1. Introduction
In the era of information explosion, Large Language Models (LLMs) have achieved remarkable success in a wide array of general-knowledge reasoning tasks owing to their deep training on vast, publicly available datasets. This progress marks a pivotal milestone in the field of artificial intelligence [
1]. Currently, pre-training LLMs in large-scale text corpora has become an industry standard, forming a solid foundation for deploying these models across diverse application domains [
2].
Despite their extraordinary capabilities, LLMs are not without limitations. In particular, in high-stakes fields such as medicine and law, the phenomenon of hallucination in LLM presents significant risks, drawing increasing attention to another critical aspect of LLM performance, reading comprehension, particularly in tasks such as evidence-based question answering (QA). In evidence-based QA, LLMs must respond to queries using the knowledge sources provided and accurately citing these sources [
3]. This ability is crucial to ensure both the accuracy and professionalism of the model’s output. However, standalone LLMs often face challenges related to hallucinations and knowledge gaps, especially when dealing with ,complex domain-specific problems [
4,
5].
To address these issues, Retrieval-Augmented Generation (RAG) has emerged as a promising strategy. By integrating the robust processing power of LLMs with the vastness of external knowledge sources, RAG enhances the reliability and traceability of model-generated responses, offering a more robust solution for specialized domains [
6]. This approach significantly mitigates hallucinations and knowledge limitations common in standalone LLMs, while also improving the traceability and professionalism of the answers. However, RAG also introduces new challenges, particularly for LLMs with smaller parameter sizes. When tasked with processing multiple non-contiguous retrieval segments, these models exhibit a marked decline in reading comprehension, making it difficult to accurately identify pertinent information and synthesize coherent analyses.
In the medical domain, LLMs are increasingly becoming indispensable tools for both research and clinical practice. These models typically rely on open-source medical data for fine-tuning, aiming to achieve a higher degree of specialization. Models such as Huatuo [
7] and Bianque [
8] exemplify this trend. However, despite some advancements in their ability to respond to user queries, the performance of these models remains less than fully satisfactory. A primary contributing factor to this limitation is the lack of traceability in the generated answers, alongside the inconsistent quality of the open-source medical data used for training, which presents a significant barrier to further improvement [
9]. Current data construction methodologies primarily focus on enriching LLMs with medical data derived from either real or synthetic dialogues. While these datasets offer valuable resources, they also introduce potential human errors, diminishing the credibility of model outputs and presenting challenges to the professionalism of the generated answers.
Therefore, we propose a more robust data construction approach complemented by effective filtering and verification mechanisms to enhance the quality and reliability of the training data. By continuously refining the RAG strategy and integrating it with high-quality, curated medical data, we introduce a novel approach: full-parameter fine-tuning of LLMs within the medical domain using the RAG framework. This strategy not only improves the model’s ability to accurately identify relevant retrieval segments but also empowers LLMs to play an increasingly vital role in the medical field, offering more reliable and professional outputs. Our contributions can be summarized as follows:
We have developed an automated framework for generating diverse and high-quality RAG data tailored to the geriatric medicine domain. This framework leverages publicly available disease encyclopedia information from authoritative Chinese medical websites, resulting in the creation of xywyRAGQA, the Chinese medical knowledge RAG QA dataset;
We have applied the RAG strategy to conduct full-parameter fine-tuning of LLMs for the geriatric medicine domain. By integrating external knowledge sources, our approach significantly enhances the model’s ability to accurately identify and utilize the correct retrieved segments;
We have designed evaluation metrics to assess the professionalism and accuracy of the model. Experimental results demonstrate that, compared to "general LLM+RAG" strategy and "domain-finetuned LLM+RAG" strategy, our proposed method achieves notable improvements in geriatric medical QA tasks while also delivering outstanding performance in general domain QA tasks.
2. Related Work
2.1. Domain Adaptation Strategies for LLM
Although large language models (LLMs) have achieved remarkable success in general domains, their performance often falls short in fields requiring deep expertise, such as geriatric medicine. This limitation primarily arises from the insufficient domain-specific knowledge embedded within LLMs [
13]. To address this challenge, researchers have explored various approaches to adapt LLMs to specialized fields. One strategy involves pre-training models on medical corpora, enabling them to acquire specialized vocabulary and achieve more precise knowledge representation [
11]. Another approach infuses medical knowledge directly into LLMs to enhance their understanding and application of domain-specific concepts [
11]. Additionally, fine-tuning using synthetic medical dialogues or real clinical conversations further improves the adaptability of larger models to medical scenarios [
12]. These advancements demonstrate the potential of LLMs in healthcare, emphasizing that the accuracy and professionalism of their responses depend heavily on the quality of their embedded knowledge.
However, instruction-tuned LLMs face notable limitations in the medical domain. They struggle to address outdated or inaccurate internal knowledge, often failing to provide reliable information on recent research or specific cases. Moreover, even after fine-tuning, these models may underperform on rare or complex professional queries, a critical issue in high-stakes fields like medicine where incorrect information can have severe consequences.
To overcome these challenges, the Retrieval-Augmented Generation (RAG) strategy has been introduced. RAG combines the generative capabilities of LLMs with real-time retrieval from external knowledge bases, dynamically incorporating relevant information to enhance the accuracy and timeliness of responses. This integration offers a promising solution for improving the reliability and professionalism of LLMs in specialized domains like medicine.
2.2. Enhancing Domain QA with RAG
Retrieval-Augmented Generation (RAG) combines retrieval mechanisms with generative models, dynamically incorporating external knowledge to enhance the quality of generation tasks [
13]. In evidence-based QA, RAG retrieves relevant documents from large-scale knowledge bases and generates answers based on cited evidence, ensuring both coherence and factual accuracy [
14].
In the medical domain, RAG has shown promise in improving information retrieval and clinical decision support. BioReader [
15] enhances model inputs by retrieving scientific literature from a PubMed-based database of 60 million entries, enabling efficient fine-tuning and accurate predictions across diverse tasks. BEEP integrates patient-specific records with relevant medical literature to improve clinical outcome predictions, such as in-hospital mortality [
16]. Almanac provides real-time access to the latest medical guidelines, supporting clinicians in making informed decisions [
17].
Despite its potential, RAG models depend on high-quality evidence-based QA datasets, which are scarce due to the labor-intensive annotation process. Existing datasets, like MedMCQA [
18] and PubMedQA [
19], often suffer from noise and inconsistency, limiting their reliability and the performance of RAG models. Addressing these challenges is essential to fully harness RAG’s capabilities in healthcare applications.
2.3. Optimizing Fine-Tuning and Data Quality for Domain-Specific Models
Fine-tuning LLM for specific tasks often requires substantial amounts of manually labeled data, which is typically unavailable for many downstream applications due to high annotation costs. This data scarcity presents a significant challenge for task-specific model optimization. To mitigate this, researchers have explored approaches such as knowledge distillation [
20,
21,
22], data augmentation [
23,
24], module replacement [
25], semi-supervised learning [
26], and data synthesis [
27], aiming to reduce the reliance on large annotated datasets.
In the medical domain, data availability faces additional challenges. Much of the medical data is siloed within independent systems of various institutions, resulting in a pervasive “data island” phenomenon. Furthermore, real-world medical datasets are limited, and open-source Chinese medical Q&A datasets often suffer from uniform and simplistic question formats. Current strategies primarily rely on providing LLMs with medical data derived from real or synthetic dialogues [
28]. However, these methods are prone to human error, and solely relying on supervised fine-tuning to train LLMs for accurate, consistent, and non-hallucinatory responses remains a complex challenge.
RAG models, which leverage both retrieval and generation, show great potential in addressing these challenges. However, their performance heavily depends on the availability of high-quality training datasets. The scarcity of domain-specific RAG datasets has significantly limited the application of these models in professional fields. Addressing this issue requires increased investment in constructing and annotating high-quality datasets and exploring novel methods for data synthesis and enhancement to support more robust applications.
