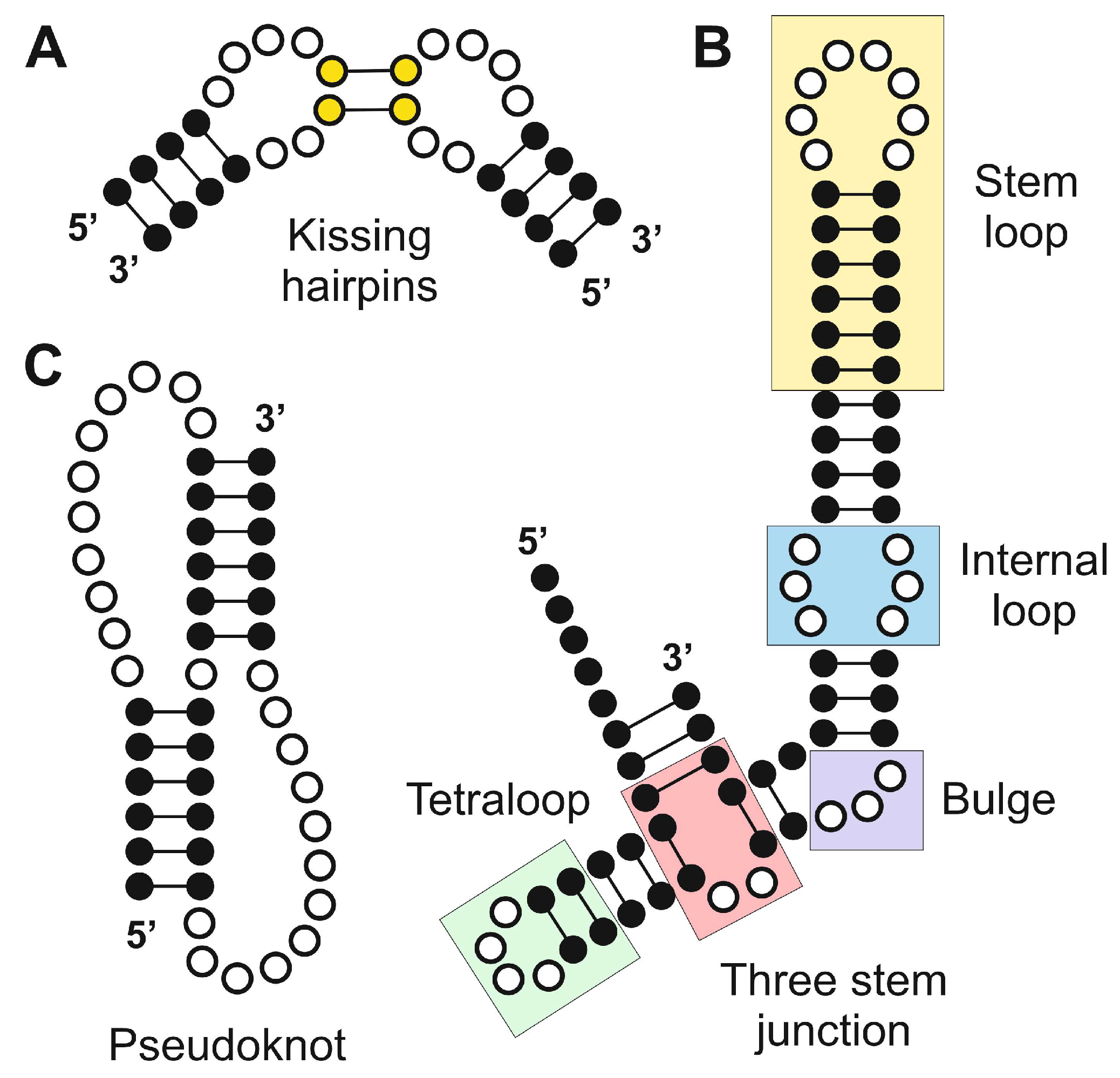
3.1. Hairpin Loops
3.2. Hairpin Loops
3.3. Pseudoknots
3.4. Kissing Hairpins
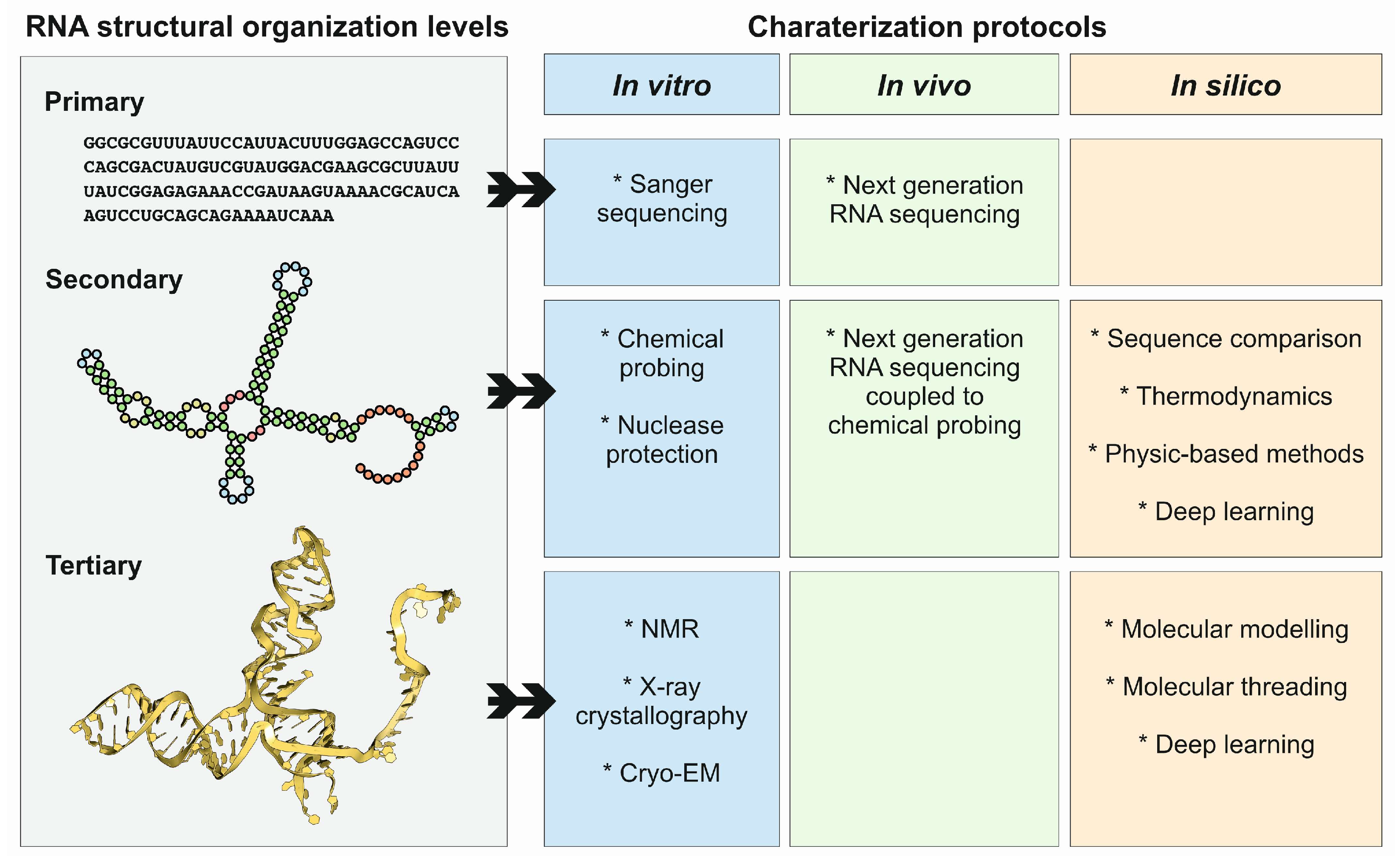
4. Methods and Protocols to Study ncRNA Structures
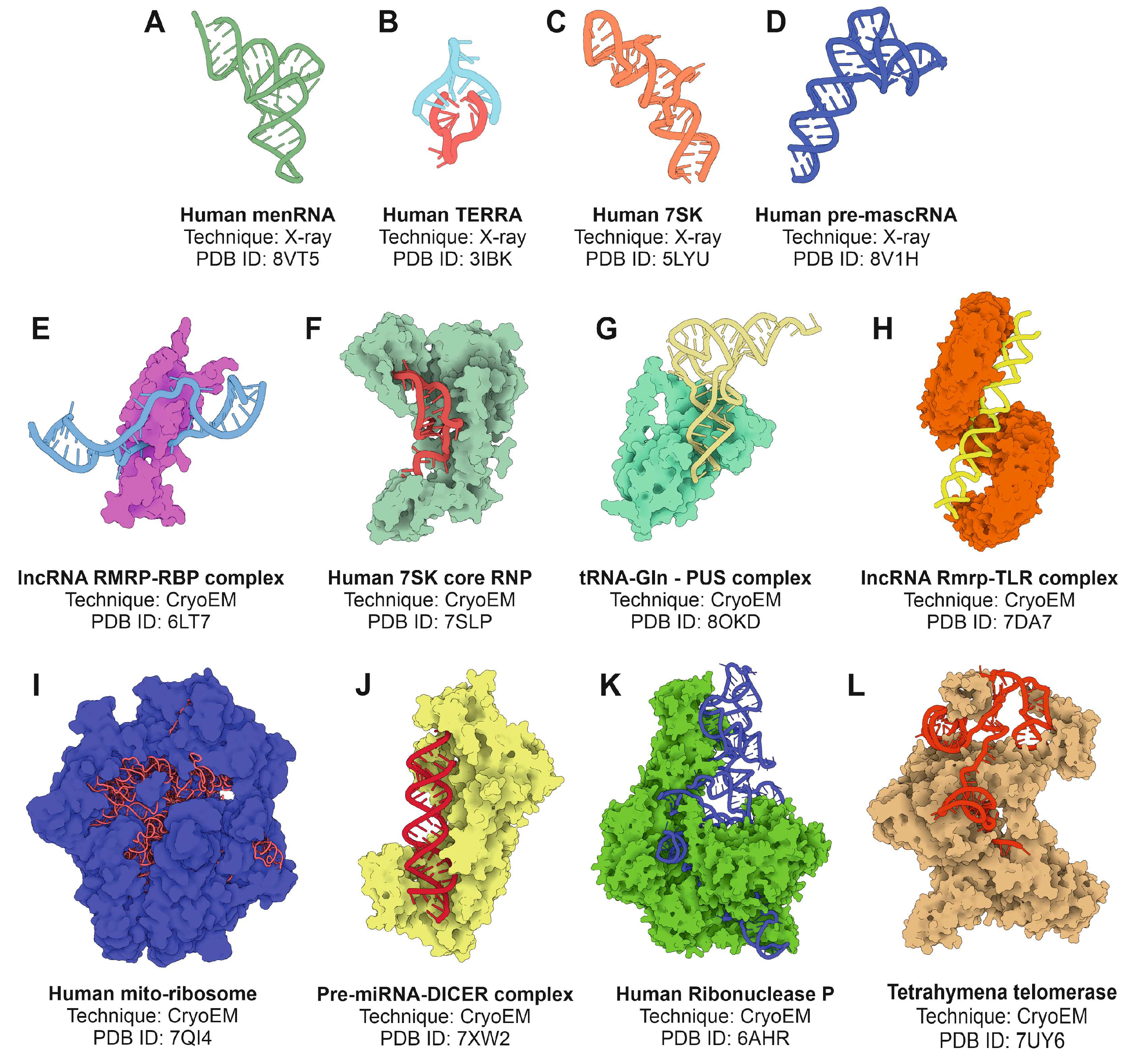
4.1. X-ray Crystallography
4.2. Cryo-Electron Microscopy
4.3. In Vivo Methods
4.4. In Silico Methods
4.4.1. Methods for RNA Secondary Structure Prediction
4.4.2. Methods for RNA Tertiary Structure Prediction
5. Perspectives and Further Developments
- Integration of multiscale data: combining atomic-level structural information with systemic data on ncRNA localization, interaction networks, and dynamics will enable a holistic understanding of ncRNA function. Hybrid approaches integrating cryo-EM and chemical probing with single-cell RNA sequencing and spatial transcriptomics are particularly promising.
- High-throughput structural characterization: automation in cryo-EM and microfluidics-based methods for RNA crystallography could facilitate the high-throughput determination of ncRNA structures, accelerating the discovery of novel functional motifs.
- Dynamic and contextual studies: capturing ncRNA structures in their native cellular environment remains a significant challenge. Emerging techniques, such as cryo-electron tomography (cryo-ET) and in situ structural studies, aim to bridge this gap by visualizing ncRNAs within intact cells.
- Functional modulation and rational drug design: structural insights into ncRNAs have profound implications for drug discovery. Small molecules targeting ncRNA structures or their interactions with proteins could provide new therapeutic avenues for diseases associated with dysregulated ncRNAs, such as cancer and neurodegenerative disorders.
- Evolutionary perspectives: structural comparisons across species can reveal conserved motifs and inform functional hypotheses. Integrating structural biology with evolutionary genomics will help identify universally important ncRNA structures and their roles in diverse organisms.
- Artificial intelligence: the use of deep-learning approaches to infer ncRNA structure and function will increase our understanding of their functions, increasing the knowledge about the molecular players related to cell physiology and human disease.
