1. Introduction
In the field of modern agricultural research, structural analysis of tomato plants is important for understanding their growth habit, optimizing cultivation management, and improving crop yield. Using point cloud data collected by depth cameras, the three-dimensional morphology of tomato plants can be accurately depicted, including the spatial layout of stems, leaves, branches, and other parts, as well as their geometric properties. However, the processing of point cloud data also suffers from data occlusion, noise interference, and sparsity, all of which may lead to missing information and segmentation errors. It is also more difficult to analyze the point cloud in the complex geometric structures of tomato plants at different stages of growth and development. With the development of deep learning technology, plant stem and leaf segmentation based on point cloud data how to accurately segment tomato point cloud data and extract useful geometric information has become a research hotspot.
Currently, the common stem and leaf segmentation in the plant domain can be categorized into three types: supervised learning, unsupervised learning and weakly supervised learning. Supervised learning achieves efficient feature extraction and fine segmentation of plant structure point cloud data through deep learning models that rely on fully labeled data for training. Researchers achieved automatic identification of key features in the input point cloud and accomplished accurate segmentation of plant organs by deep neural network training.Lou L et al. proposed a novel weakly-supervised framework Eff-3DPSeg for 3D plant bud segmentation[
1]. This method combines deep learning and point cloud techniques to address the challenges of 3D plant bud segmentation.Wang, Y et al. achieved an effective segmentation of stems and leaves of tomato plants by combining skeleton extraction with up-pixel clustering techniques to achieve a certain segmentation accuracy[
2]. Xusheng Zhong used Graph Convolutional Neural Network (GCNN) as a basic framework for plant point cloud segmentation research. The study used 864 plant point cloud data collected as a dataset and verified the performance of the model[
3]. These methods are based on the capability of deep learning in feature learning and model optimization to handle complex data and improve segmentation refinement.
Unsupervised learning does not use any labeled data but looks for patterns in the data[
4]. Pre-trained models or machine learning methods can autonomously extract features from large-scale unlabeled point cloud data and process and analyze the point cloud data using algorithms such as clustering and dimensionality reduction[
5,
6,
7,
8].Miao,T et al. proposed an automated segmentation method for maize shoot stems and leaves based on 3D point cloud. The method used skeleton information as a priori knowledge to assist point cloud segmentation and realized an automated segmentation process[
9]. Weakly supervised learning uses partially labeled data to guide the model for training, which is suitable for those scenarios where labeling resources are limited but supervised information is still needed[
10].Wu J et al. proposed a weakly supervised 3D point cloud semantic segmentation (WS3DSS) method, which is to develop a robust model that is able to learn from limited labeled data and effectively utilize the unlabeled data to enhance segmentation performance[
11]. However, unsupervised learning has no labeling information, and the learned model may not be directly applicable to specific classification or regression tasks with limited performance enhancement and low accuracy. Weakly supervised learning may not be sufficient to effectively guide model learning due to incomplete or uncertainty in the labeling information, thus affecting the accuracy and stability of segmentation results [
12,
13,
14].
Aiming at the challenges faced by the current segmentation, this paper proposes an optimization algorithm based on the improved red-billed blue magpie, which aims to solve the following problems:
1. deep learning models generally have the problem of large parameter sizes, although the demand for computational resources can be significantly reduced through model lightweighting techniques, this process often leads to a decline in model performance, i.e., loss of accuracy, which affects the accuracy of the final segmentation results.
2. The segmentation accuracy of some deep learning models shows a certain degree of instability in complex environments, and this instability limits the applicability of the model in practical application scenarios, making it difficult to meet the demand for highly reliable segmentation results in agricultural production.
Aiming at the current problems, this paper aims to propose a method that can still obtain high segmentation accuracy with fewer convolutional layers and fewer number of parameters. To achieve this goal, the following strategies are adopted in this study:
1. the Red-billed Blue Magpie Optimization Algorithm (RBMO) is optimized using an elite strategy to improve the stability and search capability of the algorithm. The elite strategy improves the overall performance of the algorithm by retaining the best individuals in successive generations and guiding the evolution of the algorithm towards better solutions.
2. the optimized red-billed blue magpie algorithm is fused with a deep learning network model to optimize the effect of stem and leaf segmentation of tomato plants. This fusion strategy aims to ensure the robustness of the model when dealing with complex point cloud data, while reducing the dependence on a large number of parameters.
3. encapsulate customized convolutional layers by combining geometric and curvature features of the point cloud data. This approach makes the network model more attuned to the processing characteristics of 3D spatial data, thus improving the ability to recognize the details of plant structures while keeping the model lightweight.