3. Methodology
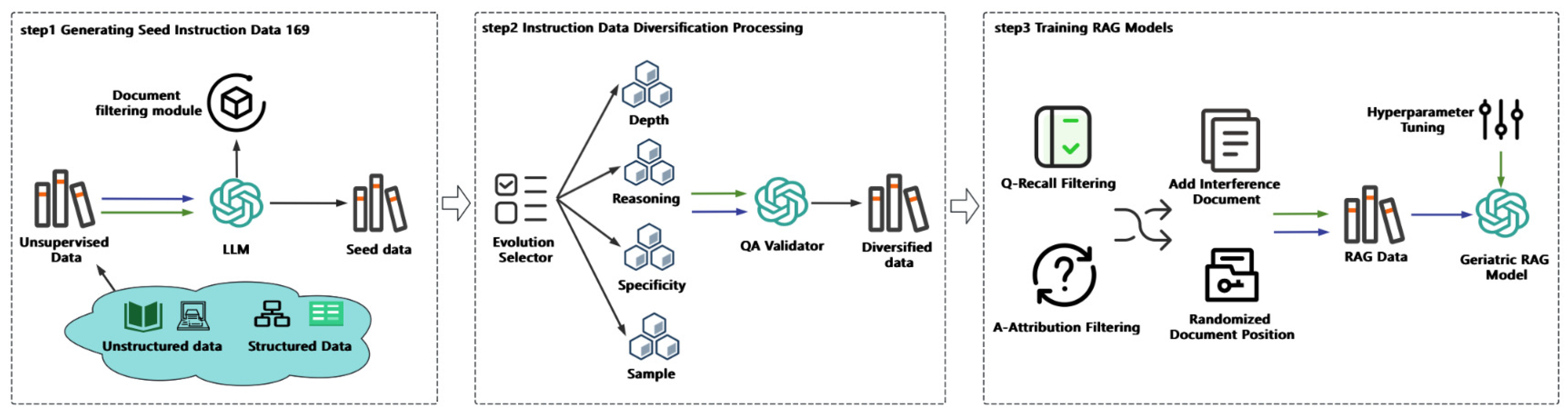
In this paper, we begin by introducing the unsupervised medical data utilized in our study. Next, we transform this unsupervised knowledge into training data optimized for the RAG framework. Finally, we fine-tune a large language model (LLM) tailored for the RAG strategy, enabling it to generate responses based on retrieved, domain-relevant medical knowledge. The overall workflow is depicted in
Figure 1.
3.1. Generating Seed Instruction Data
We collected the latest geriatric medical data from authoritative medical websites to construct a high-quality unsupervised dataset. Leveraging the advanced generative capabilities of large language models (LLMs), we implemented a meticulously designed guidance mechanism to generate medical instruction data closely aligned with real-world clinical scenarios. This approach not only significantly reduces the workload associated with manual annotation but also substantially enhances the practicality and reliability of the resulting data.
3.1.1. Unsupervised Knowledge Acquisition
Medical knowledge encompasses both structured formats, such as medical knowledge graphs and databases, and unstructured formats, such as medical guidelines and literature. In this study, we focus on the disease encyclopedia section related to geriatric medicine from an authoritative medical website. This data source combines both structured and unstructured knowledge relevant to elderly healthcare. First, the structured medical knowledge is transformed into unstructured text, converting the structured content into a free-text format. Next, we apply tailored filtering rules to remove garbled or incomplete data, ensuring the resulting medical text is accurate, comprehensive, and suitable for downstream applications in geriatric medicine.
3.1.2. Geriatric Medical Seed Instruction Data Generation
Based on unsupervised data from the field of geriatric medicine, we harnessed the powerful generative capabilities of large language models (LLMs) to create seed instruction data tailored to geriatric healthcare. To achieve this, we designed a systematic prompting strategy that explicitly assigns the model the role of a “physician.” By embedding professional medical contexts and well-defined task objectives into the prompts, the model is guided to understand task requirements from a specialized perspective and generate instruction data aligned with medical standards.
To further enhance the quality and logical depth of the generated data, we integrated the Chain-of-Thought (COT) [
29] mechanism into the prompting process. This approach encourages the model to engage in step-by-step reasoning, significantly improving its comprehension and handling of complex tasks. By structuring the reasoning process into a series of sequential thought steps, COT ensures the generated instructions exhibit clear logical progression, maintaining both rigor and scientific accuracy. Moreover, this incremental reasoning framework enhances the model’s reliability and consistency in generating instructions for multi-step tasks, making it particularly effective for sophisticated scenarios in geriatric medicine.
3.2. Instruction Data Diversification Processing
The quality and realism of synthetic data are critical for model performance. Synthetic data often diverges from real-world distributions, leading to high training accuracy but poor performance on real-world tasks. In contrast, models trained on manually annotated data exhibit smaller discrepancies between training and testing accuracy [?]. To mitigate this gap, we use the seed instruction data generated in last step and further diversify it to ensure alignment with human preferences.
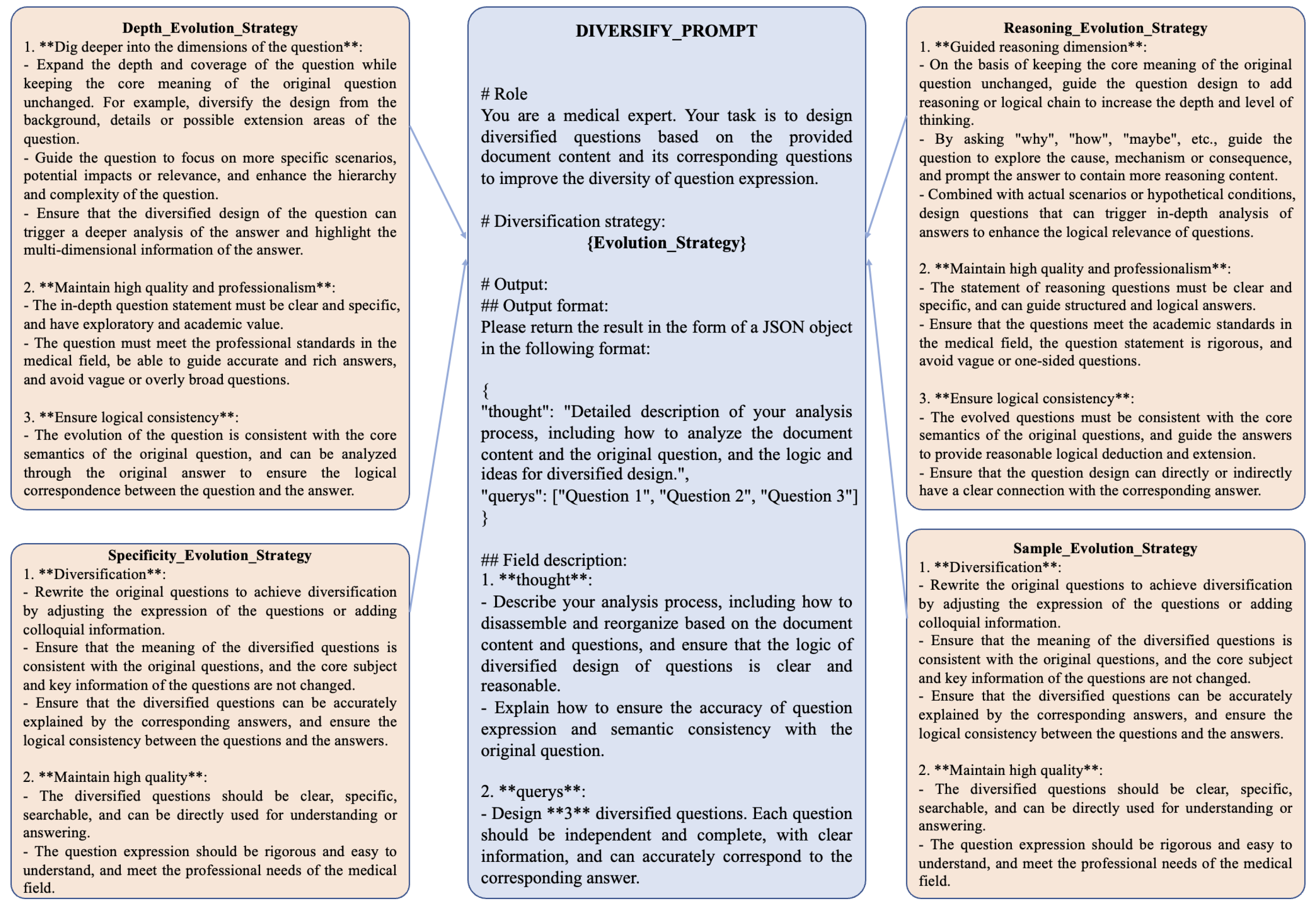
In real-world patient-doctor interactions, patients typically pose detailed, specific, and complex questions about their symptoms, rather than generalized or simplistic inquiries. Manually creating diverse and intricate instructional data is both time-consuming and labor-intensive, particularly when crafting highly nuanced instructions. To address this challenge, we have developed a method that harnesses the generative capabilities of large language models (LLMs) to produce diversified instructional data across varying levels of complexity. This approach is guided by four targeted evolutionary strategies designed to enhance the depth, specificity, and diversity of the instructional dataset. The four strategies are described as follows:
Depth Evolution Strategy: This strategy increases the depth of questions by introducing more intricate scenarios or requiring multi-step reasoning for responses;
Reasoning Evolution Strategy: This approach emphasizes logical progression and causal relationships, encouraging the generation of questions that require comprehensive inferential reasoning;
Specificity Evolution Strategy: This method focuses on creating highly specific questions tailored to individual conditions or unique patient scenarios, moving away from generic templates;
Sample Evolution Strategy: This strategy diversifies the dataset by introducing variations in patient demographics, symptom descriptions, or contextual settings, simulating a wide range of real-world medical cases.
