Scores represent the projections of the original data points (observations) onto a reduced number of components or latent variables. Loadings indicate the contributions of the original variables to these components, showing how each variable relates to the reduced dimensions.
In a biplot, observations (samples) are often represented by points, while the loadings (variables) are shown as vectors (arrows). The angles and lengths of the arrows help interpret how variables are correlated and how they contribute to the variation in the data.
This decomposition approximates the original matrix as accurately as possible based on specific criteria, with and capturing the main structure and representing the residuals or error. In fact, a biplot decomposes the expected approximate structure of the data matrix into the product of two other matrices. The matrix captures the majority of the data’s characteristics based on a predefined criterion.
Most biplots are based on the assumption of linear relationships between observed and latent variables and are primarily designed for continuous data. For binary data, however, these representations are not ideal, much like how linear regression struggles to accurately model relationships with a binary response.
3.1. Logistic Biplots
Linear biplots decompose the expected values of the approximation into a product of two lower-rank matrices. In contrast, the logistic biplot decomposes the expected probabilities using the logit link function, similar to how generalized linear models operate.
For any binary data matrix
, if we call
the decomposition, using the
logit link, is
The constants
have to be included because data can not be centered beforehand as in the continuous case. With this decomposition, the inner product of the biplot markers (coordinates) is the
logit of a expected probability, except for a constant,
The logits are easily converted into probabilities
Then, by projecting a row marker onto a column marker we obtain, except for a constant, the expected logit and thus the expected probability for the entry of . The constant serves to determine the exact point where the logit is zero or any other value.
Due to the generalized nature of the model, the geometric interpretation closely resembles that of linear biplots. Computational procedures are analogous to those employed in previous cases, with the addition of the constant term. For example, in two dimensions, we can determine, on the direction of the vector