2. Materials and Methods
2.1. Data Sources and Collection
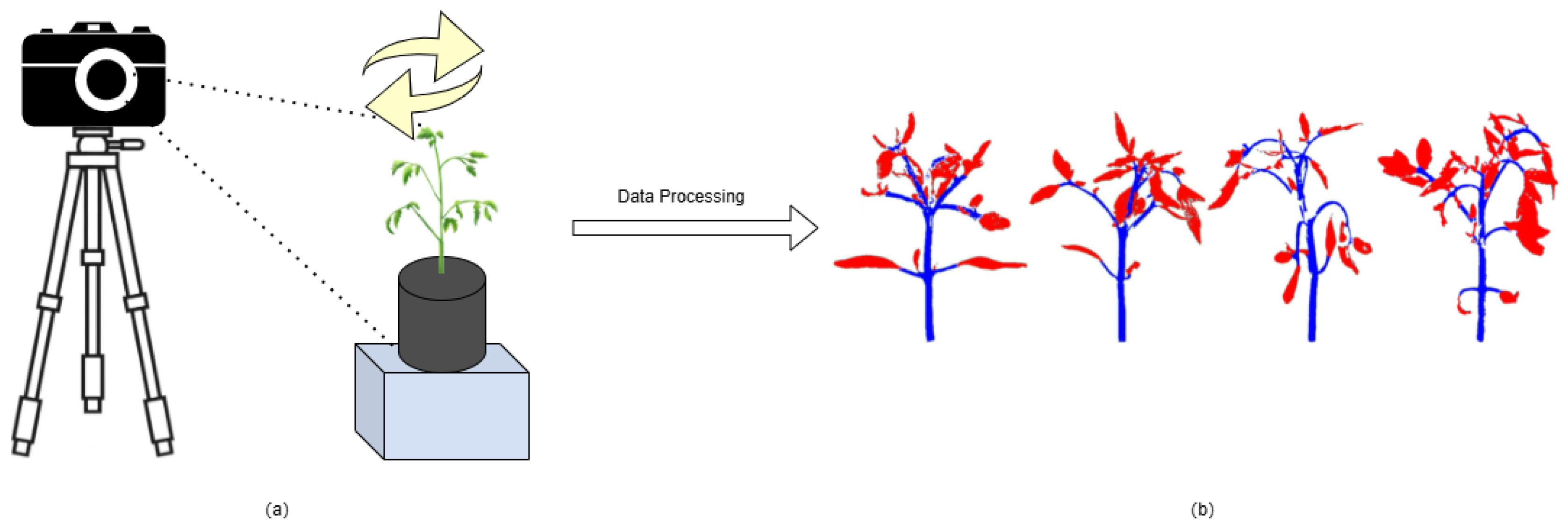
In the fourth greenhouse of Jilin Vegetable and Flower Scientific Research Institute, five independent planting blocks were set up in this study, and 96 tomato plants were cultivated in each block, with the overall sample size reaching 480 plants. Within these blocks, plants were arranged in two rows with row spacing maintained at 0.1 m, while the row length extended to 2.4 m. The plants were planted in two rows with the same spacing. Within the same row, the spacing between plants was approximately 0.05 meters. In order to obtain the 3D structural data of tomato plants, a 3DScanner-630w device was used for point cloud data acquisition (
Figure 1a), and special attention was paid to avoiding the effects of shadows and reflections during the acquisition process to ensure the integrity of the data. CloudCompare software was utilized to perform pre-processing operations on the acquired image data. Finally, the pre-processed image data were visualized in three dimensions through the Python programming language (
Figure 1b).
2.2. Methodologies
2.2.1. Algorithmic Principles of the Red-Billed Blue Magpie Algorithm for Elite Strategy Optimization
The Elite Strategy-based Improved Red-billed Blue Magpie Optimization Algorithm (ES-RBMO) optimizes the red-billed blue magpie by simulating its foraging behavior in the hyper-parameter space of deep learning networks [
15,
16,
17]. The elite strategy effectively avoids falling into a local optimum by retaining the current optimal solution and prioritizing the use of its information for subsequent searches.
Each individual (bird) represents a configuration of a deep learning model. Assuming that the deep learning model has multiple hyperparameters,
the position of each individual can be represented v as.
where
denotes the value of dimension
of the
th individual (solution) in the hyperparameter space (e.g., learning rate, number of network layers, etc.).
The fitness function is a quantitative form of the optimization objective, and we compute the fitness of an individual based on accuracy, recall, precision, F1 score, and loU. Let
denote the fitness value of the
th individual, we define the weighted fitness function as.
where,
,
,
,
,
denotes each performance indicator of the model corresponding to the
th individual, respectively Individual corresponds to each performance metric of the model.
,
,
,
,
is the weight of each indicator,and it satisfies
.
The core of the red-billed blue magpie optimization algorithm is to search for the best solution by updating the individuals with the following update rules:
indicates the th Individuals in position . indicates the th Individuals in position . is the best adapted elite individual in the current population. is the positional mean of all individuals in the current population. and are the step coefficients for global and local search, respectively.
Elite strategy by selecting the best adapted individuals,preserves its solution and passes it on directly to the next generation.This mechanism helps the algorithm to keep the search in the neighborhood of the optimal solution and avoid falling into local optima.
While local search performs a meticulous search in the vicinity of the current solution to optimize the solution quality through minor adjustments, jump search boldly explores a larger search space by randomly adjusting the position of individuals in order to find global optimal solutions that may have been overlooked [
18,
19].With this strategy, the Red-billed Blue Magpie algorithm is not only able to accurately perform local optimization when dealing with point cloud data, but also maintains the vitality of the global search, ensuring that the algorithm performs well in point cloud alignment, shape recognition, and other complex tasks[
20]. By retaining the elite individuals with the highest fitness, the algorithm ensures that the optimal solution is always retained during the iteration process, which improves the overall search efficiency and the quality of the solution[
21].
Local search optimizes the solution by making small adjustments based on the fitness of the current individual position. For example, better solutions are explored within a neighborhood through mutation operations[
22]:
is a smaller constant that controls the trim step.
Jump search[
23] appears to be more aggressively applied in the Red-billed Blue Magpie algorithm, which explores domains in the solution space far from the current population location by increasing the search step size, thus potentially discovering a better solution[
24]. In each generation of the algorithm, we update the position of the population based on the fitness value of each individual and generate new individuals.In this process, we use an elite strategy, i.e., we keep the individuals with the best fitness as the elite, and these elite individuals will not be eliminated to ensure that the algorithm can maintain the inheritance of the optimal solution during the iteration process[
25].By applying this elite strategy-optimized red-billed blue magpie algorithm to point cloud data processing, the solution space can be explored and utilized more efficiently, and the efficiency and accuracy of point cloud alignment and shape optimization can be improved [
26,
27,
28,
29,
30].