Through iterative application of these strategies, we enhance the complexity, richness, and diversity of the instruction dataset, ensuring it better aligns with the variability and specificity found in real-world medical scenarios. Furthermore, we have designed dynamic prompt templates capable of switching between strategies in a randomized manner. These templates employ diverse question diversification techniques, enabling the generation of highly varied instruction data. This approach introduces randomness and adaptability into the instruction generation process, ensuring a broader representation of potential patient inquiries.The prompt templates used for generating diversified instructional data are illustrated in
Figure 2.
3.3. Instruction Data Quality Filtering
Designing automated evaluation metrics for instruction data quality is a critical task to ensure data consistency and maintain high standards. To achieve this, we established two key metrics: question recall rate and answer attributability, which are used to filter and identify high-quality data for RAG tasks. These metrics play a pivotal role in enhancing the overall reliability and effectiveness of the dataset.
3.3.1. Question Recall Rate
To ensure that the questions generated in the question-answer pairs are relevant to the document content, we follow these steps for quality control of the questions:
First, we convert all document content into vector representations by using an embedding model. Suppose there are N documents, where each document is vectorized as . Similarly, a question is vectorized as . The process for calculating the question recall rate for each document is as follows:
For the set of questions
Q generated from
, we use the vector representation of the question
to retrieve the most relevant documents from the vector library. The cosine similarity between the question
and each document vector
is computed using the following equation:
The top 10 documents with the highest similarity are selected as the retrieval results, denoted as
. Next, we check the position of the correct document
within the retrieved results:
If the correct document appears in the top 3 positions, i.e., , then the question is retained. Otherwise, the question is filtered out.
3.3.2. Answer Attributability
Using an NLI (Natural Language Inference) model for quality control is an effective method, as NLI models validate the logical relationship between an answer and its corresponding document content, thereby assessing the attributability of the answer. The scoring for answer attributability is defined as follows:
where:
: Total number of sentences in the answer.
: Subset of sentences in the answer identified as entailing the document content.
: Subset of sentences in the answer identified as contradicting the document content.
: Subset of sentences in the answer identified as neutral to the document content.
To achieve high-precision entailment detection, we utilize the top-performing attributability prediction models. If both models predict a sentence as “attributable,” the sentence is considered to be fact-supported and is included in . We set an attributability threshold to filter out synthetic data and retain only high-quality RAG data. This approach ensures that the generated answers are logically consistent with the document content, improving the reliability of the dataset.
3.4. Training RAG Models
To effectively train a high-performing RAG model, traditional reading comprehension datasets, while improving a model’s ability to answer questions based on single documents, are insufficient for the demands of RAG tasks. RAG typically requires retrieving the top 50 or more documents, making it essential to construct multi-document reading comprehension datasets, or RAG-specific datasets. These datasets train the model to first identify the correct document, then perform reading comprehension on it, and finally generate an accurate answer.
Several key considerations must be addressed when constructing RAG datasets. First, since retrieved documents are ranked based on their similarity to the query, correct-answer documents are often placed in the top positions. This creates a misleading assumption that answers are always found in the leading documents, limiting the model’s performance. LLMs naturally prioritize highly ranked documents and may overlook answers located in middle or lower-ranked documents. However, in real-world scenarios, answers can appear randomly within the retrieved documents. Therefore, constructing multi-document datasets must deliberately randomize the placement of correct-answer documents to better reflect practical situations.Second, regarding the selection of distractor documents, a common approach is to use documents entirely unrelated to the correct-answer document to minimize interference and improve the model’s ability to identify the correct document. However, this method is not aligned with the requirements of RAG tasks, where retrieved documents are typically highly relevant to the query. Instead, distractor documents should be selected to be closely related to the correct-answer document but without containing the correct answer. This strategy enhances the model’s ability to distinguish the correct document within highly similar contexts, while also exposing it to domain-relevant distractors, thereby deepening the model’s understanding of the knowledge field.
In summary, constructing multi-document reading comprehension datasets for RAG training requires adherence to the following principles:
Randomized placement of correct-answer documents to prevent the model from developing position-based biases;
Selection of relevant distractor documents to simulate realistic retrieval scenarios, improve discrimination in challenging contexts, and enhance the model’s domain knowledge.
3.4.1. Set Relevant Distractor Documents
The selection of distractor documents is based on a retrieval database of geriatric medical documents. Using an embedding model, documents are randomly selected from a preprocessed and cleaned corpus, ensuring semantic similarity scores between 0.5 and 0.9 relative to the correct-answer document. These distractor documents must exhibit significant content differences from the correct-answer document to increase the difficulty of the retrieval task, thereby effectively enhancing the model’s retrieval accuracy. Additionally, distractor documents should avoid duplication and maintain high quality to prevent introducing noise that could disrupt model training. The primary goal of this step is to incorporate negative samples into the dataset, creating a contrastive learning scenario. This enables the model to accurately identify and retrieve relevant information from semantically similar but content-wise unrelated documents, improving its robustness and precision in complex retrieval tasks.
The semantic similarity between texts, denoted as , is calculated for document pairs and . Using an embedding model, the vector representations of documents, and , are computed. For the correct-answer document , candidate distractor documents are selected from the document corpus based on the following conditions: . Here, represents documents within the corpus that serve as potential distractor documents. For the filtered distractor document set , content redundancy is further eliminated by removing document pairs where the similarity exceeds: . Finally, the correct-answer document is combined with the selected distractor documents to construct the RAG dataset, enabling effective training for retrieval-augmented generation tasks.
3.4.2. Randomly Place Correct Documents
To enhance the model’s ability to perceive the position of correct documents in RAG data, we designed a targeted distribution strategy for correct document placement. Given that correct documents are typically retrieved in the first position in most cases, we allocated 50% of the correct documents to the top position. To address potential decreases in retrieval accuracy, the remaining 50% of the correct documents were distributed as follows: 40% were randomly placed within the top 10 retrieved documents, while the remaining 10% were randomly distributed across all retrieved documents. This strategy aims to balance the model’s adaptability to both high-accuracy and low-accuracy retrieval scenarios, enhancing its robustness and overall performance in practical applications.
3.5. Evaluation Metrics
3.5.1. Domain Metric
In the medical domain, to evaluate the effectiveness of various models in responding to user queries, we propose a comprehensive evaluation metric that incorporates both Answer Correctness and Semantic Similarity. This metric is designed to provide a holistic assessment of the quality of model-generated responses, ensuring that they meet the necessary standards of accuracy and contextual relevance.