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Davis, C.A.; Hitz, B.C.; Sloan, C.A.; Chan, E.T.; Davidson, J.M.; Gabdank, I.; Hilton, J.A.; Jain, K.; Baymuradov, U.K.; Narayanan, A.K.; et al. The Encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res 2018, 46, D794–D801. [Google Scholar] [CrossRef] [PubMed]
- Mattick, J.S.; Gagen, M.J. The evolution of controlled multitasked gene networks: the role of introns and other noncoding RNAs in the development of complex organisms. Mol Biol Evol 2001, 18, 1611–1630. [Google Scholar] [CrossRef] [PubMed]
- Villa, T.; Porrua, O. Pervasive transcription: a controlled risk. FEBS J 2023, 290, 3723–3736. [Google Scholar] [CrossRef] [PubMed]
- Carroll, S.B. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 2008, 134, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Levine, M.; Tjian, R. Transcription regulation and animal diversity. Nature 2003, 424, 147–151. [Google Scholar] [CrossRef]
- Mudge, J.M.; Carbonell-Sala, S.; Diekhans, M.; Martinez, J.G.; Hunt, T.; Jungreis, I.; Loveland, J.E.; Arnan, C.; Barnes, I.; Bennett, R.; et al. GENCODE 2025: reference gene annotation for human and mouse. Nucleic Acids Res 2024. [Google Scholar] [CrossRef]
- Chen, L.L.; Kim, V.N. Small and long non-coding RNAs: Past, present, and future. Cell 2024, 187, 6451–6485. [Google Scholar] [CrossRef] [PubMed]
- Assmann, S.M.; Chou, H.L.; Bevilacqua, P.C. Rock, scissors, paper: How RNA structure informs function. Plant Cell 2023, 35, 1671–1707. [Google Scholar] [CrossRef]
- Tinoco, I., Jr.; Bustamante, C. How RNA folds. J Mol Biol 1999, 293, 271–281. [Google Scholar] [CrossRef]
- Mathews, D.H.; Turner, D.H. Prediction of RNA secondary structure by free energy minimization. Curr Opin Struct Biol 2006, 16, 270–278. [Google Scholar] [CrossRef]
- Juan, V.; Wilson, C. RNA secondary structure prediction based on free energy and phylogenetic analysis. J Mol Biol 1999, 289, 935–947. [Google Scholar] [CrossRef] [PubMed]
- Lemieux, S.; Major, F. RNA canonical and non-canonical base pairing types: a recognition method and complete repertoire. Nucleic Acids Res 2002, 30, 4250–4263. [Google Scholar] [CrossRef] [PubMed]
- Brion, P.; Westhof, E. Hierarchy and dynamics of RNA folding. Annu Rev Biophys Biomol Struct 1997, 26, 113–137. [Google Scholar] [CrossRef]
- Westhof, E.; Fritsch, V. RNA folding: beyond Watson-Crick pairs. Structure 2000, 8, R55–65. [Google Scholar] [CrossRef] [PubMed]
- Tinoco, I., Jr.; Li, P.T.; Bustamante, C. Determination of thermodynamics and kinetics of RNA reactions by force. Q Rev Biophys 2006, 39, 325–360. [Google Scholar] [CrossRef] [PubMed]
- Liphardt, J.; Onoa, B.; Smith, S.B.; Tinoco, I., Jr.; Bustamante, C. Reversible unfolding of single RNA molecules by mechanical force. Science 2001, 292, 733–737. [Google Scholar] [CrossRef]
- Vieregg, J.; Cheng, W.; Bustamante, C.; Tinoco, I., Jr. Measurement of the effect of monovalent cations on RNA hairpin stability. J Am Chem Soc 2007, 129, 14966–14973. [Google Scholar] [CrossRef]
- Guckian, K.M.; Schweitzer, B.A.; Ren, R.X.; Sheils, C.J.; Paris, P.L.; Tahmassebi, D.C.; Kool, E.T. Experimental Measurement of Aromatic Stacking Affinities in the Context of Duplex DNA. J Am Chem Soc 1996, 118, 8182–8183. [Google Scholar] [CrossRef]
- Serra, M.J.; Turner, D.H. Predicting thermodynamic properties of RNA. Methods Enzymol 1995, 259, 242–261. [Google Scholar] [CrossRef]
- Broyde, S. Setting the stage for predicting RNA thermodynamic properties and their structural components. Biophys J 1996, 70, 1571–1572. [Google Scholar] [CrossRef]
- Lilley, D.M. Structures of helical junctions in nucleic acids. Q Rev Biophys 2000, 33, 109–159. [Google Scholar] [CrossRef] [PubMed]
- Herschlag, D. RNA chaperones and the RNA folding problem. J Biol Chem 1995, 270, 20871–20874. [Google Scholar] [CrossRef] [PubMed]
- Lilley, D.M. The origins of RNA catalysis in ribozymes. Trends Biochem Sci 2003, 28, 495–501. [Google Scholar] [CrossRef] [PubMed]
- Draper, D.E.; Grilley, D.; Soto, A.M. Ions and RNA folding. Annu Rev Biophys Biomol Struct 2005, 34, 221–243. [Google Scholar] [CrossRef] [PubMed]
- Draper, D.E. RNA folding: thermodynamic and molecular descriptions of the roles of ions. Biophys J 2008, 95, 5489–5495. [Google Scholar] [CrossRef]
- Denesyuk, N.A.; Hori, N.; Thirumalai, D. Molecular Simulations of Ion Effects on the Thermodynamics of RNA Folding. J Phys Chem B 2018, 122, 11860–11867. [Google Scholar] [CrossRef]
- dell’Erba, M.G.; Zemba, G.R. Thermodynamics of a model for RNA folding. Phys Rev E Stat Nonlin Soft Matter Phys 2009, 79, 011913. [Google Scholar] [CrossRef]
- Leontis, N.B.; Lescoute, A.; Westhof, E. The building blocks and motifs of RNA architecture. Curr Opin Struct Biol 2006, 16, 279–287. [Google Scholar] [CrossRef]