, what point predicts a given probability
p. Let’s
that point, satisfying the equation
Prediction also verifies that
The point in the direction of
that predicts
(
), is
Using Equations (
33), we can position markers for different probabilities along the direction of
, creating a graded scale similar to a coordinate axis. The interpretation remains fundamentally similar to that of linear biplots; however, markers representing equidistant probabilities may not be spaced equally in the graph.
As an illustrative simple example with data taken from [
42]. The data companies collect can include the expected, such as your name, date of birth, and email address, as well as more unusual details, such as your pets, hobbies, height, weight, and even personal preferences in the bedroom. They may also store your banking information and links to your social media accounts, along with any data you share there.
How companies use this data varies depending on their business, but it often leads to targeted advertising and optimizing website management.
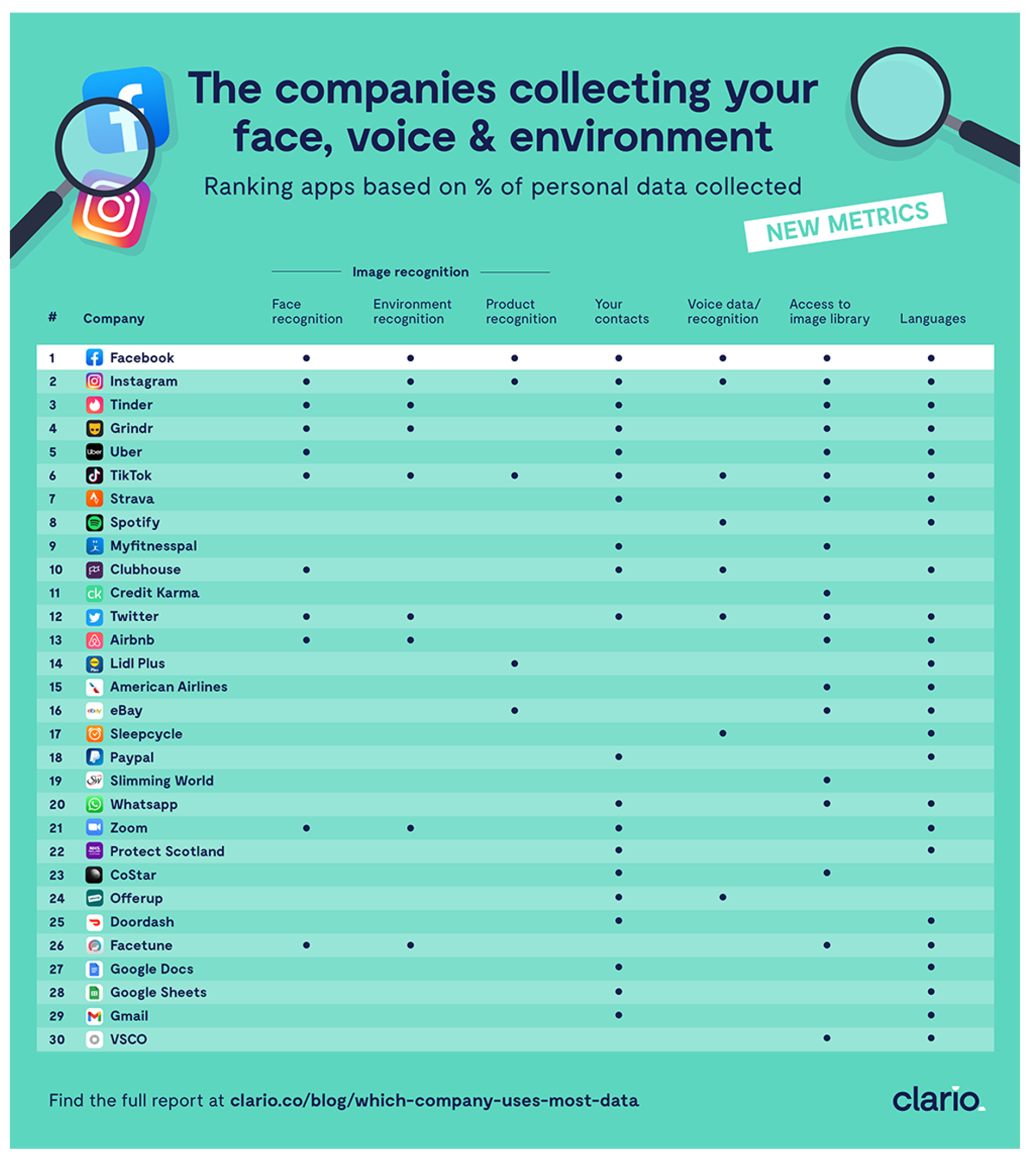
The infographics taken from the site is shown in