The metric integrates Answer Correctness, which evaluates the classification accuracy of the model, and Semantic Similarity, which assesses the degree of alignment in linguistic expression and content coverage between the generated answers and standard reference answers. Specifically, Answer Correctness is evaluated using GPT-4, which provides a robust assessment of factual accuracy by leveraging its advanced reasoning and comprehension capabilities. Semantic Similarity, on the other hand, is evaluated using the pre-trained embedding model bge-large-zh-v1.5, which calculates the degree of alignment between generated and reference answers by analyzing linguistic expression and content representation.
The calculation of the Answer Correctness is defined as follows:
where:
: Number of true positives.
: Number of false positives.
: Number of false negatives.
The overall metric is calculated as a weighted sum of the Answer Correctness and semantic similarity:
where:
: Weight assigned to the Answer Correctness, with a default value of 0.75.
: Weight assigned to Semantic Similarity, with a default value of 0.25.
This comprehensive metric ensures that the evaluation framework is both robust and practical. It measures the ability of the model to deliver accurate responses while maintaining a high degree of semantic alignment, thereby reflecting the model’s overall effectiveness in real-world applications. This comprehensive framework is particularly suited for the medical domain, where the reliability and contextual appropriateness of responses are critical. By employing this metric, we provide a systematic and reliable tool for assessing and improving the quality of model-generated answers in medical settings.
3.5.2. General Metric
To evaluate the general capabilities of the model in the general domain, we utilized the Chinese tasks from the Longbench [
31] benchmarking suite. Longbench offers a diverse collection of tasks and datasets, making it an ideal framework for assessing the multifaceted competencies of language models across various dimensions.
We primarily utilized the following tasks from Longbench for our evaluation:
LSHT: A Chinese classification task that involves categorizing news articles into 24 distinct categories;
DuReader: A task requiring the answering of relevant Chinese questions based on multiple retrieved documents;
MultiFieldQA_ZH: A question-answering task based on a single document, where the documents span diverse domains;
VCSum: A summarization task that entails generating concise summaries of Chinese meeting transcripts;
Passage_Retrieval_ZH: A retrieval task where, given several Chinese passages from the C4 dataset, the model must identify which passage corresponds to a given summary.
4. Results and Discussion
4.1. Data Diversity
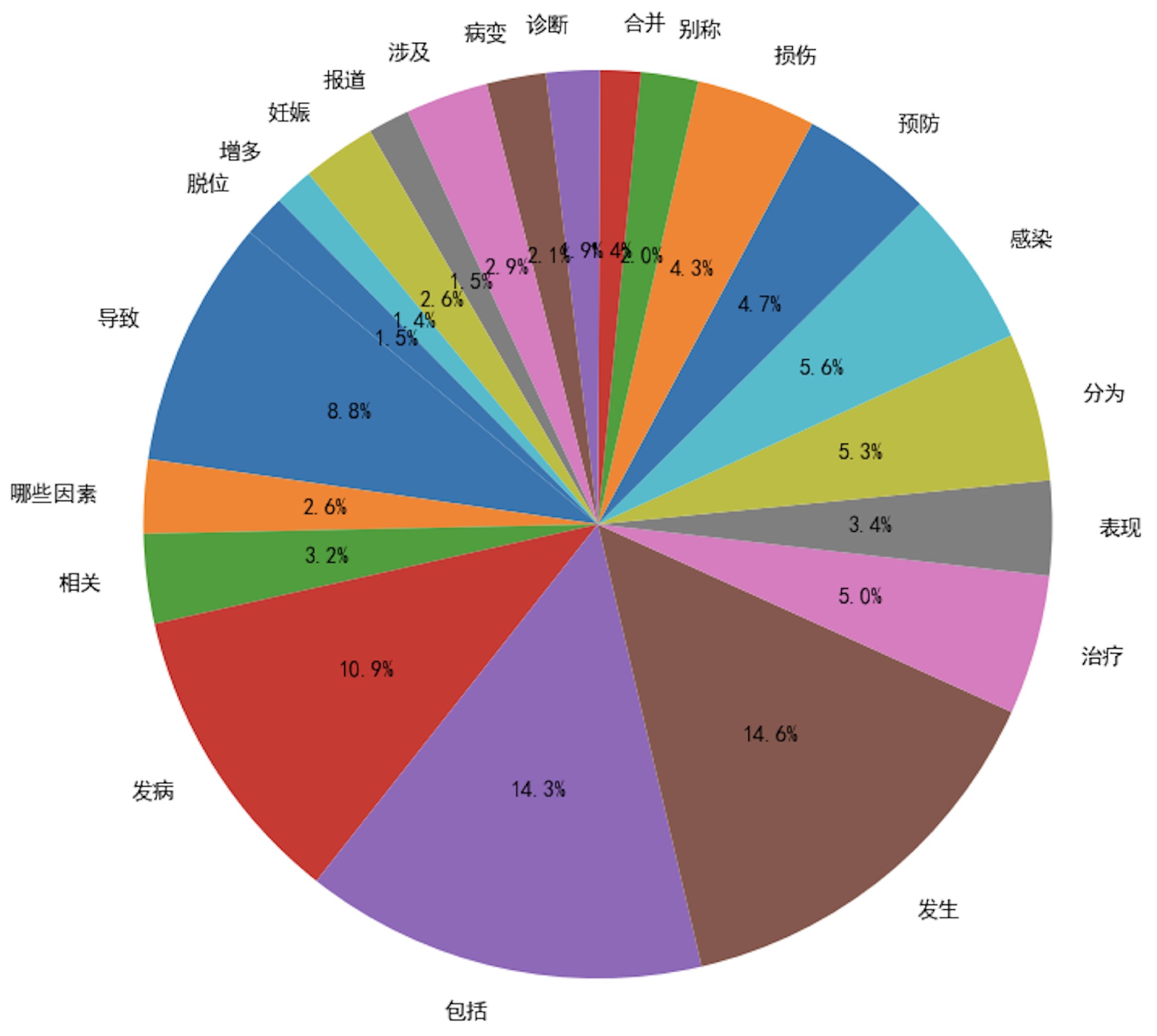
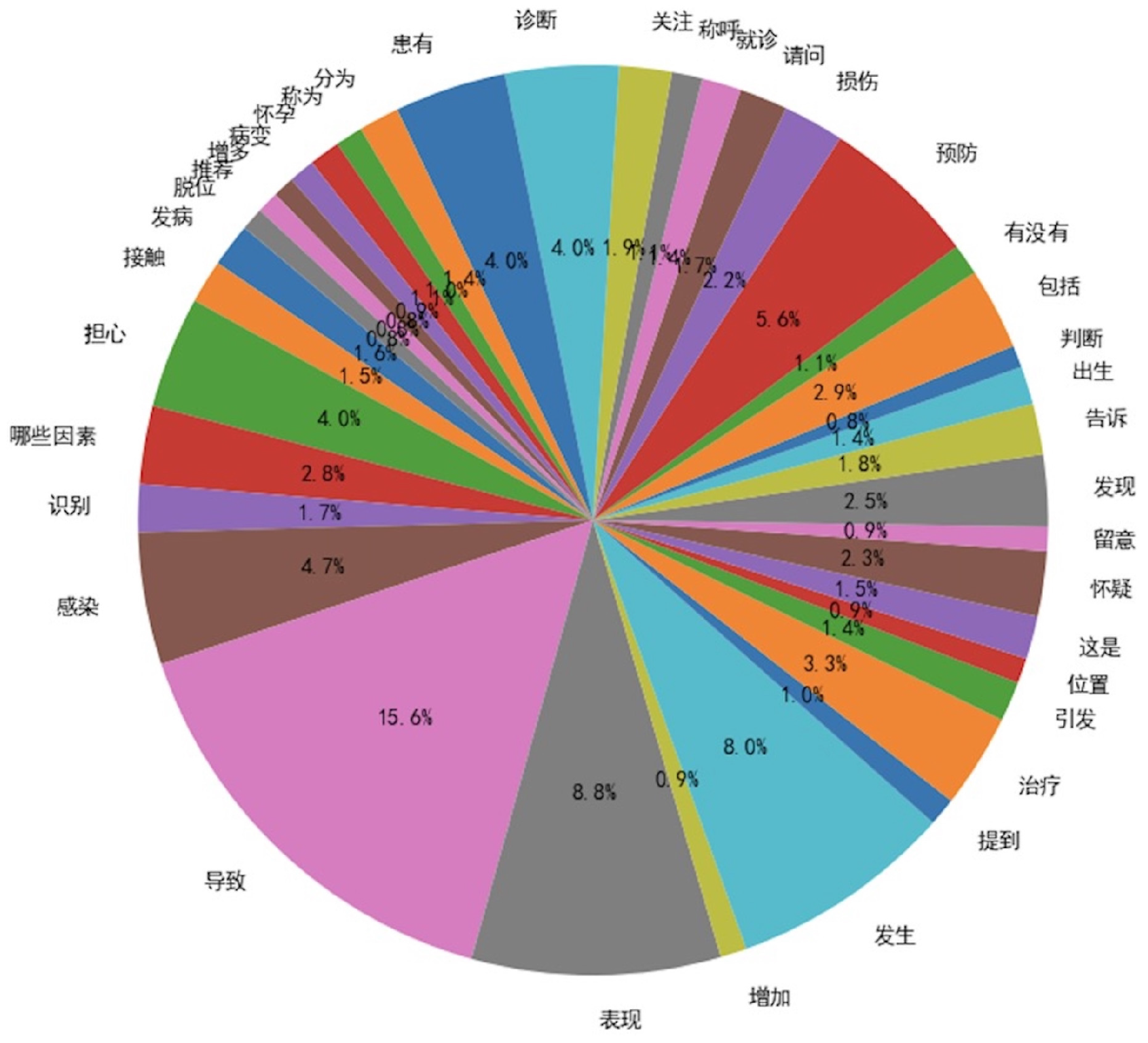
To validate the effectiveness of our approach in generating diversified instructional data, we introduced two metrics for diversity assessment:
Verb Usage Frequency: A higher number of verbs exceeding a predefined frequency threshold indicates greater diversity;
ROUGE-L: A lower average ROUGE-L score within the same dataset signifies higher diversity.