The termination conditions of the algorithm include the following three aspects: firstly, when the algorithm reaches the predetermined maximum number of iterations, the search process will be stopped automatically; secondly, if the change of the fitness of the optimal solution is less than a certain set threshold in a number of consecutive generations, it indicates that the algorithm has converged, and the iteration will be terminated at this time as well [
31,
32,
33,
34]; lastly, if the value of the fitness of the optimal solution exceeds the predetermined threshold, it means that the algorithm has found a satisfactory solution and therefore will stop early [
35,
36]. With this optimization strategy, we apply the red-billed blue magpie algorithm to point cloud data processing to achieve more efficient and accurate search and matching.
2.2.2. Algorithmic Principles of 3DCNN
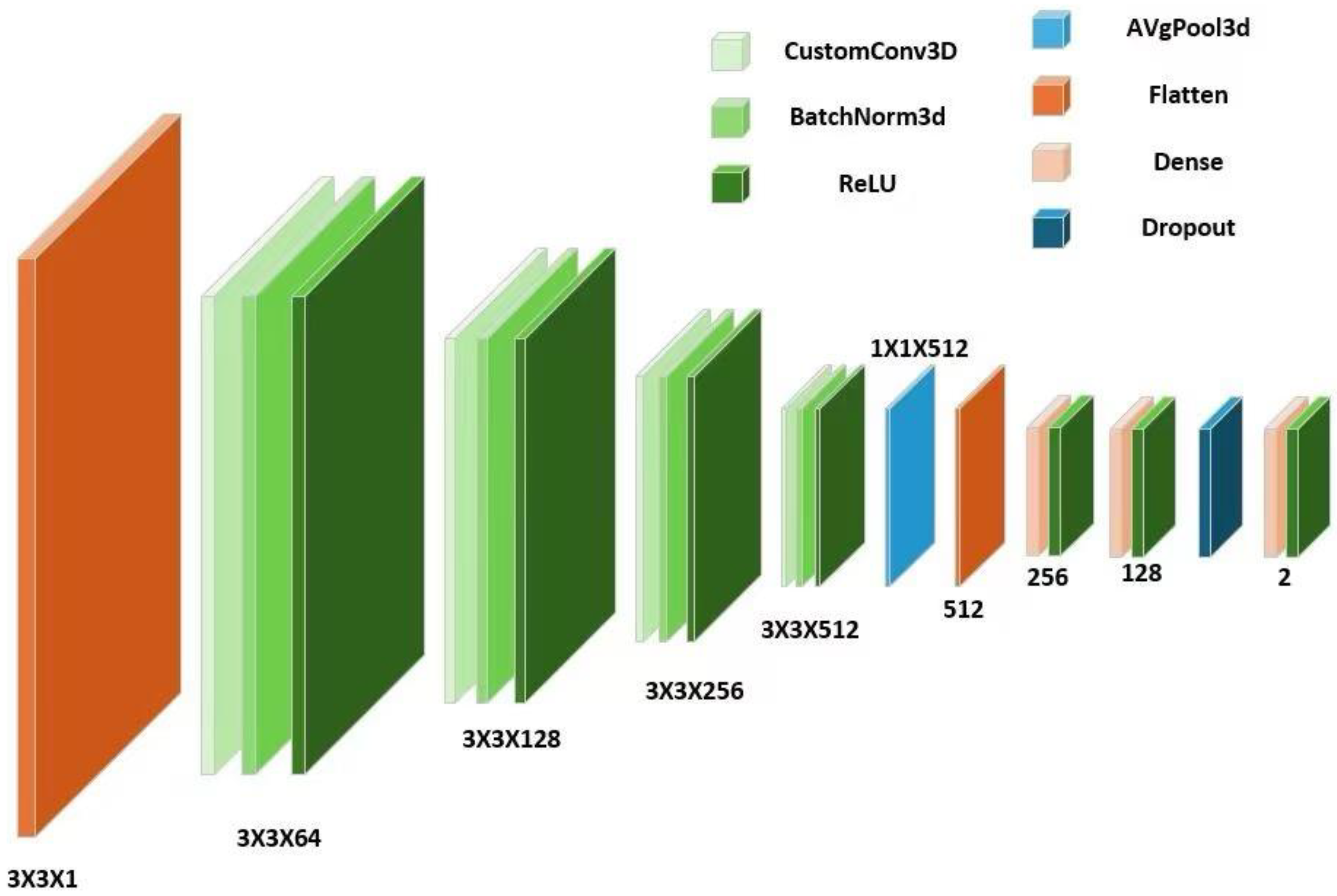
The 3DCNN incorporates the geometric and curvature properties of point cloud data to construct a four-layer deep learning architecture by encapsulating customized convolutional layers[
37]. The first convolutional layer of this network, conv1, uses a 3x3x3 convolutional kernel for feature extraction from single-channel input data, which achieves the mapping from raw data to 64-dimensional feature space, and accelerates convergence and stabilizes the training through a batch normalization operation in the bn1 layer[
38]. conv2 with the bn2 layer extends the feature dimensions to 128 dimensions, which enhances the abstract representation of the features. conv3 with the bn3 layers map features to 256 dimensions to capture higher-level structural information. The optional conv4 & bn4 layers achieve deeper feature extraction, enhancing 256-dimensional features to 512 dimensions, which significantly improves the model's ability to model and recognize complex spatial features[
39], as shown in
Figure 2. This method effectively extracts and abstracts rich geometric and structural information from the point cloud data by increasing the feature dimensions layer by layer, which significantly improves the model's modeling and recognition ability of complex 3D shapes; meanwhile, combined with batch normalization, it optimizes the training process, accelerates the convergence speed, and enhances the model's generalization performance.