Figure 1. The picture shows information on your face, voice, and environment, that some internet companies collect.
The original data has been transformed into a binary matrix, where the rows represent internet companies, the columns represent the types of information collected, and each entry is marked as 1 if the company collects that information, and 0 if it does not. The data matrix is shown in
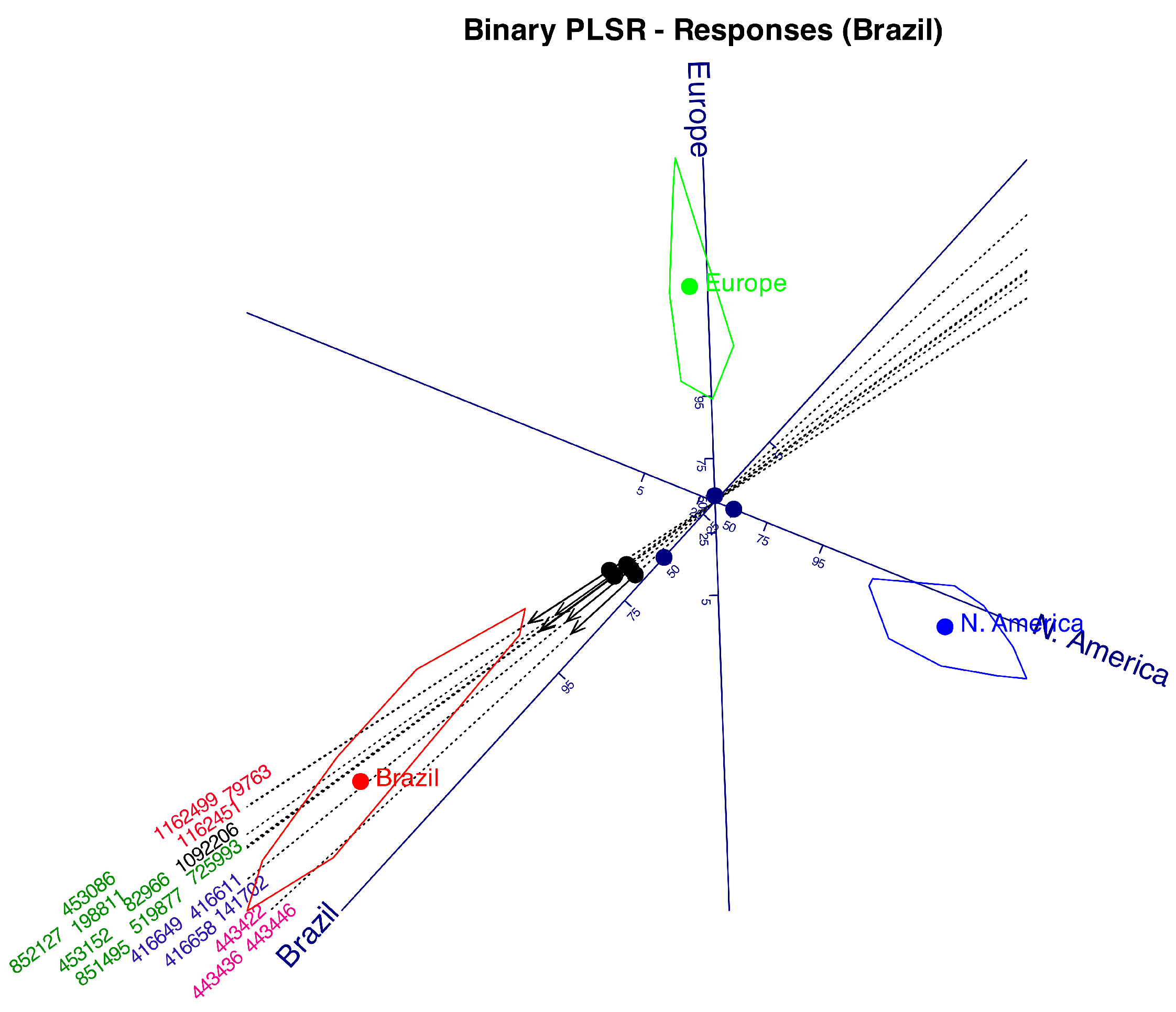
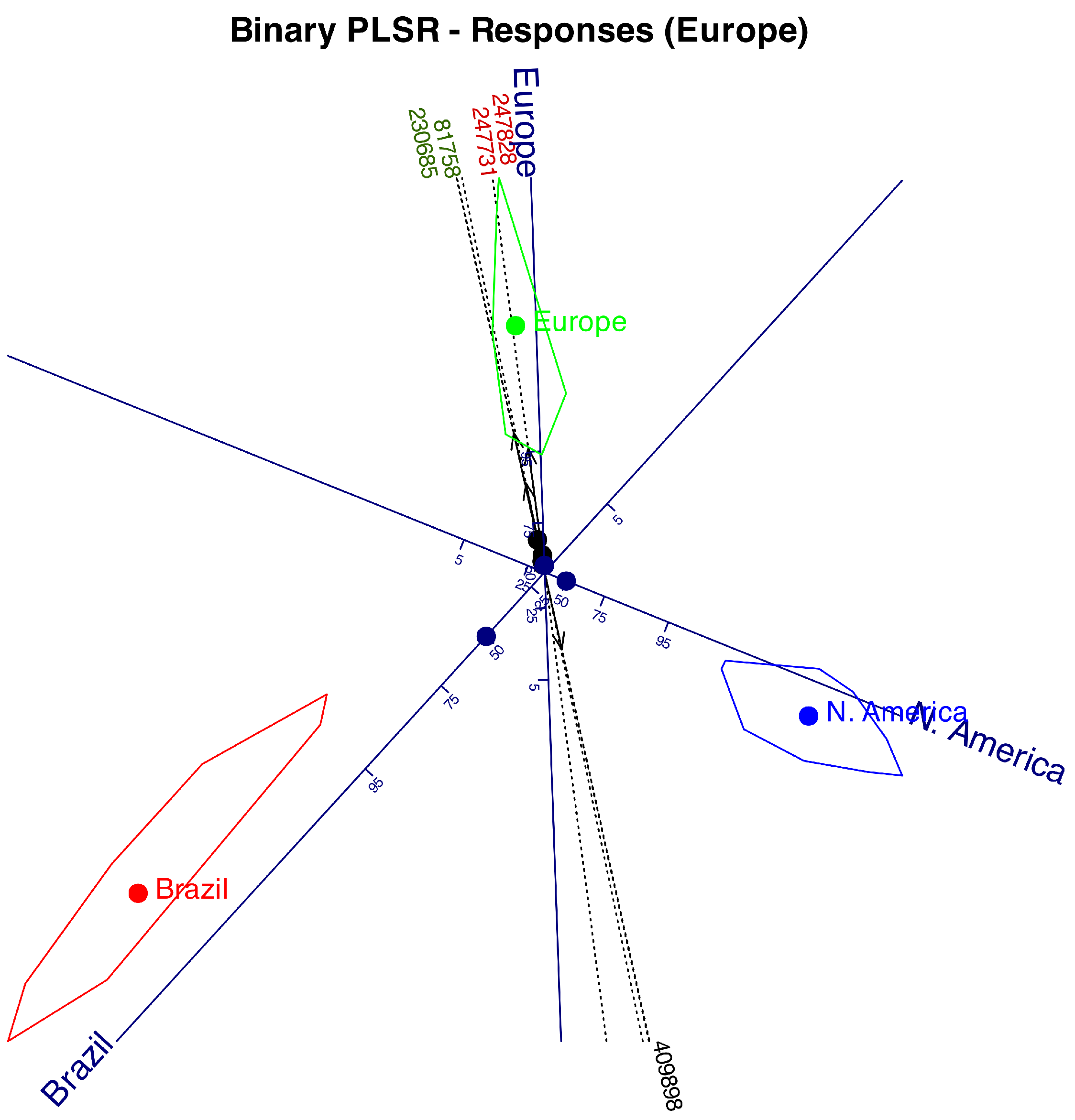
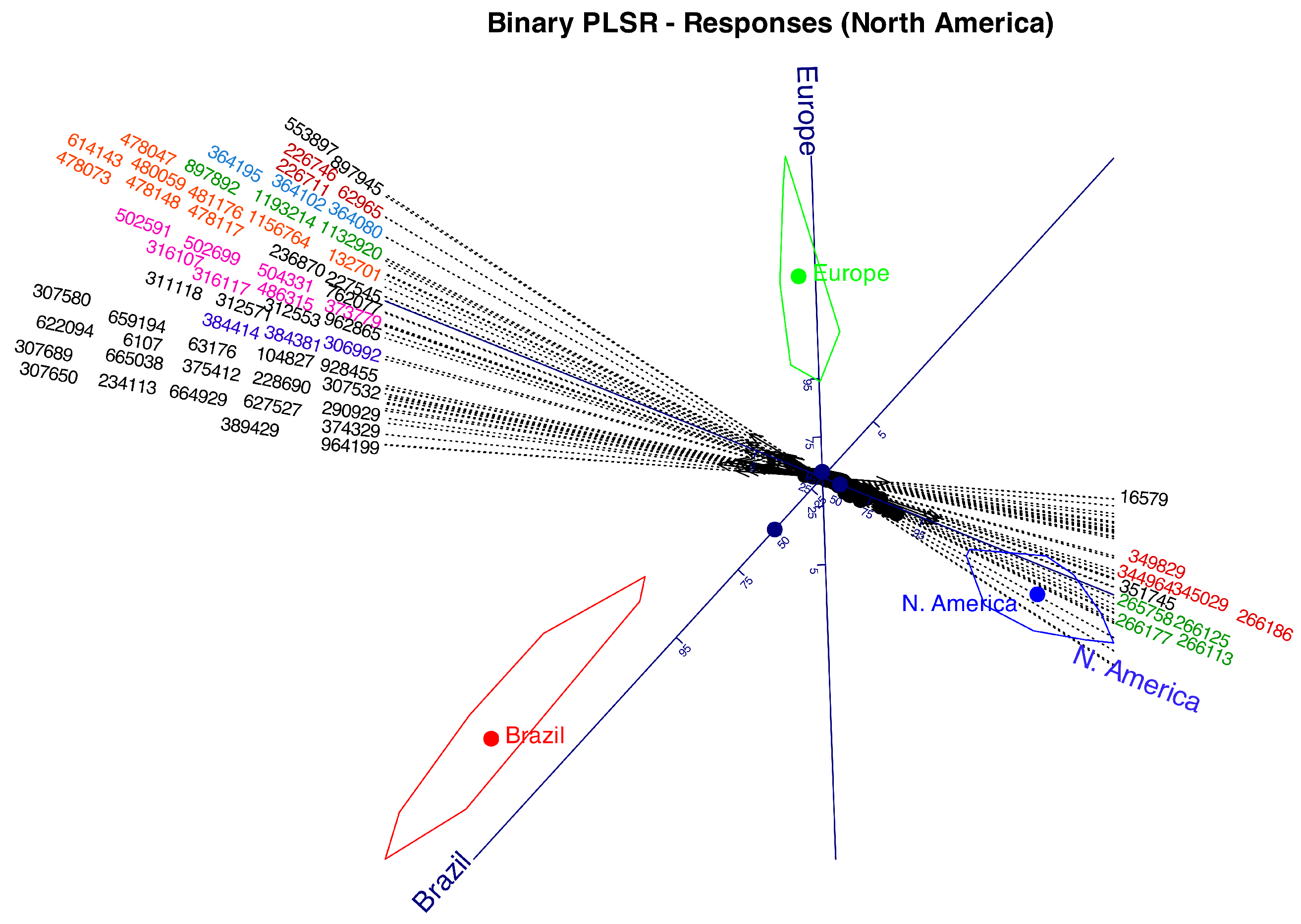
Table 1. With this data we have performed a Logistic Biplot that summarizes the information in a two dimensional graph. We obtain a joint representation of rows and columns of the data matrix. Distances among companies will be interpreted as similarities, i. e., companies lying near on the biplot display have similar profiles in relation to the collected information. Angles between variables (kind of information) is interpreted as correlations. Acute small angles mean strong positive correlations, near straight angles, strong negative correlations and almost rights angles mean no correlation. Projecting the row markers onto column markers, we have the expected probability for each entry.