- Gregorian, R.S., Jr.; Crothers, D.M. Determinants of RNA hairpin loop-loop complex stability. J Mol Biol 1995, 248, 968–984. [Google Scholar] [CrossRef]
- Dale, T.; Smith, R.; Serra, M.J. A test of the model to predict unusually stable RNA hairpin loop stability. RNA 2000, 6, 608–615. [Google Scholar] [CrossRef]
- Krol, J.; Sobczak, K.; Wilczynska, U.; Drath, M.; Jasinska, A.; Kaczynska, D.; Krzyzosiak, W.J. Structural features of microRNA (miRNA) precursors and their relevance to miRNA biogenesis and small interfering RNA/short hairpin RNA design. J Biol Chem 2004, 279, 42230–42239. [Google Scholar] [CrossRef] [PubMed]
- Rice, G.M.; Shivashankar, V.; Ma, E.J.; Baryza, J.L.; Nutiu, R. Functional Atlas of Primary miRNA Maturation by the Microprocessor. Mol Cell 2020, 80, 892–902. [Google Scholar] [CrossRef] [PubMed]
- Garg, A.; Shang, R.; Cvetanovic, T.; Lai, E.C.; Joshua-Tor, L. The structural landscape of Microprocessor-mediated processing of pri-let-7 miRNAs. Mol Cell 2024, 84, 4175–4190. [Google Scholar] [CrossRef]
- Xiong, X.; Kang, X.; Zheng, Y.; Yue, S.; Zhu, S. Identification of loop nucleotide polymorphisms affecting microRNA processing and function. Mol Cells 2013, 36, 518–526. [Google Scholar] [CrossRef]
- Dang, T.L.; Le, C.T.; Le, M.N.; Nguyen, T.D.; Nguyen, T.L.; Bao, S.; Li, S.; Nguyen, T.A. Select amino acids in DGCR8 are essential for the UGU-pri-miRNA interaction and processing. Commun Biol 2020, 3, 344. [Google Scholar] [CrossRef]
- McIntyre, G.J.; Yu, Y.H.; Lomas, M.; Fanning, G.C. The effects of stem length and core placement on shRNA activity. BMC Mol Biol 2011, 12, 34. [Google Scholar] [CrossRef]
- Zhang, D.; Jue, D.; Smith, N.; Zhong, C.; Finnegan, E.J.; de Feyter, R.; Wang, M.B.; Greaves, I. Asymmetric bulges within hairpin RNA transgenes influence small RNA size, secondary siRNA production and viral defence. Nucleic Acids Res 2024, 52, 9904–9916. [Google Scholar] [CrossRef]
- Wang, C.; Wang, L.; Ding, Y.; Lu, X.; Zhang, G.; Yang, J.; Zheng, H.; Wang, H.; Jiang, Y.; Xu, L. LncRNA Structural Characteristics in Epigenetic Regulation. Int J Mol Sci 2017, 18. [Google Scholar] [CrossRef]
- Cerase, A.; Tartaglia, G.G. Long non-coding RNA-polycomb intimate rendezvous. Open Biol 2020, 10, 200126. [Google Scholar] [CrossRef]
- Brown, J.A.; Bulkley, D.; Wang, J.; Valenstein, M.L.; Yario, T.A.; Steitz, T.A.; Steitz, J.A. Structural insights into the stabilization of MALAT1 noncoding RNA by a bipartite triple helix. Nat Struct Mol Biol 2014, 21, 633–640. [Google Scholar] [CrossRef]
- Walter, A.E.; Turner, D.H.; Kim, J.; Lyttle, M.H.; Muller, P.; Mathews, D.H.; Zuker, M. Coaxial stacking of helixes enhances binding of oligoribonucleotides and improves predictions of RNA folding. Proc Natl Acad Sci U S A 1994, 91, 9218–9222. [Google Scholar] [CrossRef] [PubMed]
- Mondal, M.; Mukherjee, S.; Halder, S.; Bhattacharyya, D. Stacking geometry for non-canonical G:U wobble base pair containing dinucleotide sequences in RNA: dispersion-corrected DFT-D study. Biopolymers 2015, 103, 328–338. [Google Scholar] [CrossRef]
- Hermann, T.; Patel, D.J. RNA bulges as architectural and recognition motifs. Structure 2000, 8, R47–54. [Google Scholar] [CrossRef] [PubMed]
- Correll, C.C.; Munishkin, A.; Chan, Y.L.; Ren, Z.; Wool, I.G.; Steitz, T.A. Crystal structure of the ribosomal RNA domain essential for binding elongation factors. Proc Natl Acad Sci U S A 1998, 95, 13436–13441. [Google Scholar] [CrossRef] [PubMed]
- Sargsyan, K.; Lim, C. Arrangement of 3D structural motifs in ribosomal RNA. Nucleic Acids Res 2010, 38, 3512–3522. [Google Scholar] [CrossRef]
- Macrae, I.J.; Zhou, K.; Li, F.; Repic, A.; Brooks, A.N.; Cande, W.Z.; Adams, P.D.; Doudna, J.A. Structural basis for double-stranded RNA processing by Dicer. Science 2006, 311, 195–198. [Google Scholar] [CrossRef]
- Zhao, J.; Sun, B.K.; Erwin, J.A.; Song, J.J.; Lee, J.T. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 2008, 322, 750–756. [Google Scholar] [CrossRef]
- Tripathi, V.; Ellis, J.D.; Shen, Z.; Song, D.Y.; Pan, Q.; Watt, A.T.; Freier, S.M.; Bennett, C.F.; Sharma, A.; Bubulya, P.A.; et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 2010, 39, 925–938. [Google Scholar] [CrossRef]
- Staple, D.W.; Butcher, S.E. Pseudoknots: RNA structures with diverse functions. PLoS Biol 2005, 3, e213. [Google Scholar] [CrossRef]
- Huang, F.W.; Li, L.Y.; Reidys, C.M. Sequence-structure relations of pseudoknot RNA. BMC Bioinformatics 2009, 10 Suppl 1, S39. [Google Scholar] [CrossRef]