3. Results
3.1. Comparison Experiment
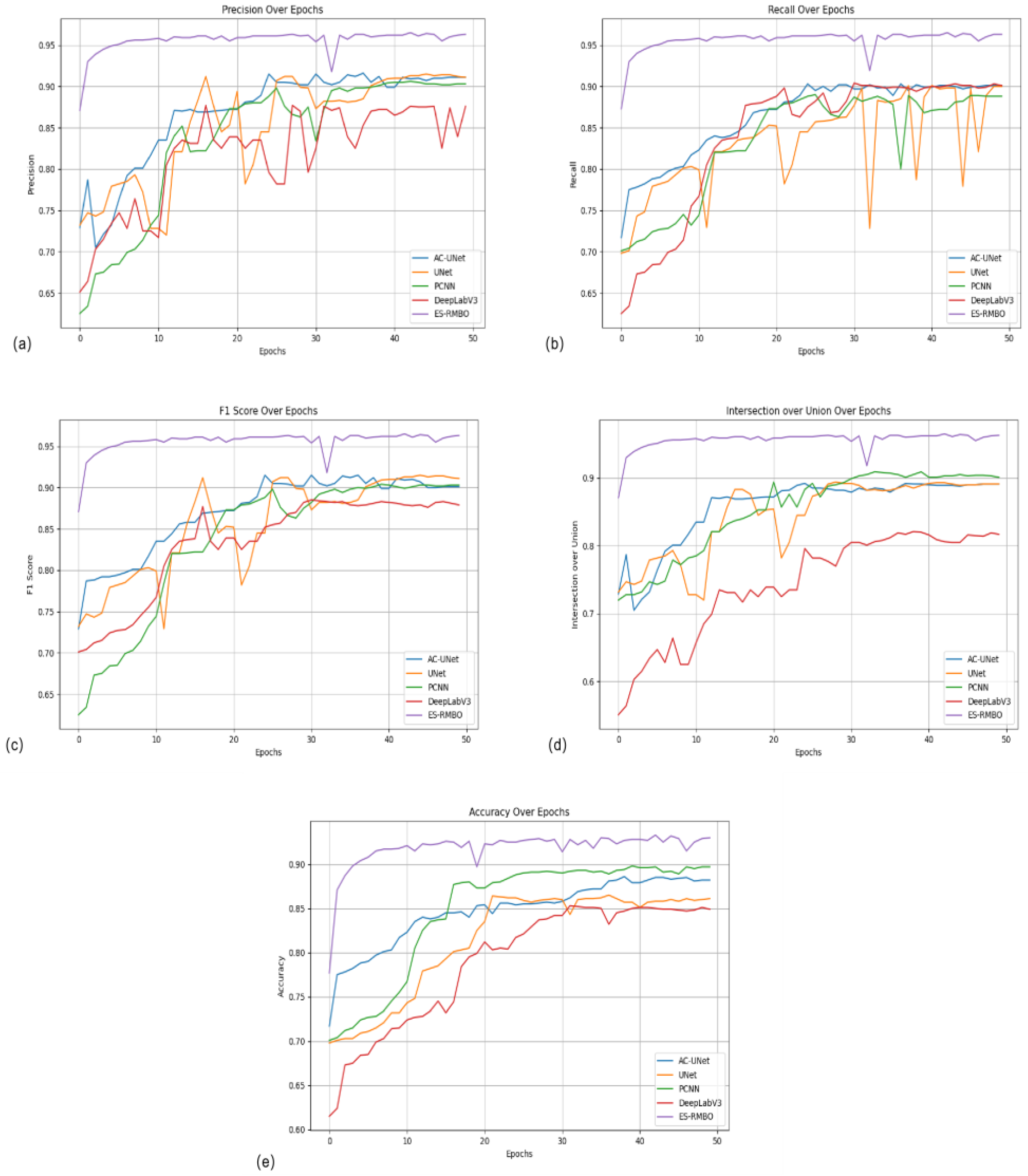
The ES-RMBO algorithm proposed in this paper shows significant superiority in stem-and-leaf segmentation tasks. In comparison with other mainstream algorithms, ES-RMBO achieves the optimal level in key performance metrics such as precision (0.965), recall (0.965), F1 score (0.965), IoU (0.965), and ACC (0.933), demonstrating higher segmentation accuracy and stability (
Table 1). In contrast, although the UNet algorithm[
40], AC-UNet algorithm[
41], PCNN algorithm [
42]and DeepLabV3 algorithm[
43] also demonstrated better performance in the segmentation task, they were slightly lower in several metrics, such as precision (
Figure 3a), recall (
Figure 3b), F1 scores (
Figure 3c), IoU (
Figure 3d) and ACC (
Figure 3e), than the Tomato plants have unique growth characteristics, in which the stems are more upright and regular, while the leaves are dense and diverse, with relatively complex structures.The RMBO algorithm can more accurately identify and distinguish these complex structural features by effectively integrating the geometric information of the point cloud data.The RMBO algorithm strikes a better balance between precision and recall, which effectively reduces the occurrence of misclassification, and further reduces the incidence of misclassification, and further reduces the incidence of misclassification. reduces the occurrence of misclassification and further improves the reliability and accuracy of segmentation results.
3.2. Ablation Experiment
In order to verify the effectiveness of ES-RMBO algorithm in tomato stem and leaf segmentation task, we conducted ablation experiments to compare ES-RMBO with other unoptimized algorithms, including the original 3DCNN and the addition of the unoptimized RMBO algorithm.