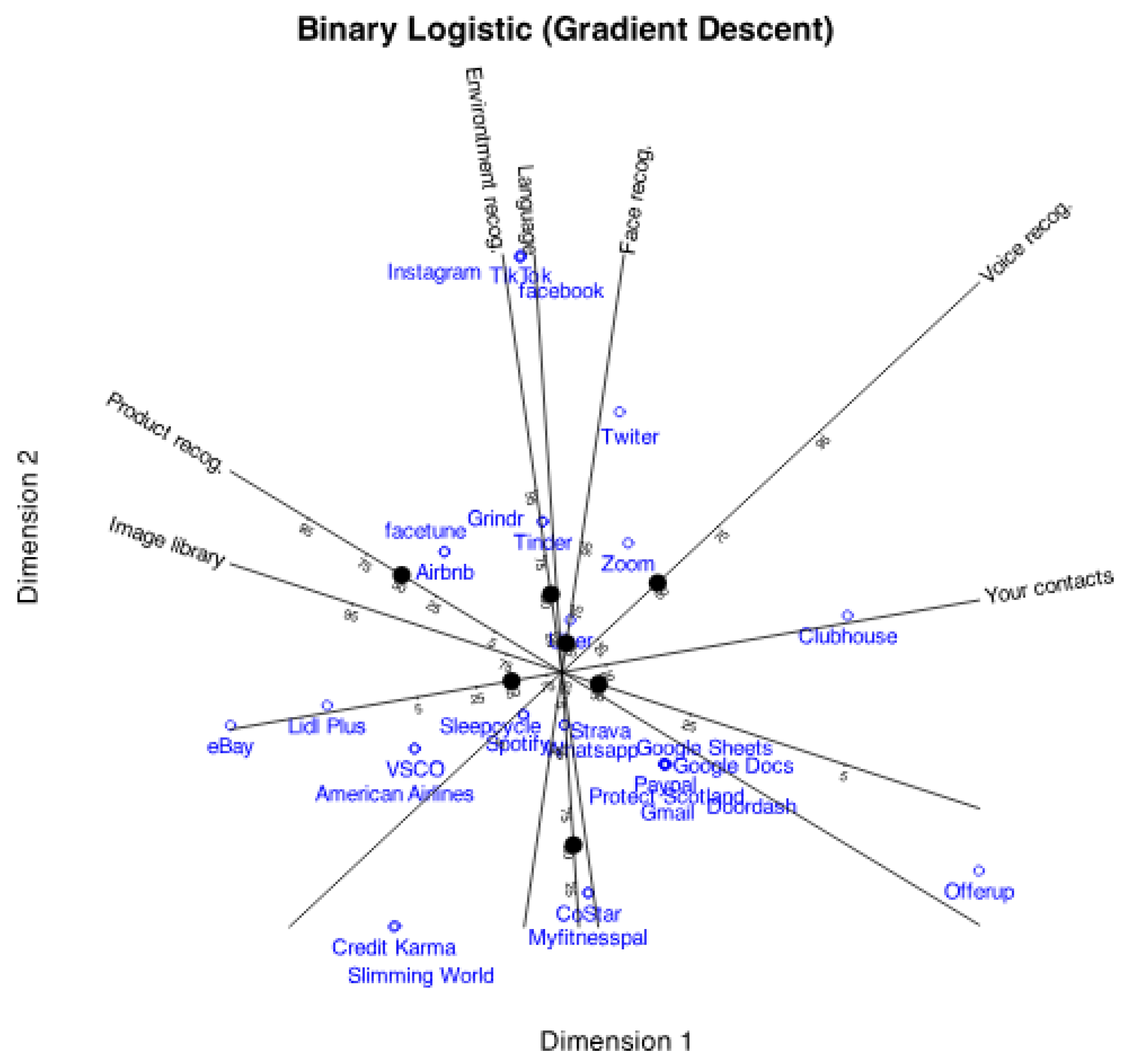
Figure 2 shows the graphical results as a typical logistic biplot with scales for probabilities 0.05, 0.25, 0.5, 0.75 and 0.95. We have used percents (5, 25, 50, 75, 95) rather than probabilities to simplify the plot. We could have used any other values for the probabilities.
The plot displays a two-dimensional scattergram, where points represent the rows (companies) of the data matrix, and vectors or directions correspond to the variables. Distances among points for companies are inversely related to their similarity, for example, points representing Facebook, Instagram and Tik-Tok coincide, indicating that the three companies have exactly the same profiles (the three collect all the available information). Companies with a single point have unique patterns, for example, Offerup, Uber or eBay, and groups with close points have several characteristics in common.
As previously mentioned, the angles between the variable directions provide an approximation of the correlations between them. For example, face recognition, environment recognition and languages have high positive correlations because the increasing probabilities point into the same direction, i. e. have small acute angles between them. Collecting the contacts is not correlated to this group because they form an straight angle with it. Contacts and image library are negatively correlations because the probabilities increase in opposite directions.
The angles are connected to tetrachoric correlation due to the close relationship between this representation and the factorization of the tetrachoric correlation matrix, as we will explain later.