In the evaluation process, we analyzed and compared the verb usage frequency in the seed instruction data and the diversified instruction data, using a frequency threshold of 50. As illustrated in
Figure 3 and
Figure 4, the diversified instruction data incorporates a significantly greater variety of verbs compared to the seed instruction data. Furthermore, we examined the ROUGE-L score distributions of the two datasets. As depicted in
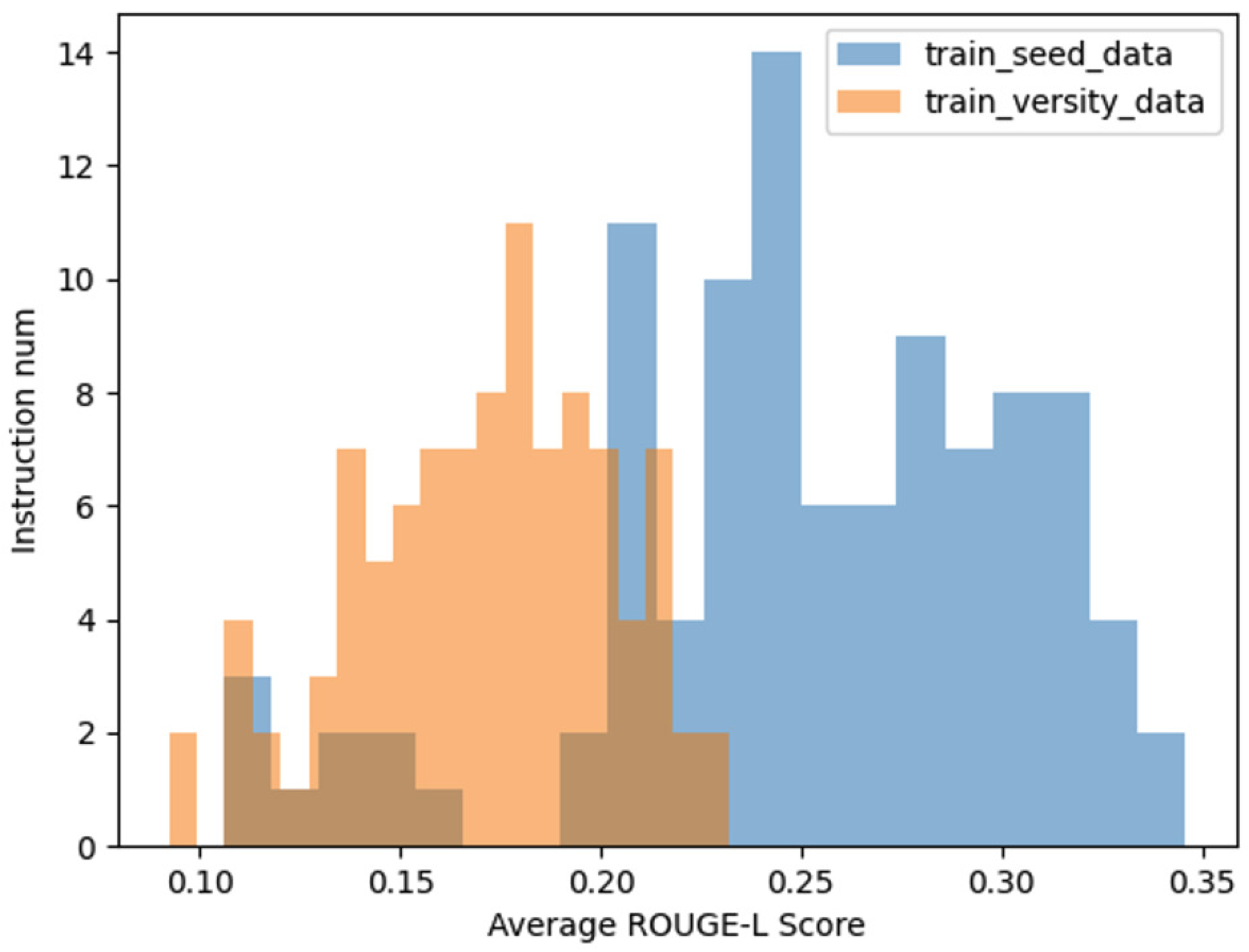
Figure 5, the average ROUGE-L score for the diversified instruction data is notably lower than that of the seed instruction data, reinforcing the conclusion that our method successfully enhances diversity.