- Green, L.; Kim, C.H.; Bustamante, C.; Tinoco, I., Jr. Characterization of the mechanical unfolding of RNA pseudoknots. J Mol Biol 2008, 375, 511–528. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.F.; Chang, K.C.; Chen, Y.L.; Hsieh, P.S.; Lee, A.I.; Tu, J.Y.; Chen, Y.T.; Wen, J.D. Formation of frameshift-stimulating RNA pseudoknots is facilitated by remodeling of their folding intermediates. Nucleic Acids Res 2021, 49, 6941–6957. [Google Scholar] [CrossRef] [PubMed]
- Su, L.; Chen, L.; Egli, M.; Berger, J.M.; Rich, A. Minor groove RNA triplex in the crystal structure of a ribosomal frameshifting viral pseudoknot. Nat Struct Biol 1999, 6, 285–292. [Google Scholar] [CrossRef]
- Theimer, C.A.; Blois, C.A.; Feigon, J. Structure of the human telomerase RNA pseudoknot reveals conserved tertiary interactions essential for function. Mol Cell 2005, 17, 671–682. [Google Scholar] [CrossRef]
- Uroda, T.; Anastasakou, E.; Rossi, A.; Teulon, J.M.; Pellequer, J.L.; Annibale, P.; Pessey, O.; Inga, A.; Chillon, I.; Marcia, M. Conserved Pseudoknots in lncRNA MEG3 Are Essential for Stimulation of the p53 Pathway. Mol Cell 2019, 75, 982–995. [Google Scholar] [CrossRef]
- Turner, D.H. Thermodynamics of base pairing. Curr Opin Struct Biol 1996, 6, 299–304. [Google Scholar] [CrossRef]
- Wilusz, J.E.; Freier, S.M.; Spector, D.L. 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell 2008, 135, 919–932. [Google Scholar] [CrossRef]
- Tsai, M.C.; Manor, O.; Wan, Y.; Mosammaparast, N.; Wang, J.K.; Lan, F.; Shi, Y.; Segal, E.; Chang, H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010, 329, 689–693. [Google Scholar] [CrossRef]
- Yang, L.; Froberg, J.E.; Lee, J.T. Long noncoding RNAs: fresh perspectives into the RNA world. Trends Biochem Sci 2014, 39, 35–43. [Google Scholar] [CrossRef]
- Naganuma, T.; Nakagawa, S.; Tanigawa, A.; Sasaki, Y.F.; Goshima, N.; Hirose, T. Alternative 3′-end processing of long noncoding RNA initiates construction of nuclear paraspeckles. EMBO J 2012, 31, 4020–4034. [Google Scholar] [CrossRef]
- Rivas, E.; Clements, J.; Eddy, S.R. A statistical test for conserved RNA structure shows lack of evidence for structure in lncRNAs. Nat Methods 2017, 14, 45–48. [Google Scholar] [CrossRef] [PubMed]
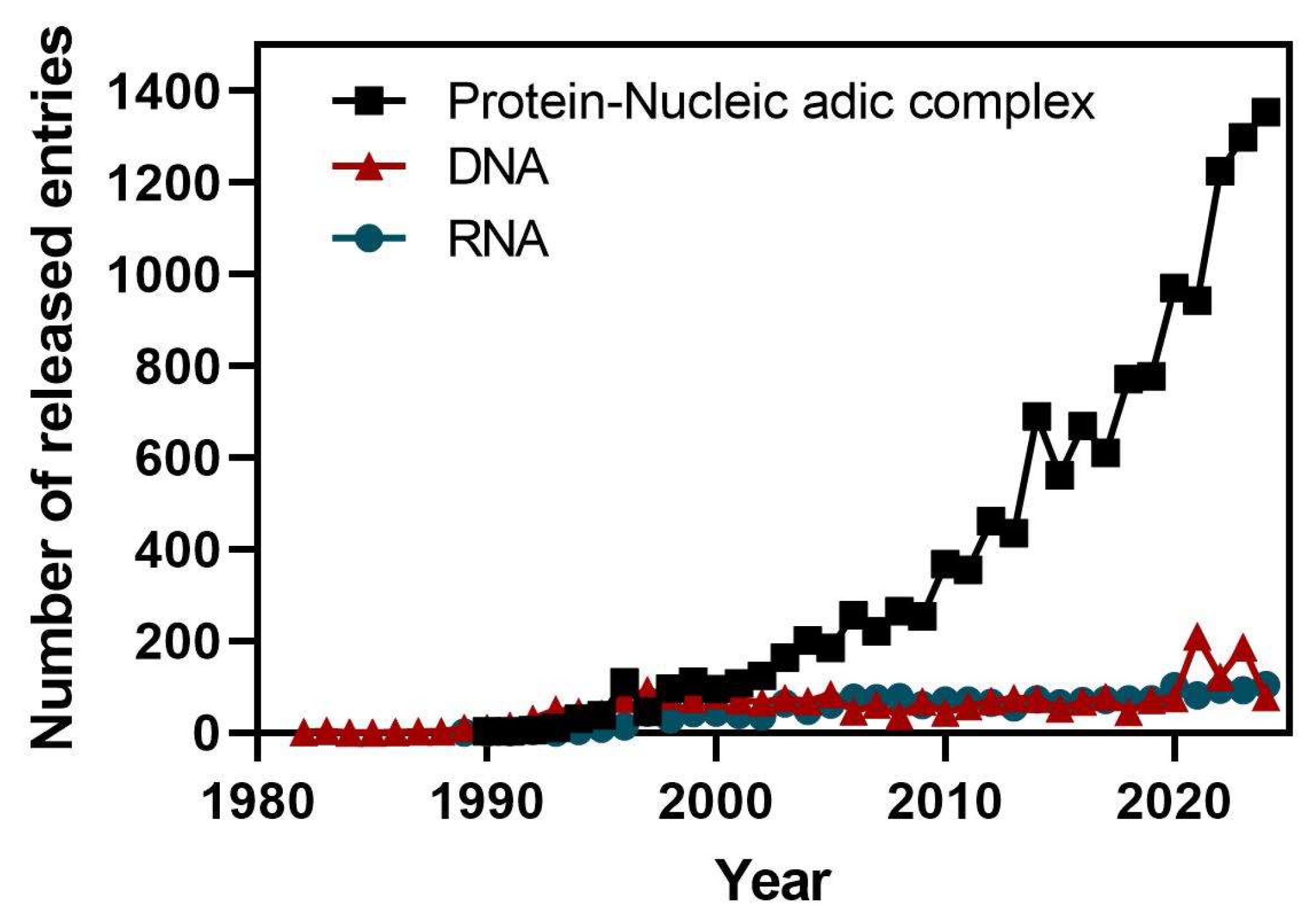
- Sussman, J.L.; Lin, D.; Jiang, J.; Manning, N.O.; Prilusky, J.; Ritter, O.; Abola, E.E. Protein Data Bank (PDB): database of three-dimensional structural information of biological macromolecules. Acta Crystallogr D Biol Crystallogr 1998, 54, 1078–1084. [Google Scholar] [CrossRef] [PubMed]
- Lawson, C.L.; Berman, H.M.; Chen, L.; Vallat, B.; Zirbel, C.L. The Nucleic Acid Knowledgebase: a new portal for 3D structural information about nucleic acids. Nucleic Acids Res 2024, 52, D245–D254. [Google Scholar] [CrossRef]
- Neidle, S. Beyond the double helix: DNA structural diversity and the PDB. J Biol Chem 2021, 296, 100553. [Google Scholar] [CrossRef]
- Lietzke, S.E.; Barnes, C.L.; Kundrot, C.E. Crystallization and structure determination of RNA. Curr Opin Struct Biol 1995, 5, 645–649. [Google Scholar] [CrossRef]
- Ferre-D’Amare, A.R.; Zhou, K.; Doudna, J.A. A general module for RNA crystallization. J Mol Biol 1998, 279, 621–631. [Google Scholar] [CrossRef]
- Golden, B.L.; Kundrot, C.E. RNA crystallization. J Struct Biol 2003, 142, 98–107. [Google Scholar] [CrossRef]
- Vicens, Q.; Gooding, A.R.; Laederach, A.; Cech, T.R. Local RNA structural changes induced by crystallization are revealed by SHAPE. RNA 2007, 13, 536–548. [Google Scholar] [CrossRef]
- Zhang, J.; Ferre-D’Amare, A.R. Post-crystallization Improvement of RNA Crystal Diffraction Quality. Methods Mol Biol 2015, 1316, 13–24. [Google Scholar] [CrossRef]
- Carrasco, N.; Buzin, Y.; Tyson, E.; Halpert, E.; Huang, Z. Selenium derivatization and crystallization of DNA and RNA oligonucleotides for X-ray crystallography using multiple anomalous dispersion. Nucleic Acids Res 2004, 32, 1638–1646. [Google Scholar] [CrossRef]
- Zhang, J.; Ferre-D’Amare, A.R. Post-crystallization Improvement of RNA Crystals by Synergistic Ion Exchange and Dehydration. Bio Protoc 2015, 5. [Google Scholar] [CrossRef]
- Pley, H.W.; Flaherty, K.M.; McKay, D.B. Three-dimensional structure of a hammerhead ribozyme. Nature 1994, 372, 68–74. [Google Scholar] [CrossRef]
- Scott, W.G.; Finch, J.T.; Klug, A. The crystal structure of an all-RNA hammerhead ribozyme: a proposed mechanism for RNA catalytic cleavage. Cell 1995, 81, 991–1002. [Google Scholar] [CrossRef]
- Edwards, A.L.; Garst, A.D.; Batey, R.T. Determining structures of RNA aptamers and riboswitches by X-ray crystallography. Methods Mol Biol 2009, 535, 135–163. [Google Scholar] [CrossRef]
- Serganov, A.; Polonskaia, A.; Phan, A.T.; Breaker, R.R.; Patel, D.J. Structural basis for gene regulation by a thiamine pyrophosphate-sensing riboswitch. Nature 2006, 441, 1167–1171. [Google Scholar] [CrossRef]