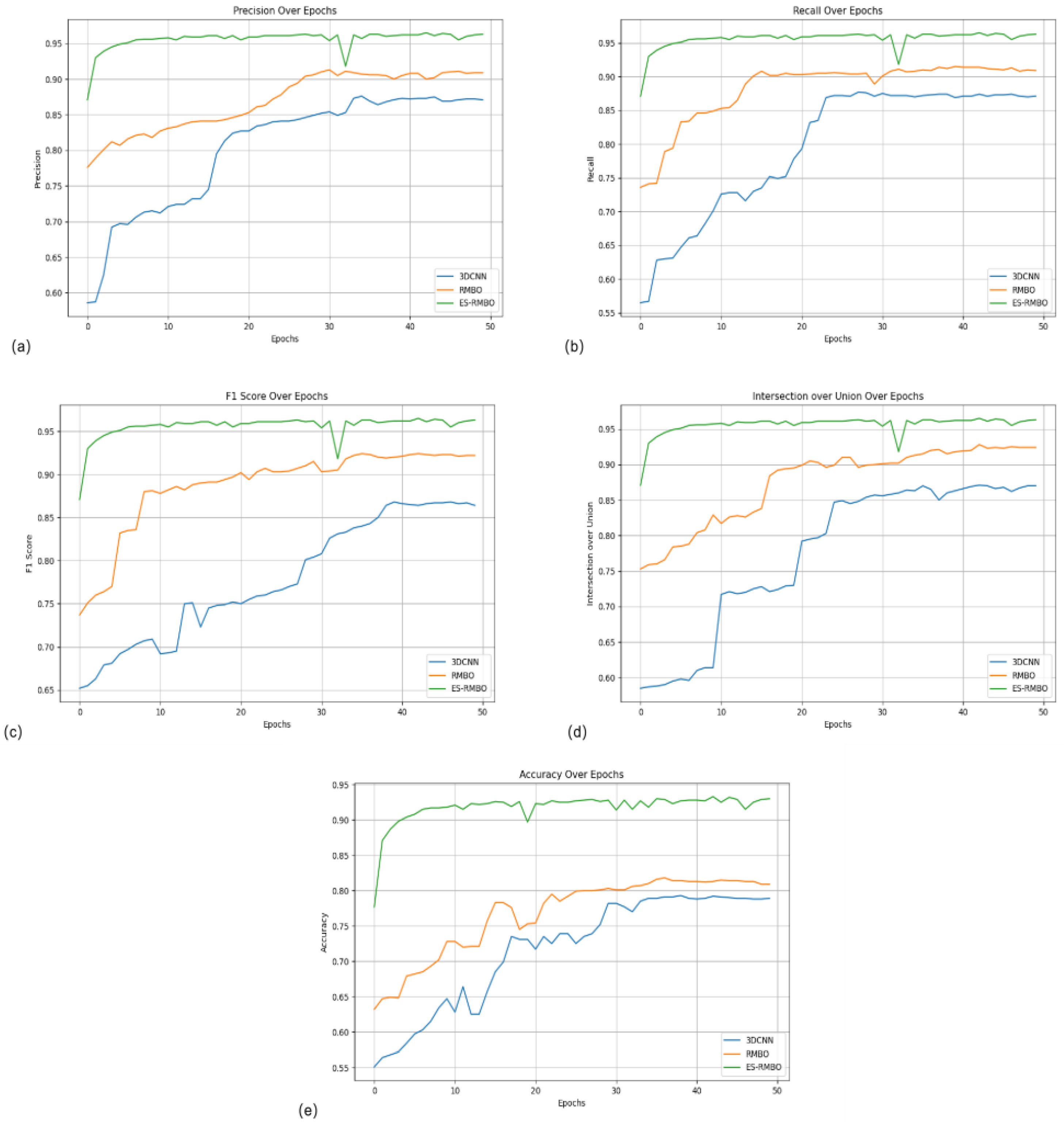
Table 2 demonstrates the comparison of the performance of different algorithms on the tomato stem and leaf segmentation task. As can be seen from the table, the ES-RMBO algorithm achieves optimal results in all key performance metrics of precision (0.965), recall (0.965), F1 score (0.965), IoU (0.965), and ACC (0.933), which proves its superiority in the tomato stem-and-leaf segmentation task. The results of the ablation experiments show that several values of precision (
Figure 4a), recall (
Figure 4b), F1 value (
Figure 4c), IoU (
Figure 4d) and ACC (
Figure 4e) of the unoptimized algorithm are not as good as the ES-RMBO algorithm in the tomato stem and leaf segmentation task, which suggests that the ES-RMBO algorithm is able to efficiently process the tomato point cloud data, improve segmentation precision and stability, providing an efficient and accurate solution for tomato growth monitoring, variety identification and phenotypic analysis.