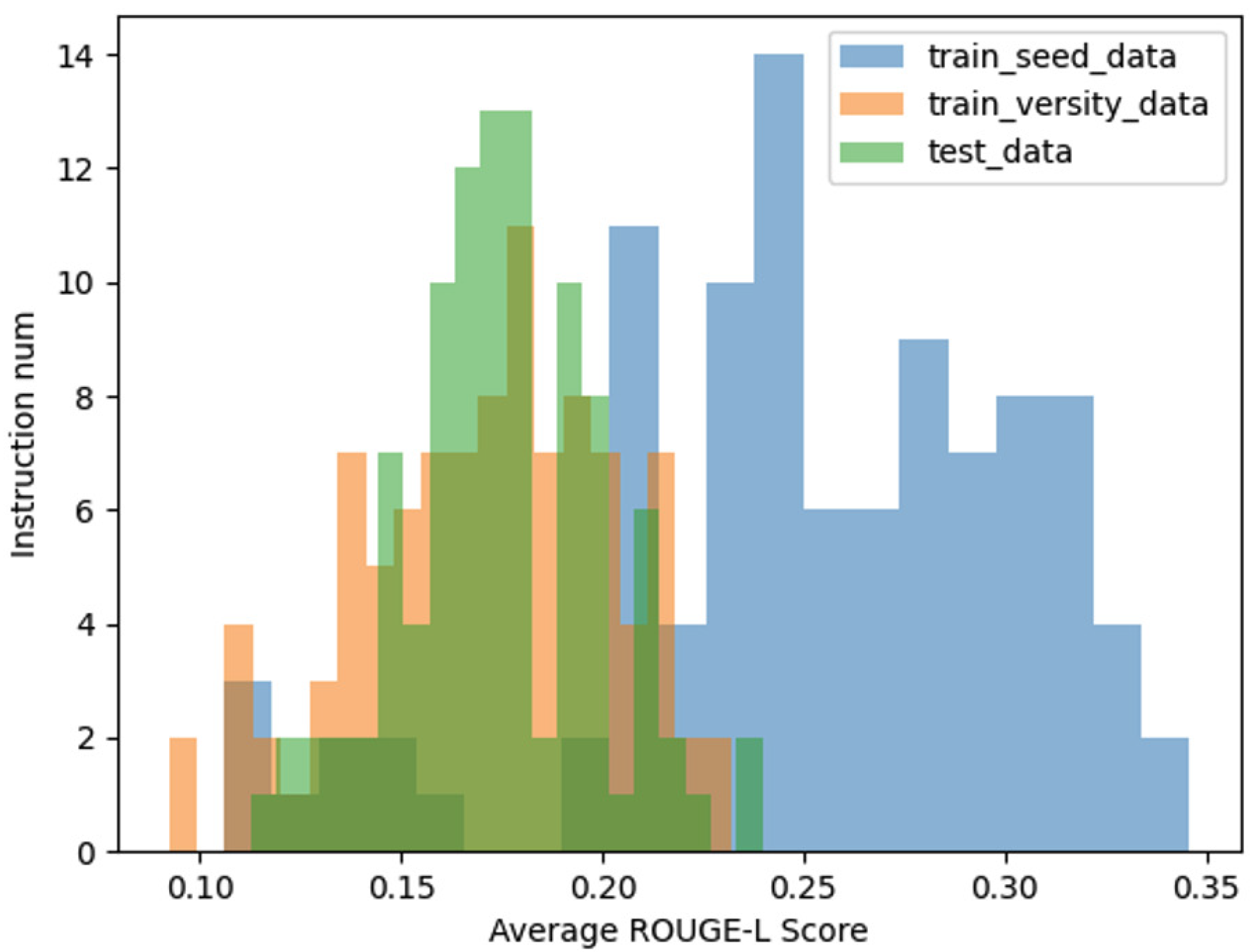
In addition to the aforementioned analyses, we further compared the average ROUGE-L score distributions among three datasets: the seed instruction data, the diversified instruction data, and a set of 100 manually curated instruction datasets, which serve as a benchmark for real-world data. As illustrated in
Figure 7 , the distribution of the ROUGE-L scores for the diversified instruction data exhibits a closer alignment with the distribution observed in the manually curated data.
This comparison underscores the effectiveness of our method in approximating the characteristics of real-world instructional data. By achieving a ROUGE-L score distribution that closely mirrors that of human-generated data, the diversified instruction data demonstrates not only increased variety but also enhanced representational fidelity to real-world scenarios. This alignment further validates the practical utility of our approach in generating high-quality, diverse instructional datasets.
4.2. Model Training
Due to computational resource constraints, we selected three models as the base models for our experiments: Qwen2.5-7B-Instruct, DeepSeek-V2-Lite-Chat, and GLM-4-9B-Chat. Qwen2.5-7B-Instruct represents the latest iteration of Alibaba’s large language model, incorporating 7 billion parameters and improved instruction-following capabilities. DeepSeek-V2-Lite-Chat is a lightweight, high-performance model optimized for conversational tasks, while GLM-4-9B-Chat is a 9-billion-parameter model designed for advanced generative language modeling.
Leveraging our constructed diversified instruction data, we applied supervised fine-tuning (SFT) and RAG instruction fine-tuning to these base models. This multi-model approach enables us to evaluate the effectiveness of our methods across varying model architectures and parameter scales, ensuring robust and comprehensive performance analysis.
The experimental results presented in
Table 1 underscore significant variations in model performance across different strategies, particularly in terms of correctness and similarity within the test set in the medical domain. The baseline model, while establishing a foundational benchmark, exhibited limitations in both correctness and similarity. However, when integrated with the RAG strategy, a marked improvement was observed, reflecting the ability of RAG to enhance both the accuracy and semantic consistency of the model’s responses. This suggests that incorporating external retrieval mechanisms, as implemented in the RAG strategy, can substantially enrich the model’s understanding and alignment with domain-specific information. Further analysis reveals that models utilizing the SFT strategy demonstrated notable advancements in correctness, surpassing the baseline model. Nevertheless, this gain in correctness was accompanied by a marginal decline in similarity, indicating a potential trade-off between precision in reasoning and semantic alignment. Importantly, when the SFT strategy was combined with RAG, the model achieved significant gains across both metrics, demonstrating the complementary nature of these approaches. This combination effectively balances domain-specific fine-tuning with enhanced contextual retrieval, leading to more robust performance. The final optimized model, which integrates our advanced SFT strategy, achieved the highest scores in both correctness and similarity between all configurations. This result highlights the effectiveness and superiority of our approach in addressing the complex challenges of medical test sets. By leveraging the strengths of SFT and RAG in a unified framework, the model demonstrates its capability to achieve exceptional performance in tasks requiring high accuracy and semantic alignment. These findings not only emphasize the robustness of our methodology but also underscore its potential for broader applications in domains where precision and reliability are paramount.
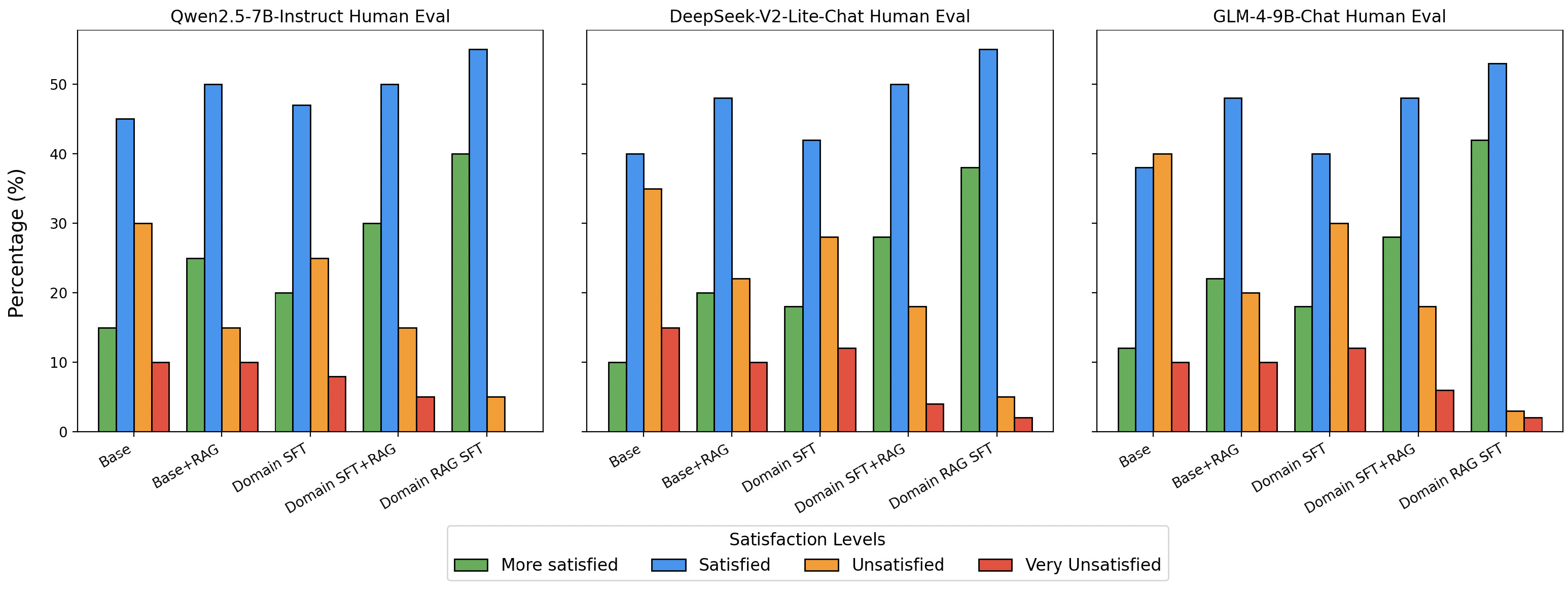
To further validate the effectiveness of our approach beyond automated metrics, we conducted human evaluations with medical domain experts.