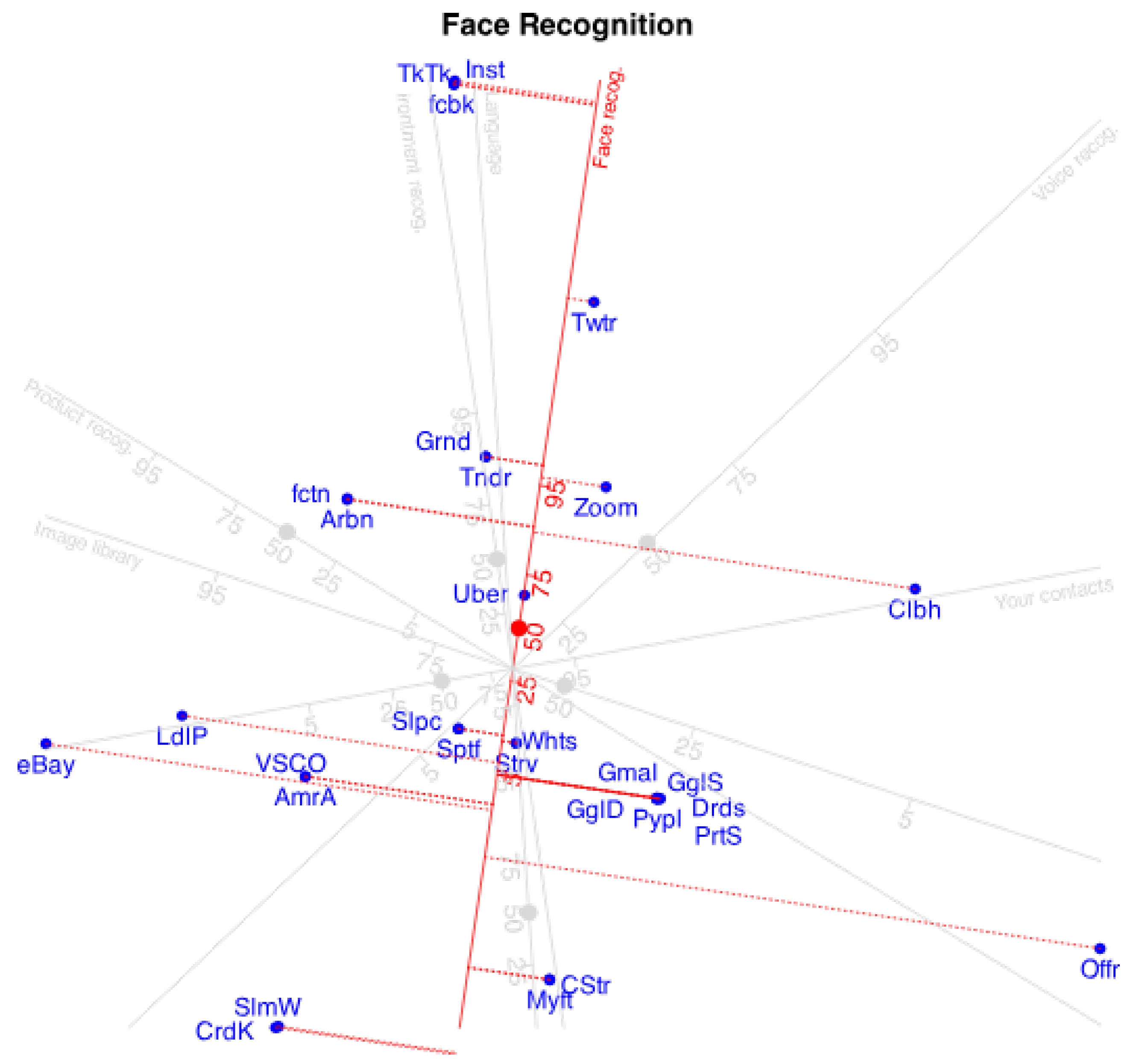
Projecting companies into the direction of a variable, we have the expected probabilities given by the approximation.
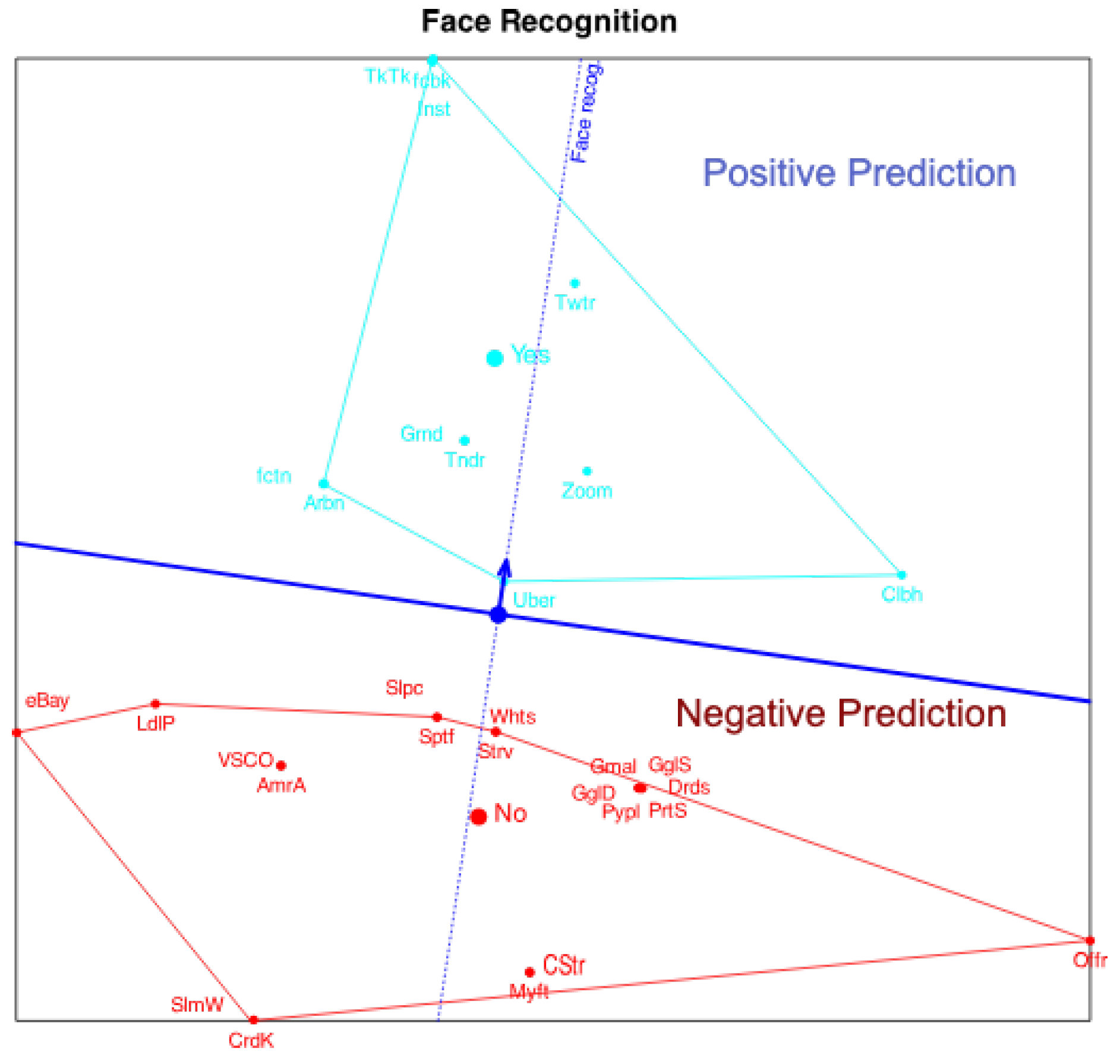
Figure 3 shows the projections onto the variable "Face Recognition". The point predicting 0.5 has been marked with a circle because is usually the cut point to predict presence or absence of the characteristic.