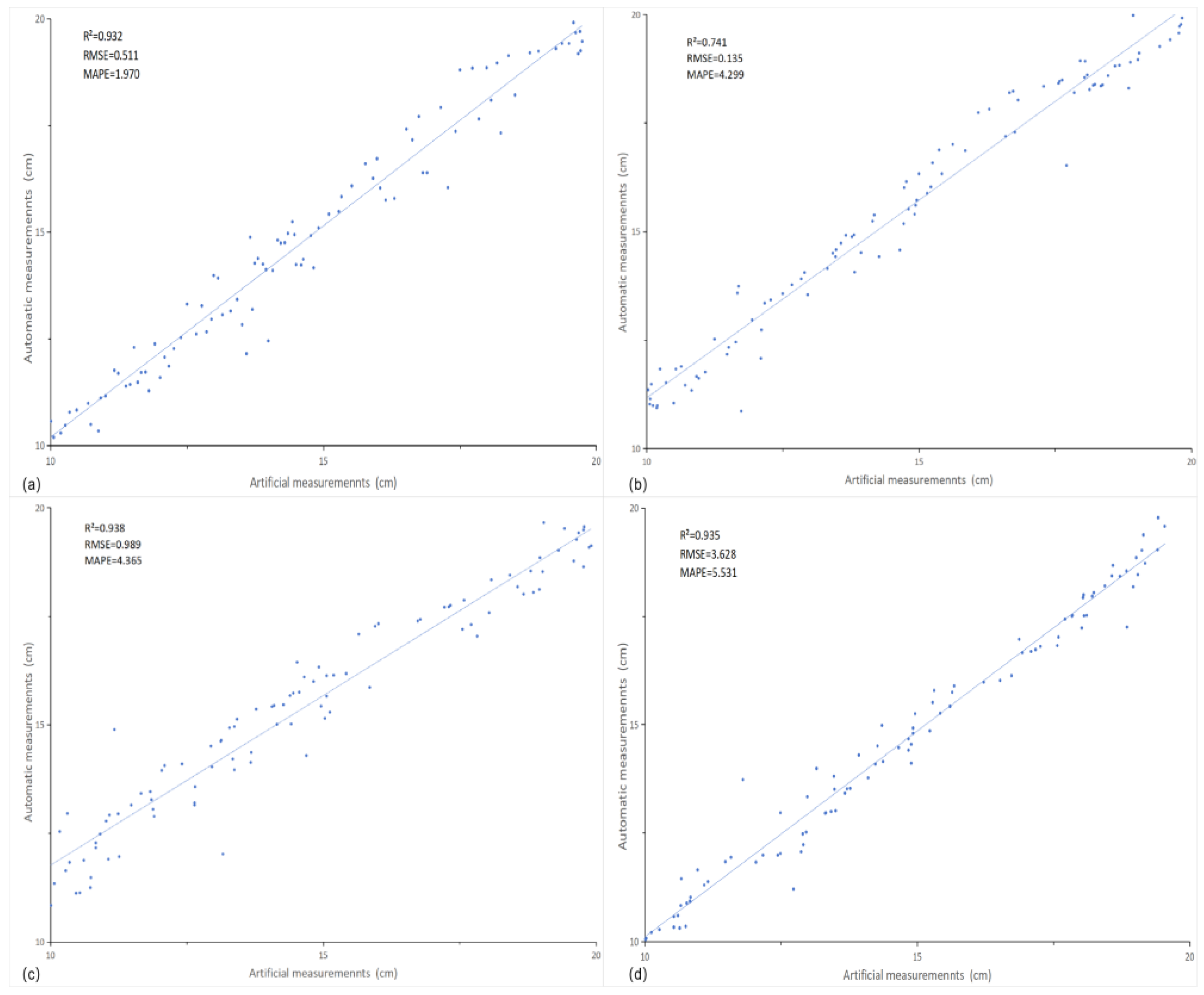
3.3. Phenotypic Parameter Measurement Results and Analysis
Measurement results and analysis were taken from 100 tomato plants, comparing the manual measurements with the extracted values of 3D point cloud parameters, including plant height R²=0.932, RMSE=0.511, MAPE=1.970, (Fig. 5a); stem thickness R²=0.741, RMSE=0.135, MAPE=4.299, (Fig. 5b); leaf area R²= 0.938, RMSE=0.989, MAPE=4.365, (
Figure 5c); leaf inclination R²=0.935, RMSE=3.628, MAPE=5.531, (
Figure 5d). The experimental data showed that the results indicated a high correlation between the measured results and the real data for the measured leaf data.
4. Discussion
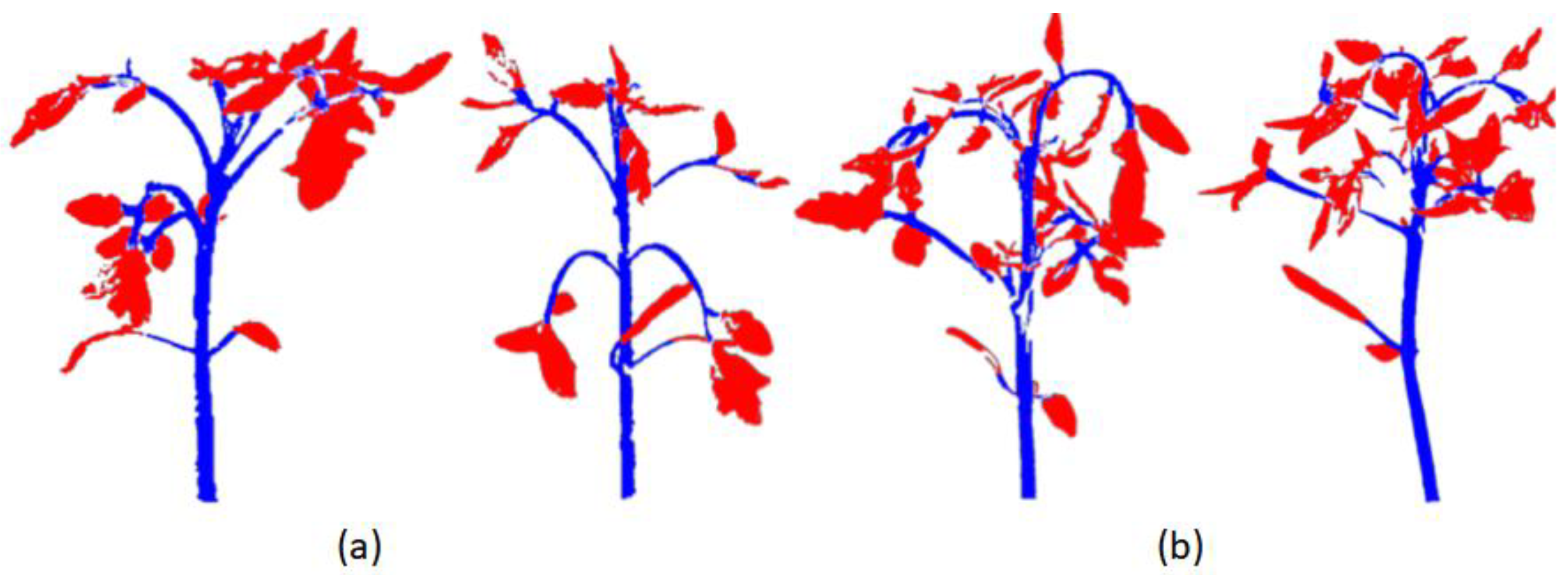
The improved red-billed blue magpie optimization algorithm in this paper has been effectively applied to the processing of tomato plant point cloud data, which demonstrates excellent adaptability and robustness to the point cloud characteristics exhibited by plants at different growth and development stages and under diverse environmental conditions. By extracting the geometric and curvature features of the point cloud in a hierarchical manner, this optimization algorithm retains the current optimal solution, accelerates the convergence speed and the quality of the algorithm, and improves the accuracy and stability of segmentation. And it can reach accurate stem and leaf separation without the support of large-scale labeled data. The experimental results show that the algorithm can efficiently perform stem and leaf segmentation on tomato point cloud data processing, and achieves the optimal level of key performance metrics, such as precision, recall, F1 score, IoU, and ACC, demonstrating higher segmentation precision and stability (
Figure 6a).