Figure 7 illustrates the human evaluation results conducted by five medical students who were tasked with assessing model-generated answers on a predefined medical test set. Each student categorized the responses into four levels of satisfaction: More Satisfied, Satisfied, Unsatisfied, and Very Unsatisfied. The baseline model exhibited moderate levels of satisfaction, indicating foundational performance but leaving room for improvement. The integration of the RAG strategy significantly improved satisfaction scores, as evidenced by a noticeable reduction in dissatisfaction rates. Models incorporating the SFT strategy further demonstrated enhanced correctness and contextual understanding, with higher proportions of responses in the “More Satisfied” and “Satisfied” categories. The combination of SFT and RAG strategies yielded the most notable results, with the final model achieving the highest satisfaction rates among all configurations. These evaluations, conducted by individuals with medical domain expertise, provide robust evidence for the effectiveness of our approach in optimizing model performance and user satisfaction in practical, domain-specific scenarios.
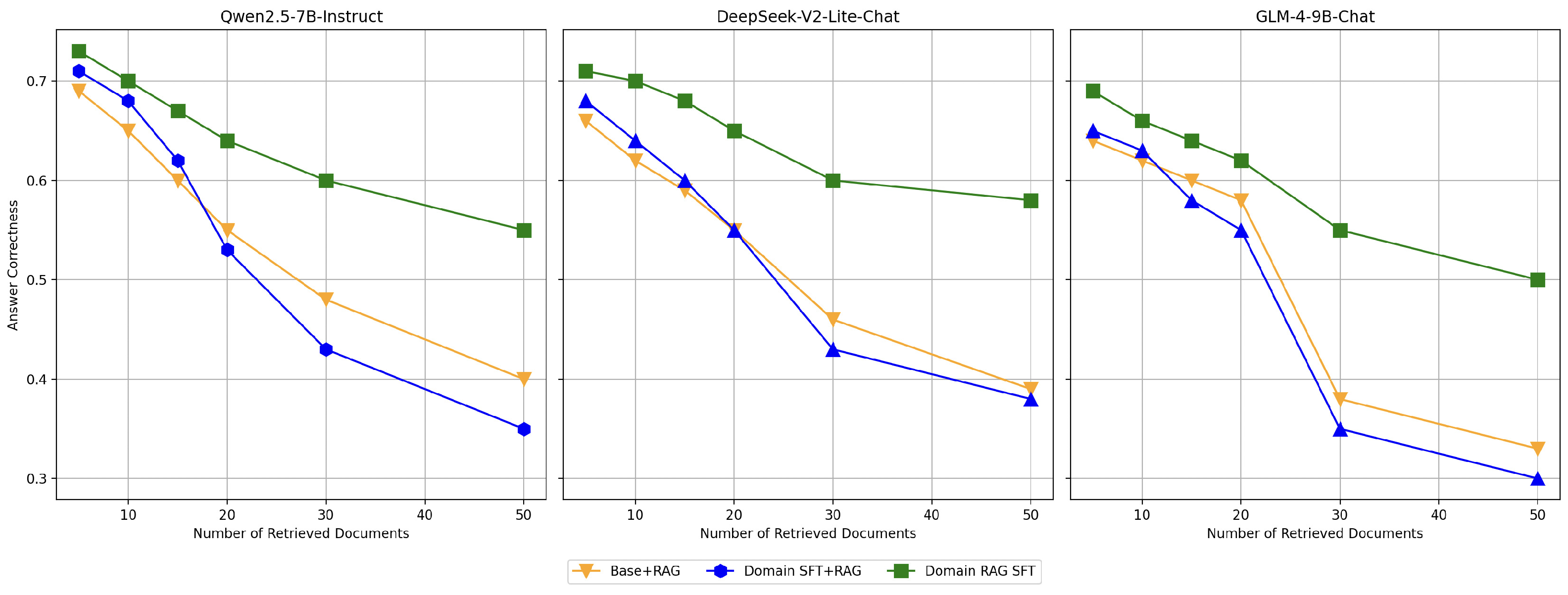
To evaluate the impact of increasing the number of retrieved documents on model performance, we conducted the following experiment: using the same test set, we varied the number of retrieved documents. The results demonstrate that as the number of retrieved documents increases, the interference from irrelevant or low-relevance documents becomes more pronounced, leading to a gradual decline in model performance. This trend highlights the challenges of maintaining correctness in scenarios with abundant retrieved information. The experimental results are shown in
Figure 8.
Among the configurations, the Domain Fine-Tuned model (Domain SFT) exhibits a significant decline in performance as the number of retrieved documents increases. Notably, when a large number of documents are retrieved, its performance even falls below that of the Base+RAG model. This indicates that while domain-specific fine-tuning enhances correctness in scenarios with fewer retrieved documents, it struggles to effectively manage the noise introduced by larger retrieval sets. In contrast, the Domain RAG Fine-Tuned model (Domain RAG SFT) demonstrates remarkable robustness across varying retrieval quantities. By applying domain-specific optimization to the retrieval-augmented generation (RAG) framework, this configuration improves the model’s ability to identify and focus on relevant documents, mitigating the negative impact of irrelevant retrievals. As a result, its performance remains relatively stable, showing only moderate decline even when the number of retrieved documents increases significantly.
These findings validate the effectiveness of the Domain RAG Fine-Tuning strategy. By enhancing the model’s sensitivity to relevant documents, this approach addresses the limitations of traditional fine-tuning methods. It ensures superior performance in retrieval-augmented generation tasks, even under challenging scenarios with a large number of retrieved documents.
To verify whether fine-tuning tailored for RAG would compromise the model’s original general capabilities, we conducted experiments on five Chinese tasks from Longbench. The experiments involved testing the Base model, the Domain SFT model, and the Domain RAG SFT model. The results, as shown in
Table 2, the Domain SFT model, which relies solely on traditional domain-specific fine-tuning, appears to harm the model’s original general capabilities due to its narrow focus. For example, its performance dropped on tasks like multifieldqa_zh and passage_retrieval_zh, suggesting that the singular nature of domain-specific fine-tuning may impair the model’s ability to generalize effectively. In contrast, the Domain RAG SFT model, designed to enhance the model’s sensitivity to relevant documents, not only avoided reducing the model’s general capabilities but also achieved modest improvements in reading comprehension tasks. This demonstrates that the Domain RAG SFT strategy effectively balances the trade-off between domain-specific enhancements and general-purpose performance.
In summary, the results highlight that while traditional domain fine-tuning may risk diminishing a model’s general capabilities, the Domain RAG SFT strategy successfully preserves these capabilities. Moreover, it introduces measurable improvements in tasks such as reading comprehension, proving its effectiveness in augmenting both specialized and general abilities.
5. Conclusions
This study explores the application of advanced retrieval-augmented generation (RAG) strategies in the field of geriatric medicine, addressing the critical need for accurate and reliable responses in medical question-answering (QA) systems. By leveraging publicly available medical knowledge, we proposed an automated method for generating high-quality RAG datasets specific to the geriatric domain. This approach enabled the creation of a specialized Chinese medical knowledge QA dataset tailored for geriatric healthcare.
The integration of RAG strategies introduced external knowledge sources into large language models, resulting in a significant improvement in answer quality, particularly in terms of authenticity and accuracy. To evaluate model performance in the medical field, we designed two tailored metrics: answer similarity and answer correctness. Experimental results consistently demonstrated the superiority of the proposed approach over baseline methods in delivering reliable and precise answers.