- Itoh, Y.; Chiba, S.; Sekine, S.; Yokoyama, S. Crystal structure of human selenocysteine tRNA. Nucleic Acids Res 2009, 37, 6259–6268. [Google Scholar] [CrossRef]
- Zong, X.; Nakagawa, S.; Freier, S.M.; Fei, J.; Ha, T.; Prasanth, S.G.; Prasanth, K.V. Natural antisense RNA promotes 3′ end processing and maturation of MALAT1 lncRNA. Nucleic Acids Res 2016, 44, 2898–2908. [Google Scholar] [CrossRef]
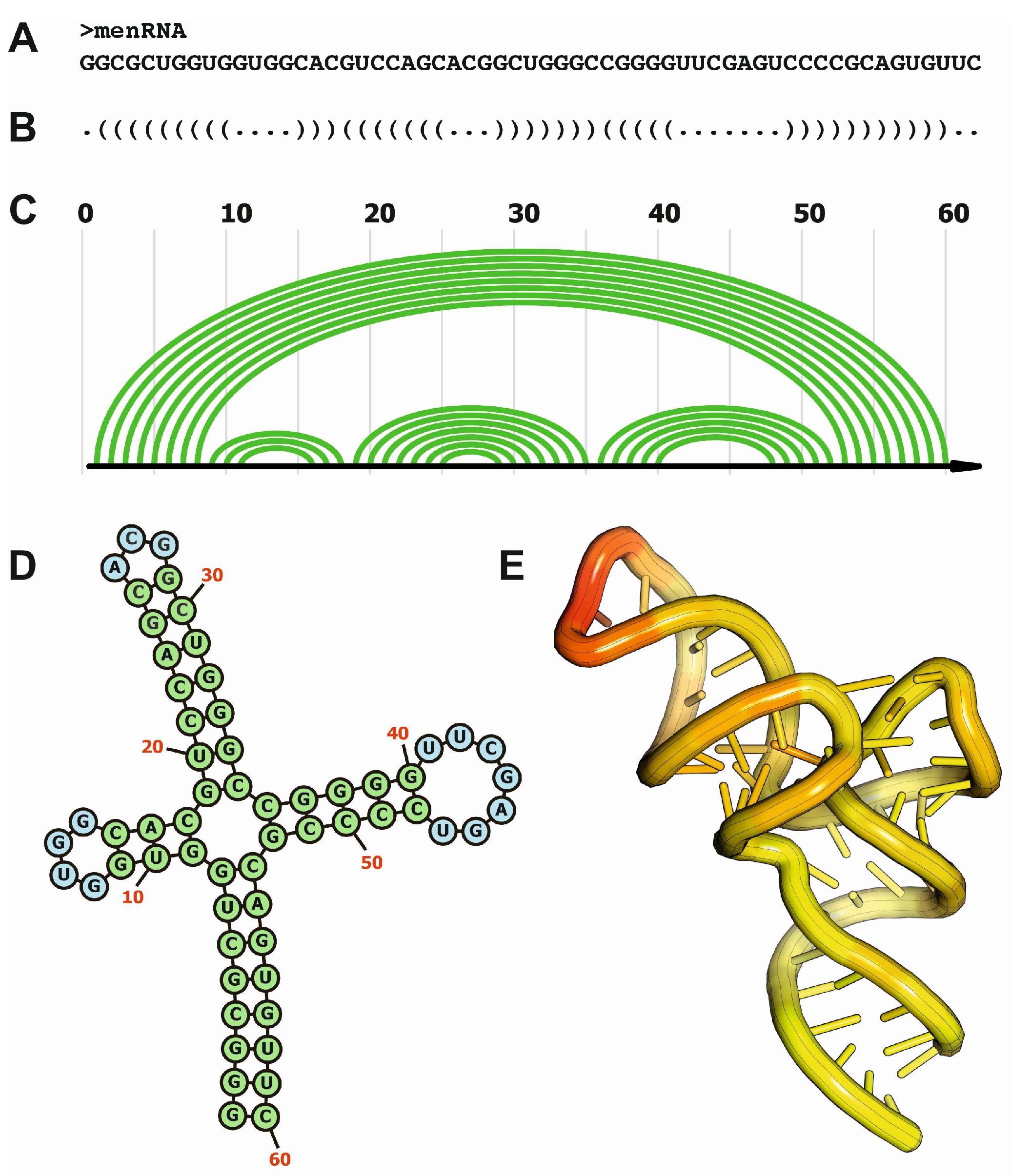
- Skeparnias, I.; Zhang, J. Structural basis of NEAT1 lncRNA maturation and menRNA instability. Nat Struct Mol Biol 2024, 31, 1650–1654. [Google Scholar] [CrossRef]
- Ma, H.; Jia, X.; Zhang, K.; Su, Z. Cryo-EM advances in RNA structure determination. Signal Transduct Target Ther 2022, 7, 58. [Google Scholar] [CrossRef]
- Bonilla, S.L.; Kieft, J.S. The promise of cryo-EM to explore RNA structural dynamics. J Mol Biol 2022, 434, 167802. [Google Scholar] [CrossRef]
- Chen, X.; Wang, L.; Xie, J.; Nowak, J.S.; Luo, B.; Zhang, C.; Jia, G.; Zou, J.; Huang, D.; Glatt, S.; et al. RNA sample optimization for cryo-EM analysis. Nat Protoc 2024. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Hang, J.; Wan, R.; Huang, M.; Wong, C.C.; Shi, Y. Structure of a yeast spliceosome at 3.6-angstrom resolution. Science 2015, 349, 1182–1191. [Google Scholar] [CrossRef]
- Yan, C.; Wan, R.; Bai, R.; Huang, G.; Shi, Y. Structure of a yeast activated spliceosome at 3.5 A resolution. Science 2016, 353, 904–911. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Wan, R.; Bai, R.; Huang, G.; Shi, Y. Structure of a yeast step II catalytically activated spliceosome. Science 2017, 355, 149–155. [Google Scholar] [CrossRef] [PubMed]
- Bertram, K.; Agafonov, D.E.; Dybkov, O.; Haselbach, D.; Leelaram, M.N.; Will, C.L.; Urlaub, H.; Kastner, B.; Luhrmann, R.; Stark, H. Cryo-EM Structure of a Pre-catalytic Human Spliceosome Primed for Activation. Cell 2017, 170, 701–713. [Google Scholar] [CrossRef]
- Bertram, K.; Agafonov, D.E.; Liu, W.T.; Dybkov, O.; Will, C.L.; Hartmuth, K.; Urlaub, H.; Kastner, B.; Stark, H.; Luhrmann, R. Cryo-EM structure of a human spliceosome activated for step 2 of splicing. Nature 2017, 542, 318–323. [Google Scholar] [CrossRef]
- Zhang, X.; Yan, C.; Hang, J.; Finci, L.I.; Lei, J.; Shi, Y. An Atomic Structure of the Human Spliceosome. Cell 2017, 169, 918–929. [Google Scholar] [CrossRef]
- Jiang, J.; Wang, Y.; Susac, L.; Chan, H.; Basu, R.; Zhou, Z.H.; Feigon, J. Structure of Telomerase with Telomeric DNA. Cell 2018, 173, 1179–1190. [Google Scholar] [CrossRef]
- Liu, B.; He, Y.; Wang, Y.; Song, H.; Zhou, Z.H.; Feigon, J. Structure of active human telomerase with telomere shelterin protein TPP1. Nature 2022, 604, 578–583. [Google Scholar] [CrossRef]
- Palka, C.; Forino, N.M.; Hentschel, J.; Das, R.; Stone, M.D. Folding heterogeneity in the essential human telomerase RNA three-way junction. RNA 2020, 26, 1787–1800. [Google Scholar] [CrossRef]
- Tomasello, G.; Armenia, I.; Molla, G. The Protein Imager: a full-featured online molecular viewer interface with server-side HQ-rendering capabilities. Bioinformatics 2020, 36, 2909–2911. [Google Scholar] [CrossRef] [PubMed]
- Leitao, A.L.; Enguita, F.J. A Structural View of miRNA Biogenesis and Function. Noncoding RNA 2022, 8. [Google Scholar] [CrossRef] [PubMed]
- Partin, A.C.; Zhang, K.; Jeong, B.C.; Herrell, E.; Li, S.; Chiu, W.; Nam, Y. Cryo-EM Structures of Human Drosha and DGCR8 in Complex with Primary MicroRNA. Mol Cell 2020, 78, 411–422. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Wang, J.; Cheng, H.; Ke, X.; Sun, L.; Zhang, Q.C.; Wang, H.W. Cryo-EM Structure of Human Dicer and Its Complexes with a Pre-miRNA Substrate. Cell 2018, 173, 1191–1203. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Roh, S.H. Cryo-EM structures of human DICER dicing a pre-miRNA substrate. FEBS J 2024, 291, 3072–3079. [Google Scholar] [CrossRef]
- Nguyen, T.D.; Trinh, T.A.; Bao, S.; Nguyen, T.A. Secondary structure RNA elements control the cleavage activity of DICER. Nat Commun 2022, 13, 2138. [Google Scholar] [CrossRef]
- Okada, C.; Yamashita, E.; Lee, S.J.; Shibata, S.; Katahira, J.; Nakagawa, A.; Yoneda, Y.; Tsukihara, T. A high-resolution structure of the pre-microRNA nuclear export machinery. Science 2009, 326, 1275–1279. [Google Scholar] [CrossRef]
- Spitale, R.C.; Flynn, R.A.; Zhang, Q.C.; Crisalli, P.; Lee, B.; Jung, J.W.; Kuchelmeister, H.Y.; Batista, P.J.; Torre, E.A.; Kool, E.T.; et al. Structural imprints in vivo decode RNA regulatory mechanisms. Nature 2015, 519, 486–490. [Google Scholar] [CrossRef]