Although the optimization algorithm proposed in this paper shows excellent performance in the tomato plant point cloud segmentation task, the characteristics of tomato plants still bring some difficulties to the segmentation task, which needs further research and optimization. It mainly stems from its complex growth morphology. The stems and leaves of tomato plants grow densely, and the leaves shade each other with fuzzy boundaries that are difficult to distinguish. This dense and interlaced growth pattern leads to the difficulty of traditional edge detection algorithms in accurately recognizing leaf edges, thus affecting the accuracy of the segmentation results (
Figure 6b).
5. Conclusions
In this paper, we propose a stem-and-leaf segmentation method based on the improved optimization algorithm for red-billed blue magpies, and verify its excellent performance and robustness in point cloud data of different densities. The method significantly improves the accuracy and stability of stem-and-leaf segmentation by fusing the geometric features, spatial distribution information and deep learning model of point cloud data. Through dynamic volume compression, the aggregation of the point cloud is enhanced, which effectively improves the accuracy of the feature extraction process. For different point cloud densities, the algorithm is able to accurately capture stem and leaf features, optimize local details, and significantly reduce misclassification. During the processing of tomato plants, ES-RMBO demonstrates strong stability and reliability, and is able to accurately distinguish stems from leaves. Through hierarchical processing and geometric feature computation, the method successfully copes with the unique hierarchical structure of tomato plants, further enhancing the segmentation effect. The algorithm achieves optimal results in key performance metrics such as precision, recall, F1 score, IoU and ACC, which verifies its high adaptability in plant growth stages. The algorithm proposed in this paper can effectively deal with complex plant structures and provides solid technical support for plant growth monitoring and automated segmentation tasks in precision agriculture, and its applicability in a wide range of plant species and agricultural application scenarios can be expanded in the future by further optimizing the algorithm's generalization ability.
Author Contributions
Conceptualization, L.Z. and Z.H.; methodology, Z.Y.; software, H.Y. and L.Z.; validation, H.Y. and L.Z.; formal analysis, S.Y.; investigation, Z.H. and B.Y.; data curation, Z.H. and S.Z.; writing—original draft preparation, H.Y. and Y.L.; writing—review and editing,,L.Z. and Z.H.; supervision,Z.Y..; project administration,L.Z.;translation,Y.L.;proofreading,X.Z.;Image beautification,X.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by science and technology development plan project of Science and Technology Department of Jilin Province, project name: A Trusted Traceability System for Smart Agriculture Based on Blockchain Technology,project No: 20220202036NC.
Data Availability Statement
For experimental data in this paper, please contact the corresponding author for assistance.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Luo L, Jiang X, Yang Y, et al. Eff-3DPSeg: 3D organ-level plant shoot segmentation using annotationefficient
point clouds[J]. arxiv preprint arxiv:2212.10263, 2022.
- Wang Y,Liu Q,Yang J, et al. A Method for Tomato Plant Stem and Leaf Segmentation and Phenotypic Extraction Based on Skeleton Extraction and Supervoxel Clustering[J]. Agronomy 2024, 14. [Google Scholar]
- Morteza G ,Kevin W ,K. M F C , et al.Deep Segmentation of Point Clouds of Wheat [J].Frontiers in Plant Science,2021,12608732-608732.
- Yonatan L ,Ofri R ,Merav A .Dissecting the roles of supervised and unsupervised learning in perceptual discrimination judgments.[J].The Journal of neuroscience : the official journal of the Society for Neuroscience,2020,41(4):.
- Yao D ,Chuanchuan Y ,Hao C , et al.Low-complexity point cloud denoising for LiDAR by PCA-based dimension reduction[J].Optics Communications,2021,482.
- Alkadri F M,Yuliana Y,Agung C R M, et al.Enhancing preservation: Addressing humidity challenges in Indonesian heritage buildings through advanced detection methods point cloud data[J].Results in Engineering,2024,24103292-103292.
- Jing R,Shao Y,Zeng Q, et al.Multimodal feature integration network for lithology identification from point cloud data[J].Computers and Geosciences,2025,194105775-105775.
- Chen T,Ying X.FPSMix: data augmentation strategy for point cloud classification[J].Frontiers of Computer Science,2024,19(2):192701-192701.
- Teng M ,Chao Z ,Tongyu X , et al.Automatic stem-leaf segmentation of maize shoots using three-dimensional point cloud[J].Computers and Electronics in Agriculture,2021,187.
- Shen,Fengli,Lu, et al.Dual semantic-guided model for weakly-supervised zero-shot semantic segmentation[J].Multimedia Tools and Applications,2021,81(4):1-16.
- Wu J,Sun M,Xu H, et al.Class agnostic and specific consistency learning for weakly-supervised point cloud semantic segmentation[J].Pattern Recognition,2025,158111067-111067.