Furthermore, we validated the generalization capabilities of the proposed model through evaluations on diverse Chinese tasks, showcasing its adaptability to broader QA scenarios beyond the medical domain. This highlights the dual advantage of RAG-based approaches in enhancing specialized domain performance while maintaining strong general-purpose capabilities.
In conclusion, our research provides an efficient and precise QA framework for geriatric medicine and offers valuable insights into the broader application of RAG strategies in specialized domains. As high-quality datasets and retrieval techniques continue to advance, we anticipate that RAG-based QA models will become indispensable tools across various fields, delivering accurate and trustworthy information to meet diverse user needs.
Author Contributions
Conceptualization, B.W.; methodology, B.W. and S.L.; software, B.W.; validation, B.W., Z.H. and C.L.; formal analysis, B.W.; investigation, B.W.; resources, S.L.; data curation, B.W. and C.L.; writing—original draft preparation, B.W. , S.L. and Z.H.; writing—review and editing, B.W. , S.L. , Z.H. and C.L.; visualization, B.W.; supervision, S.L.; project administration, B.W. , S.L. and Z.H.; funding acquisition, S.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Research on Key Technologies and System Development of Global Collaborative Cognitive Computing for Livable Cities grant number 2020YFB2104402.
Data Availability Statement
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| QA |
Question-answering |
| LLM |
Large Language Model |
| RAG |
Retrieval-augmented Generation |
| SFT |
Supervised Fine-Tuning |
References
- Brown, T.; Mann, B.; Ryder, N.; et al. Language models are few-shot learners. Adv. Neural Inf. Process. Syst. 2020, 33, 1877–1901. [Google Scholar]
- Lazaridou, A.; Gribovskaya, E.; Stokowiec, W.; et al. Internet-augmented language models through few-shot prompting for open-domain question answering. arXiv Preprint 2022, arXiv:2203.05115. [Google Scholar]
- Ni, J.; Bingler, J.; Colesanti-Senni, C.; et al. Chatreport: Democratizing sustainability disclosure analysis through LLM-based tools. arXiv Preprint 2023, arXiv:2307.15770. [Google Scholar]
- Ji, Z.; Lee, N.; Frieske, R.; et al. Survey of hallucination in natural language generation. ACM Comput. Surv. 2023, 55, 1–38. [Google Scholar] [CrossRef]
- Hu, X.; Chen, J.; Li, X.; et al. Do large language models know about facts? arXiv Preprint 2023, arXiv:2310.05177. [Google Scholar]
- Gao, Y.; Xiong, Y.; Gao, X.; et al. Retrieval-augmented generation for large language models: A survey. arXiv Preprint 2023, arXiv:2312.10997. [Google Scholar]
- Wang, H.; Liu, C.; Xi, N.; et al. Huatuo: Tuning llama model with Chinese medical knowledge. arXiv Preprint 2023, arXiv:2304.06975. [Google Scholar]
- Chen, Y.; Wang, Z.; Xing, X.; et al. Bianque: Balancing the questioning and suggestion ability of health LLMs with multi-turn health conversations polished by ChatGPT. arXiv Preprint 2023, arXiv:2310.15896. [Google Scholar]
- Li, L.; Wang, P.; Yan, J.; et al. Real-world data medical knowledge graph: Construction and applications. Artif. Intell. Med. 2020, 103, 101817. [Google Scholar] [CrossRef]
- Lewis, P.; Ott, M.; Du, J.; et al. Pretrained language models for biomedical and clinical tasks: Understanding and extending the state-of-the-art. In Proceedings of the 3rd Clinical Natural Language Processing Workshop, Online, 19 November 2020; pp. 146–157. [Google Scholar]
- Zhang, T.; Cai, Z.; Wang, C.; et al. SMedBERT: A knowledge-enhanced pre-trained language model with structured semantics for medical text mining. arXiv Preprint 2021, arXiv:2108.08983. [Google Scholar]
- Li, L.; Wang, P.; Yan, J.; et al. Real-world data medical knowledge graph: Construction and applications. Artif. Intell. Med. 2020, 103, 101817. [Google Scholar] [CrossRef] [PubMed]
- Lewis, P.; Perez, E.; Piktus, A.; et al. Retrieval-augmented generation for knowledge-intensive NLP tasks. Adv. Neural Inf. Process. Syst. 2020, 33, 9459–9474. [Google Scholar]
- Borgeaud, S.; Mensch, A.; Hoffmann, J.; et al. Improving language models by retrieving from trillions of tokens. In Proceedings of the International Conference on Machine Learning, Baltimore, MD, USA, 17–23 July 2022; PMLR. pp. 2206–2240. [Google Scholar]
- Frisoni, G.; Mizutani, M.; Moro, G.; et al. Bioreader: A retrieval-enhanced text-to-text transformer for biomedical literature. In Proceedings of the 2022 Conference on Empirical Methods in Natural Language Processing, Online, 7–11 December 2022; pp. 5770–5793. [Google Scholar]
- Naik, A.; Parasa, S.; Feldman, S.; et al. Literature-augmented clinical outcome prediction. arXiv Preprint 2021, arXiv:2111.08374. [Google Scholar]
- Zakka, C.; Shad, R.; Chaurasia, A.; et al. Almanac—retrieval-augmented language models for clinical medicine. NEJM AI 2024, 1, AIoa2300068. [Google Scholar] [CrossRef] [PubMed]
- Pal, A.; Umapathi, L.K.; Sankarasubbu, M. MedMCQA: A large-scale multi-subject multi-choice dataset for medical domain question answering. In Proceedings of the Conference on Health, Inference, and Learning, Virtual, 20–23 April 2022; PMLR; pp. 248–260. [Google Scholar]
- Jin, Q.; Dhingra, B.; Liu, Z.; et al. PubMedQA: A dataset for biomedical research question answering. arXiv Preprint 2019, arXiv:1909.06146. [Google Scholar]
- Fan, A.; Jernite, Y.; Perez, E.; et al. ELI5: Long form question answering. arXiv Preprint 2019, arXiv:1907.09190. [Google Scholar]
- Hinton, G. Distilling the knowledge in a neural network. arXiv Preprint 2015, arXiv:1503.02531. [Google Scholar]
- Beyer, L.; Zhai, X.; Royer, A.; et al. Knowledge distillation: A good teacher is patient and consistent. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022; pp. 10925–10934. [Google Scholar]
- Hsieh, C.Y.; Li, C.L.; Yeh, C.K.; et al. Distilling step-by-step! Outperforming larger language models with less training data and smaller model sizes. arXiv Preprint 2023, arXiv:2305.02301. [Google Scholar]
- DeVries, T.; Taylor, G.W. Dataset augmentation in feature space. arXiv Preprint 2017, arXiv:1702.05538. [Google Scholar]
- Shorten, C.; Khoshgoftaar, T.M. A survey on image data augmentation for deep learning. J. Big Data 2019, 6, 1–48. [Google Scholar] [CrossRef]
- Zhou, W.; Bras, R.L.; Choi, Y. Modular transformers: Compressing transformers into modularized layers for flexible efficient inference. arXiv Preprint 2023, arXiv:2306.02379. [Google Scholar]
- Chen, T.; Kornblith, S.; Swersky, K.; et al. Big self-supervised models are strong semi-supervised learners. Adv. Neural Inf. Process. Syst. 2020, 33, 22243–22255. [Google Scholar]
- Puri, R.; Spring, R.; Patwary, M.; et al. Training question answering models from synthetic data. arXiv Preprint 2020, arXiv:2002.09599. [Google Scholar]
- Wei, J.; Wang, X.; Schuurmans, D.; et al. Chain-of-thought prompting elicits reasoning in large language models. Advances in Neural Information Processing Systems 2022, 35, 24824–24837. [Google Scholar]
- Xu, C.; Sun, Q.; Zheng, K.; et al. WizardLM: Empowering large language models to follow complex instructions. arXiv Preprint 2023, arXiv:2304.12244. [Google Scholar]
- Bai, Y.; Lv, X.; Zhang, J.; et al. Longbench: A bilingual, multitask benchmark for long context understanding. arXiv Preprint 2023, arXiv:2308.14508. [Google Scholar]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).