- Baek, S.C.; Kim, B.; Jang, H.; Kim, K.; Park, I.S.; Min, D.H.; Kim, V.N. Structural atlas of human primary microRNAs generated by SHAPE-MaP. Mol Cell 2024, 84, 1158–1172. [Google Scholar] [CrossRef]
- Mustoe, A.M.; Busan, S.; Rice, G.M.; Hajdin, C.E.; Peterson, B.K.; Ruda, V.M.; Kubica, N.; Nutiu, R.; Baryza, J.L.; Weeks, K.M. Pervasive Regulatory Functions of mRNA Structure Revealed by High-Resolution SHAPE Probing. Cell 2018, 173, 181–195. [Google Scholar] [CrossRef]
- Zeller, M.J.; Favorov, O.; Li, K.; Nuthanakanti, A.; Hussein, D.; Michaud, A.; Lafontaine, D.A.; Busan, S.; Serganov, A.; Aube, J.; et al. SHAPE-enabled fragment-based ligand discovery for RNA. Proc Natl Acad Sci U S A 2022, 119, e2122660119. [Google Scholar] [CrossRef] [PubMed]
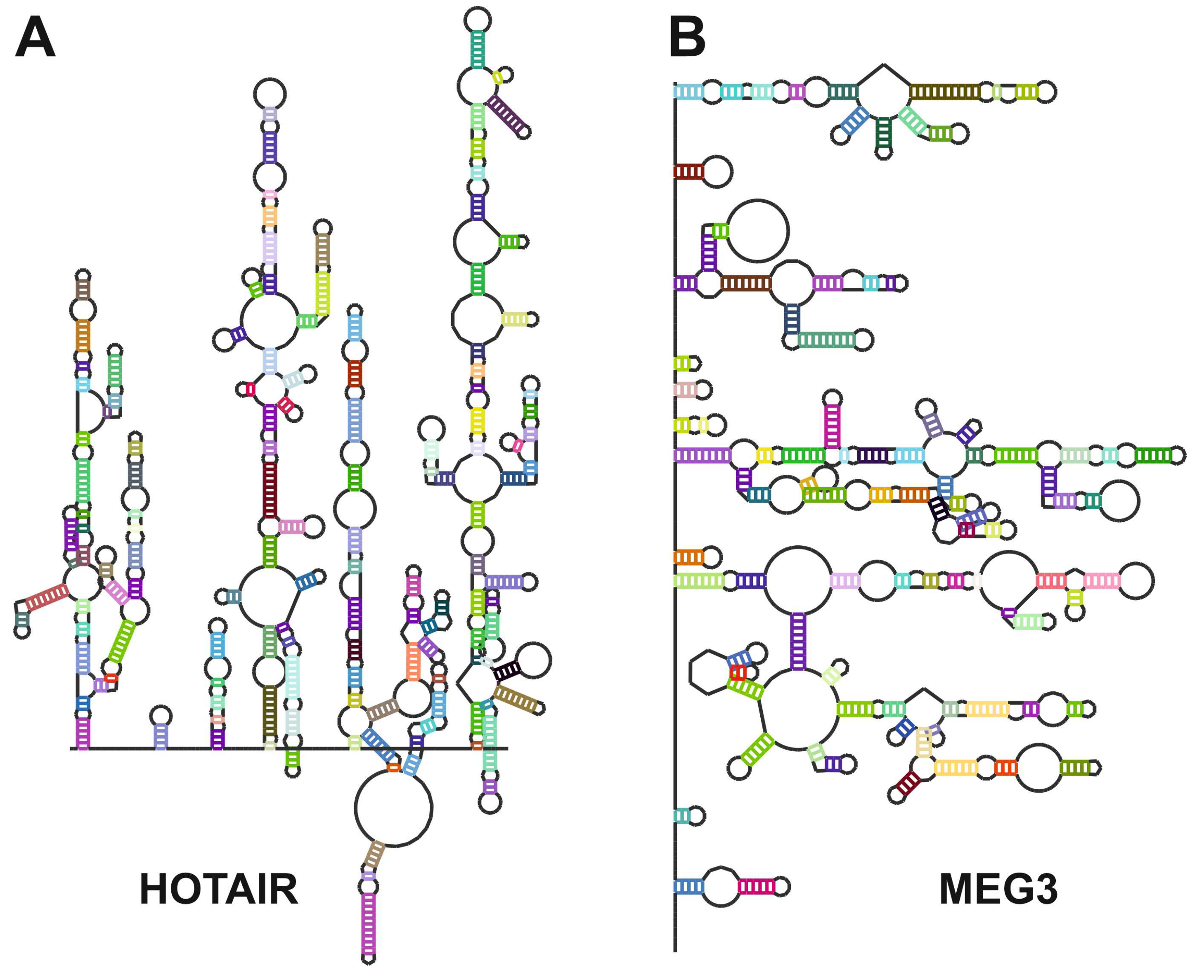
- Somarowthu, S.; Legiewicz, M.; Chillon, I.; Marcia, M.; Liu, F.; Pyle, A.M. HOTAIR forms an intricate and modular secondary structure. Mol Cell 2015, 58, 353–361. [Google Scholar] [CrossRef] [PubMed]
- Bugnon, L.A.; Edera, A.A.; Prochetto, S.; Gerard, M.; Raad, J.; Fenoy, E.; Rubiolo, M.; Chorostecki, U.; Gabaldon, T.; Ariel, F.; et al. Secondary structure prediction of long noncoding RNA: review and experimental comparison of existing approaches. Brief Bioinform 2022, 23. [Google Scholar] [CrossRef]
- Novikova, I.V.; Hennelly, S.P.; Sanbonmatsu, K.Y. Structural architecture of the human long non-coding RNA, steroid receptor RNA activator. Nucleic Acids Res 2012, 40, 5034–5051. [Google Scholar] [CrossRef]
- Monroy-Eklund, A.; Taylor, C.; Weidmann, C.A.; Burch, C.; Laederach, A. Structural analysis of MALAT1 long noncoding RNA in cells and in evolution. RNA 2023, 29, 691–704. [Google Scholar] [CrossRef]
- Clemson, C.M.; Hutchinson, J.N.; Sara, S.A.; Ensminger, A.W.; Fox, A.H.; Chess, A.; Lawrence, J.B. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell 2009, 33, 717–726. [Google Scholar] [CrossRef]
- Lin, Y.; Schmidt, B.F.; Bruchez, M.P.; McManus, C.J. Structural analyses of NEAT1 lncRNAs suggest long-range RNA interactions that may contribute to paraspeckle architecture. Nucleic Acids Res 2018, 46, 3742–3752. [Google Scholar] [CrossRef]
- Frank, F.; Kavousi, N.; Bountali, A.; Dammer, E.B.; Mourtada-Maarabouni, M.; Ortlund, E.A. The lncRNA Growth Arrest Specific 5 Regulates Cell Survival via Distinct Structural Modules with Independent Functions. Cell Rep 2020, 32, 107933. [Google Scholar] [CrossRef]
- Fang, R.; Moss, W.N.; Rutenberg-Schoenberg, M.; Simon, M.D. Probing Xist RNA Structure in Cells Using Targeted Structure-Seq. PLoS Genet 2015, 11, e1005668. [Google Scholar] [CrossRef]
- Shcherbakova, I.; Brenowitz, M. Perturbation of the hierarchical folding of a large RNA by the destabilization of its Scaffold’s tertiary structure. J Mol Biol 2005, 354, 483–496. [Google Scholar] [CrossRef]
- Reeder, J.; Hochsmann, M.; Rehmsmeier, M.; Voss, B.; Giegerich, R. Beyond Mfold: recent advances in RNA bioinformatics. J Biotechnol 2006, 124, 41–55. [Google Scholar] [CrossRef] [PubMed]
- Hofacker, I.L. RNA secondary structure analysis using the Vienna RNA package. Curr Protoc Bioinformatics 2009, Chapter 12, 12 12 11–12 12 16. [Google Scholar] [CrossRef]
- Ali, S.E.; Mittal, A.; Mathews, D.H. RNA Secondary Structure Analysis Using RNAstructure. Curr Protoc 2023, 3, e846. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Chan, C.Y.; Lawrence, C.E. Sfold web server for statistical folding and rational design of nucleic acids. Nucleic Acids Res 2004, 32, W135–141. [Google Scholar] [CrossRef]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 2003, 31, 3406–3415. [Google Scholar] [CrossRef]
- Fernandez, N.; Cordiner, R.A.; Young, R.S.; Hug, N.; Macias, S.; Caceres, J.F. Genetic variation and RNA structure regulate microRNA biogenesis. Nat Commun 2017, 8, 15114. [Google Scholar] [CrossRef]
- Long, D.; Lee, R.; Williams, P.; Chan, C.Y.; Ambros, V.; Ding, Y. Potent effect of target structure on microRNA function. Nat Struct Mol Biol 2007, 14, 287–294. [Google Scholar] [CrossRef]
- Nawrocki, E.P.; Eddy, S.R. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 2013, 29, 2933–2935. [Google Scholar] [CrossRef]
- Ontiveros-Palacios, N.; Cooke, E.; Nawrocki, E.P.; Triebel, S.; Marz, M.; Rivas, E.; Griffiths-Jones, S.; Petrov, A.I.; Bateman, A.; Sweeney, B. Rfam 15: RNA families database in 2025. Nucleic Acids Res 2024. [Google Scholar] [CrossRef]
- Sato, K.; Hamada, M. Recent trends in RNA informatics: a review of machine learning and deep learning for RNA secondary structure prediction and RNA drug discovery. Brief Bioinform 2023, 24. [Google Scholar] [CrossRef]
- Zhao, Q.; Zhao, Z.; Fan, X.; Yuan, Z.; Mao, Q.; Yao, Y. Review of machine learning methods for RNA secondary structure prediction. PLoS Comput Biol 2021, 17, e1009291. [Google Scholar] [CrossRef] [PubMed]