To simplify the plot we can place marks for just a few probabilities, for example, 0.5 and 0.75 indicating the point were expected logit is 0 (probability 0.5) and the direction of increasing probabilities.
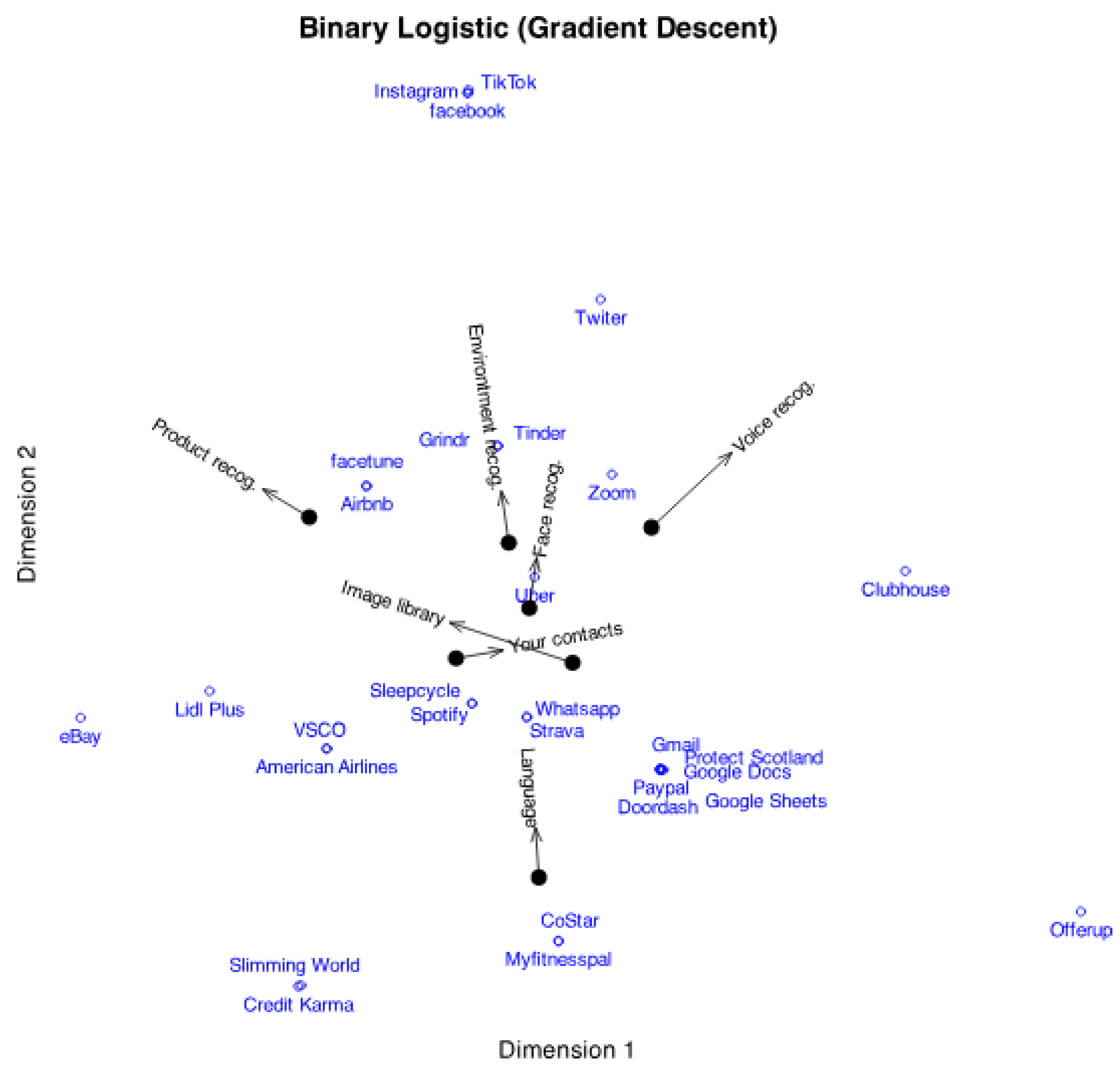
Figure 4 shows a simplified logistic biplot with probabilities 0.5 and 0.75.