- Samoaa P,Aronsson L,Longa A, et al.A unified active learning framework for annotating graph data for regression task[J].Engineering Applications of Artificial Intelligence,2024,138(PB):109383-109383.
- Bicheng S ,Peng Z ,Liang D , et al.Active deep image clustering[J].Knowledge-Based Systems,2022,252.
- Sun R,Guo S,Guo J, et al.GraphMoCo: A graph momentum contrast model for large-scale binary function representation learning[J].Neurocomputing,2024,575127273-.
- Shengwei F ,Ke L ,Haisong H , et al.Red-billed blue magpie optimizer: a novel metaheuristic algorithm for 2D/3D UAV path planning and engineering design problems[J].Artificial Intelligence Review,2024,57(6):.
- Hassen L,Ali L,Slim B, et al.Joint filter and channel pruning of convolutional neural networks as a bi-level optimization problem[J].Memetic Computing,2024,16(1):71-90.
- Baljon M .A Framework for Agriculture Plant Disease Prediction using Deep Learning Classifier[J].International Journal of Advanced Computer Science and Applications (IJACSA),2023,14(8):.
- Xiaodan L ,Zijian Z .A Whale Optimization Algorithm with Convergence and Exploitability Enhancement and Its Application[J].Mathematical Problems in Engineering,2022,2022.
- Anderson J P, Stephens D W, Dunbar S R. Saltatory search: a theoretical analysis[J]. Behavioral Ecology, 1997, 8(3): 307-317.
- Fergany E A A,Agwa M A.Red-Billed Blue Magpie Optimizer for Electrical Characterization of Fuel Cells with Prioritizing Estimated Parameters[J].Technologies,2024,12(9):156-156.
- Wang P,Liu Z,Wang Z, et al.Graph generative adversarial networks with evolutionary algorithm[J].Applied Soft Computing,2024,164111981-111981.
- Zhang M,Wang H,Cui Z, et al.Hybrid multi-objective cuckoo search with dynamical local search[J].Memetic Computing,2018,10(2):199-208.
- Li C, Priemer R, Cheng K H. Optimization by random search with jumps[J]. International Journal for Numerical Methods in Engineering, 2004, 60(7): 1301-1315.
- Shan D ,Zhang X ,Shi W , et al.Neural Architecture Search for a Highly Efficient Network with Random Skip Connections[J].Applied Sciences,2020,10(11):.
- Gan W,Li H,Hao P.Many-objective optimization algorithm based on the similarity principle and multi-mechanism collaborative search[J].The Journal of Supercomputing,2024,81(1):124-124.
- Fang G ,Weibin Z ,Guofu L , et al.A point cloud registration method based on multiple-local-feature matching[J].Optik,2023,295.
- Jingtao W,Changcai Y,Lifang W, et al.CSCE-Net: Channel-Spatial Contextual Enhancement Network for Robust Point Cloud Registration[J].Remote Sensing,2022,14(22):5751-5751.
- Wang L .High-precision point cloud registration method based on volume image correlation[J].Measurement Science and Technology,2024,35(3):.
- Chuang T Y, Jaw J J. Multi-feature registration of point clouds[J]. Remote sensing, 2017, 9(3): 281.
- Yu F,Chen Z,Cao J, et al.Redundant same sequence point cloud registration[J].The Visual Computer,2023,40(11):7719-7730.
- Xu S.An Introduction to Scientific Computing with Matlab and Python Tutorials[M].CRC Press:2021-12-24.
- Garcia H A,Zhu W.Building an Accessible and Flexible Multi-User Robotic Simulation Framework with Unity-MATLAB Bridge[J].Computers,2024,13(11):282-282.
- [1]Gasmi K,Hasnaoui S.Dataflow-based automatic parallelization of MATLAB/Simulink models for fitting modern multicore architectures[J].Cluster Computing,2024,27(5):6579-6590.
- Yang D.An improved particle swarm optimization algorithm for parameter optimization[J].Computer Informatization and Mechanical System,2022,5(4):35-38.
- Mohan B G,Kumar P R,Elakkiya R.Enhancing pre-trained models for text summarization: a multi-objective genetic algorithm optimization approach[J].Multimedia Tools and Applications,2024,(prepublish):1-17.
- Multilevel Optimization for Registration of Deformable Point Clouds.[J].IEEE transactions on image processing : a publication of the IEEE Signal Processing Society,2020,PP.
- Tianyuan L ,Jiacheng W ,Xiaodi H , et al.3DSMDA-Net: An improved 3DCNN with separable structure and multi-dimensional attention for welding status recognition[J].Journal of Manufacturing Systems,2021.
- Peyman A ,Keyhan G ,Atieh A , et al.Bayesian Multi-modeling of Deep Neural Nets for Probabilistic Crop Yield Prediction[J].Agricultural and Forest Meteorology,2022,314.
- Zhu G,0010 Z L,Shen P, et al.Continuous Gesture Segmentation and Recognition Using 3DCNN and Convolutional LSTM.[J].IEEE Trans. Multimedia,2019,21(4):1011-1021.
- Zhihua D ,Peiliang G ,Baohua Z , et al.Maize crop row recognition algorithm based on improved UNet network[J].Computers and Electronics in Agriculture,2023,210.
- Xiaomei Y ,Jiaoping W ,Peng W , et al.AC-UNet: an improved UNet-based method for stem and leaf segmentation in Betula luminifera [J].Frontiers in Plant Science,2023,14.
- Xiang R.Image segmentation for whole tomato plant recognition at night[J].Computers and Electronics in Agriculture,2018,154434-442.
- Zeng W,He M.Rice disease segmentation method based on CBAM-CARAFE-DeepLabv3+[J].Crop Protection,2024,180106665-.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).