- Singh, J.; Hanson, J.; Paliwal, K.; Zhou, Y. RNA secondary structure prediction using an ensemble of two-dimensional deep neural networks and transfer learning. Nat Commun 2019, 10, 5407. [Google Scholar] [CrossRef] [PubMed]
- Townshend, R.J.L.; Eismann, S.; Watkins, A.M.; Rangan, R.; Karelina, M.; Das, R.; Dror, R.O. Geometric deep learning of RNA structure. Science 2021, 373, 1047–1051. [Google Scholar] [CrossRef] [PubMed]
- Miao, Z.; Westhof, E. RNA Structure: Advances and Assessment of 3D Structure Prediction. Annu Rev Biophys 2017, 46, 483–503. [Google Scholar] [CrossRef]
- Thiel, B.C.; Flamm, C.; Hofacker, I.L. RNA structure prediction: from 2D to 3D. Emerg Top Life Sci 2017, 1, 275–285. [Google Scholar] [CrossRef]
- Li, J.; Chen, S.J. RNA 3D Structure Prediction Using Coarse-Grained Models. Front Mol Biosci 2021, 8, 720937. [Google Scholar] [CrossRef]
- Kryshtafovych, A.; Schwede, T.; Topf, M.; Fidelis, K.; Moult, J. Critical assessment of methods of protein structure prediction (CASP)-Round XV. Proteins 2023, 91, 1539–1549. [Google Scholar] [CrossRef]
- Miao, Z.; Adamiak, R.W.; Antczak, M.; Boniecki, M.J.; Bujnicki, J.; Chen, S.J.; Cheng, C.Y.; Cheng, Y.; Chou, F.C.; Das, R.; et al. RNA-Puzzles Round IV: 3D structure predictions of four ribozymes and two aptamers. RNA 2020, 26, 982–995. [Google Scholar] [CrossRef]
- Rother, M.; Milanowska, K.; Puton, T.; Jeleniewicz, J.; Rother, K.; Bujnicki, J.M. ModeRNA server: an online tool for modeling RNA 3D structures. Bioinformatics 2011, 27, 2441–2442. [Google Scholar] [CrossRef]
- Parisien, M.; Major, F. The MC-Fold and MC-Sym pipeline infers RNA structure from sequence data. Nature 2008, 452, 51–55. [Google Scholar] [CrossRef]
- Watkins, A.M.; Das, R. RNA 3D Modeling with FARFAR2, Online. Methods Mol Biol 2023, 2586, 233–249. [Google Scholar] [CrossRef] [PubMed]
- Watkins, A.M.; Rangan, R.; Das, R. FARFAR2: Improved De Novo Rosetta Prediction of Complex Global RNA Folds. Structure 2020, 28, 963–976. [Google Scholar] [CrossRef] [PubMed]
- Biesiada, M.; Purzycka, K.J.; Szachniuk, M.; Blazewicz, J.; Adamiak, R.W. Automated RNA 3D Structure Prediction with RNAComposer. Methods Mol Biol 2016, 1490, 199–215. [Google Scholar] [CrossRef]
- Biesiada, M.; Pachulska-Wieczorek, K.; Adamiak, R.W.; Purzycka, K.J. RNAComposer and RNA 3D structure prediction for nanotechnology. Methods 2016, 103, 120–127. [Google Scholar] [CrossRef]
- Purzycka, K.J.; Popenda, M.; Szachniuk, M.; Antczak, M.; Lukasiak, P.; Blazewicz, J.; Adamiak, R.W. Automated 3D RNA structure prediction using the RNAComposer method for riboswitches. Methods Enzymol 2015, 553, 3–34. [Google Scholar] [CrossRef]
- Sarzynska, J.; Popenda, M.; Antczak, M.; Szachniuk, M. RNA tertiary structure prediction using RNAComposer in CASP15. Proteins 2023, 91, 1790–1799. [Google Scholar] [CrossRef]
- Palermo, G.; Casalino, L.; Magistrato, A.; Andrew McCammon, J. Understanding the mechanistic basis of non-coding RNA through molecular dynamics simulations. J Struct Biol 2019, 206, 267–279. [Google Scholar] [CrossRef]
- Kuhrova, P.; Mlynsky, V.; Zgarbova, M.; Krepl, M.; Bussi, G.; Best, R.B.; Otyepka, M.; Sponer, J.; Banas, P. Improving the Performance of the Amber RNA Force Field by Tuning the Hydrogen-Bonding Interactions. J Chem Theory Comput 2019, 15, 3288–3305. [Google Scholar] [CrossRef]
- Tan, D.; Piana, S.; Dirks, R.M.; Shaw, D.E. RNA force field with accuracy comparable to state-of-the-art protein force fields. Proc Natl Acad Sci U S A 2018, 115, E1346–E1355. [Google Scholar] [CrossRef]
- Ebrahimi, P.; Kaur, S.; Baronti, L.; Petzold, K.; Chen, A.A. A two-dimensional replica-exchange molecular dynamics method for simulating RNA folding using sparse experimental restraints. Methods 2019, 162-163, 96–107. [Google Scholar] [CrossRef]
- Bell, D.R.; Cheng, S.Y.; Salazar, H.; Ren, P. Capturing RNA Folding Free Energy with Coarse-Grained Molecular Dynamics Simulations. Sci Rep 2017, 7, 45812. [Google Scholar] [CrossRef] [PubMed]
- Dawson, W.K.; Maciejczyk, M.; Jankowska, E.J.; Bujnicki, J.M. Coarse-grained modeling of RNA 3D structure. Methods 2016, 103, 138–156. [Google Scholar] [CrossRef] [PubMed]
- Paciello, G.; Acquaviva, A.; Ficarra, E.; Deriu, M.A.; Macii, E. A molecular dynamics study of a miRNA:mRNA interaction. J Mol Model 2011, 17, 2895–2906. [Google Scholar] [CrossRef] [PubMed]
- Xia, Z.; Clark, P.; Huynh, T.; Loher, P.; Zhao, Y.; Chen, H.W.; Ren, P.; Rigoutsos, I.; Zhou, R. Molecular dynamics simulations of Ago silencing complexes reveal a large repertoire of admissible ‘seed-less’ targets. Sci Rep 2012, 2, 569. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