We can use the expected probabilities from equation (
32) to generate predicted binary values:
if
and 0 otherwise. This yields an expected binary matrix
. In the plot, this indicates that for each variable, the plane is divided into two prediction regions by a line perpendicular to the variable’s arrow and passing through the point predicting 0.5. The region on the side of the arrow predicts the presence of the characteristic, while the opposite region predicts its absence.
Figure 5 illustrates this statement.
A guide for the interpretation of the biplot can be found also in the supplementary material by Demey et al. [
11].
A common overall goodness-of-fit measure is the percentage of correctly classified entries of the data matrix. To assess the quality of representation for individual rows (individuals) and columns (variables), we can calculate these percentages separately. We can also calculate the percentage of true positives (sensitivity) and true negatives (specificity).
To evaluate the quality of representation for each binary variable, we need a suitable measure that generalizes the measures of predictiveness for continuous data as in [
43]. Equation (
32) essentially defines a logistic regression model for each variable when the row coordinates are fixed, that allows to evaluate goodness of fit using pseudo-
measures commonly associated to that model, for example, McFaden, Cox-Snell or Nagelkerke.
Table 2 contains fit measures for the Internet Companies Data. It contains fit measures for each column separately and for the complete table.
94% of all entries in the data table are correctly predicted by the previously described procedure, as shown in
Figure 5. However, it is more insightful to examine the indices for each individual variable. This is because, in some cases, only a few variables are accurately represented. Such a situation can arise when dealing with a large set of variables, where only a small subset is actually relevant to the problem. For instance, in a genetic polymorphisms matrix, only certain variables may be of real interest.
In our example, all variables show a reasonable percentage of correct classifications, except for "Image Library," which correctly classifies only 80% of all values and 66.67% of the negatives. Additionally, we included the deviance for each variable in the model, using the latent dimensions as predictors, along with a p-value as an indicator of model fit. This measure, potentially adjusted for multiple comparisons, could be used to select the most significant variables, as demonstrated in [
11]. We also show pseudo-
measures. All have high values except for "Image Library" and "Voice Recog." that are worse predicted or represented on the graph.
By assuming that the observed categorical responses are discretized versions of continuous processes, we can leverage the close relationship between the presented model and the factorization of tetrachoric correlations to perform a factor analysis.
Consider a binary variable . This variable is assumed to arise from an underlying continuous variable that follows a standard normal distribution. There is a threshold, , which divide the continuous variable into two categories.
The relationship between
and
can be expressed as:
The tetrachoric correlations are the correlations among the . Let a matrix containing the tetrachoric correlations among the J binary variables and let the thresholds.
We can factorize the matrix
as
where contains the loadings of a linear factor model for the underlying variables.
It can be shown that there is a close relation between the factor model in Equation (
34) and the model in Equation (
32). You can find the details in [
9]. The model in Equation (
32) is actually equivalent to the multidimensional logistic two parameter model in Item Response Theory (IRT).
The factor model loadings
and he thresholds
can be calculated from the parameters for the variables in our model as:
In this way we provide a classical factor interpretation to our model adding the loadings and communalities (sum of squares of all dimension loadings of each variable). Loadings measure the correlation among the dimensions or latent factors and the communalities the amount of variance of each variable explained by the factors. For our data, the loadings and communalities are in
Table 3.
The two dimensional solution explains the 88% of the variance. All the comunalities are higher than 0.71 meaning that a good amount of the variance is explained by the dimensions. The communalities serve also to select important variables. Variables with low communalities may be explained by other dimension or have little importance for the description of the rows.
We can see also that Face Recognition, Environment Recognition and Language, are mostly related to the first dimension while, Product Recognition, Your contacts are related to the first dimension. Voice Recognition is related to both.