Submitted:
02 December 2024
Posted:
03 December 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
1.1. Sub Types of Ischemic Strokes
1.2. Alpha-1 Antitrypsin Role in Ischemic Stroke
1.3. Serpina1 Variants (Modified from MedlinePlus [Internet]. Bethesda (MD): National Library of Medicine (US)
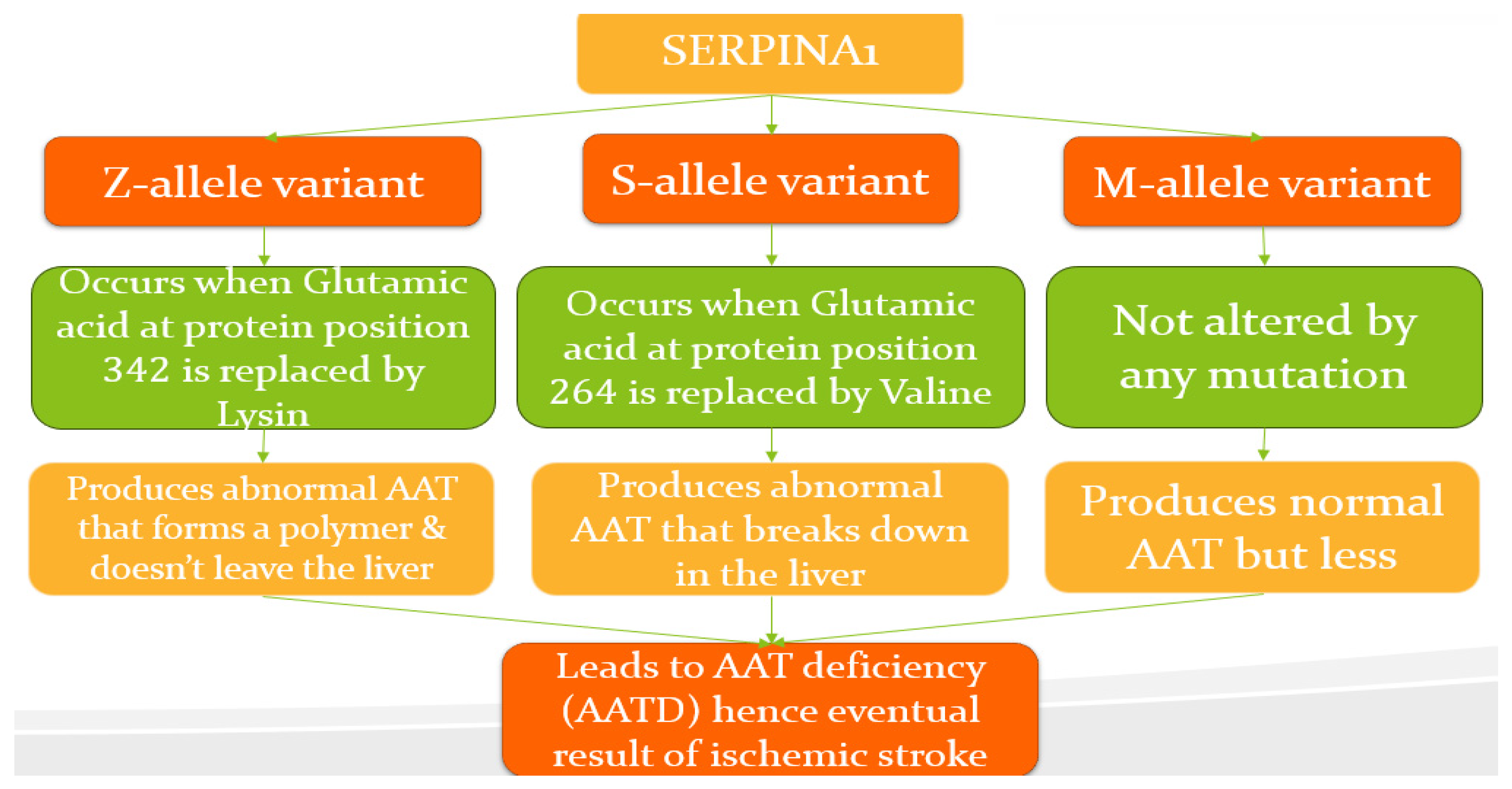
2. Literature Review
3. Methodology and Tools Used
3.1. Materials
3.2. Tools Used
3.3. Methods and Protocol
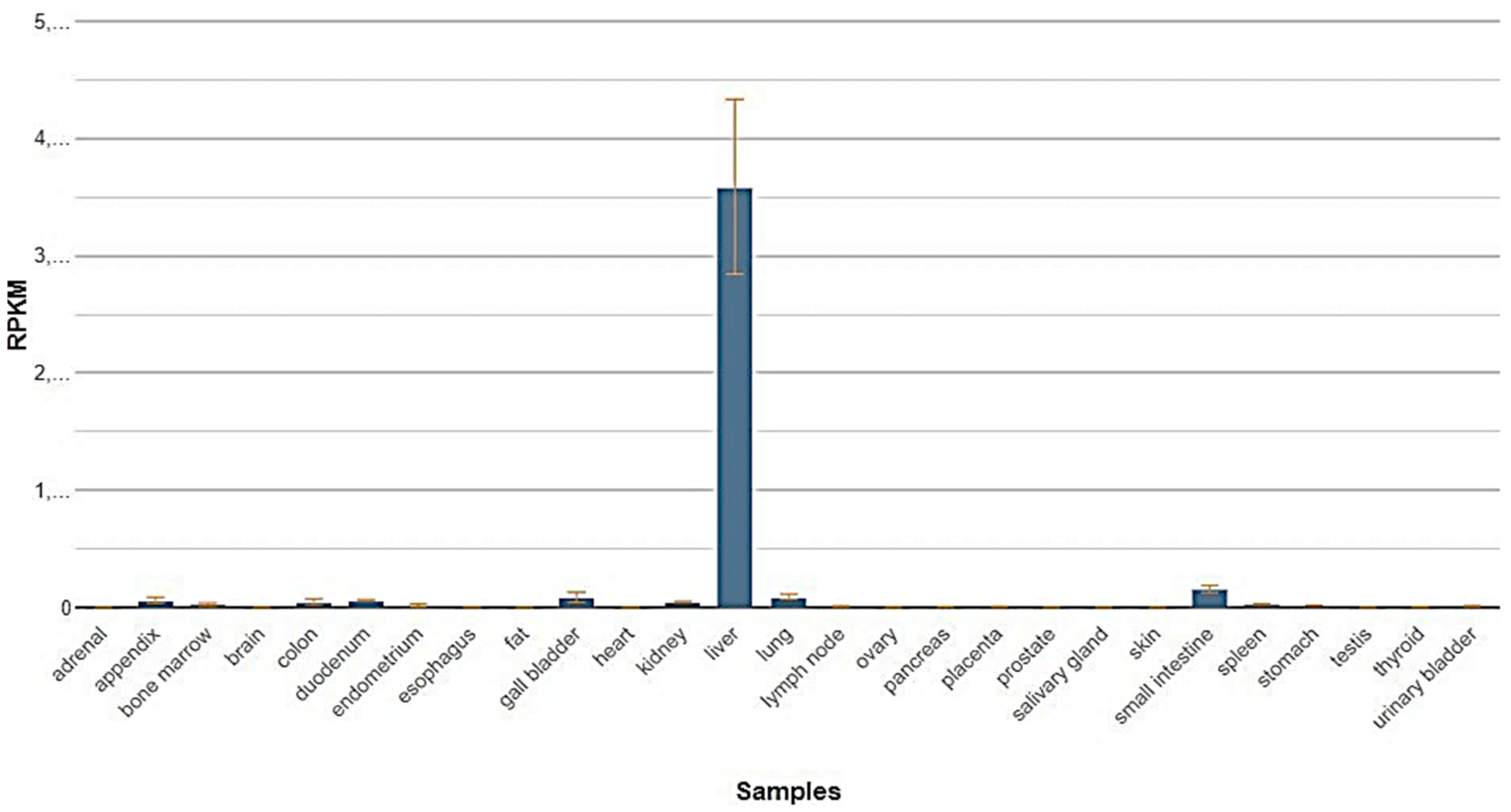
4. Results

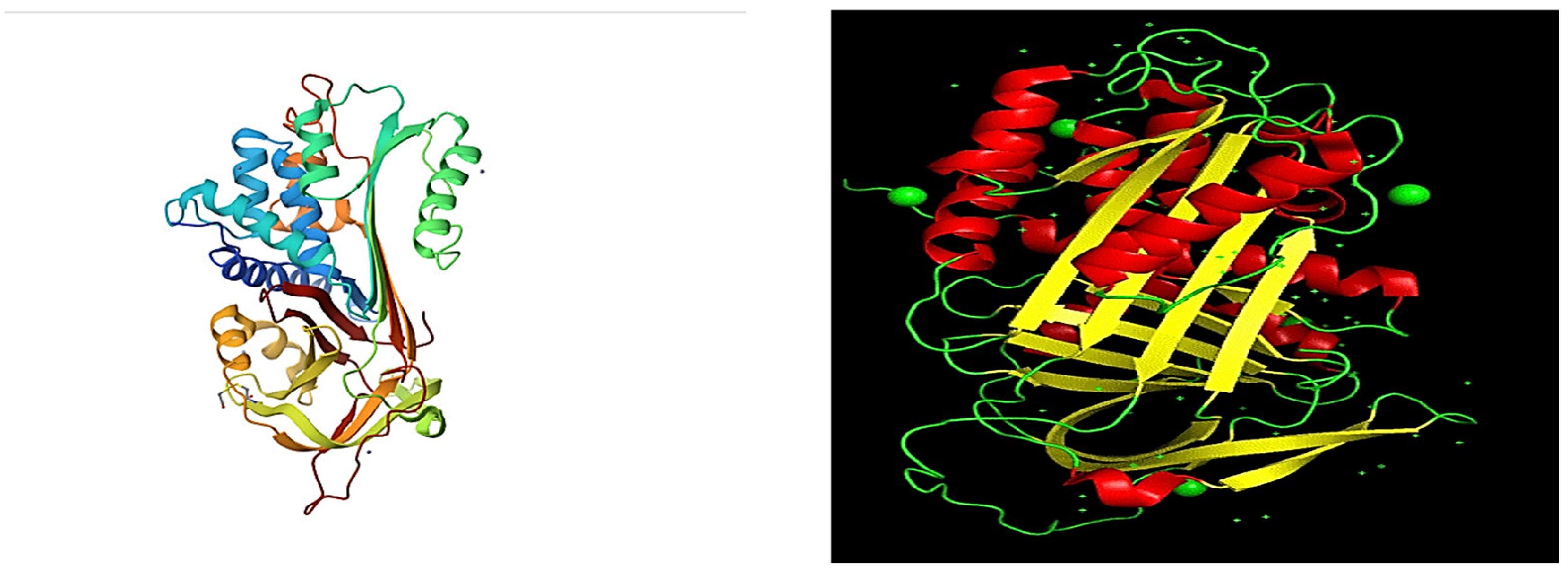
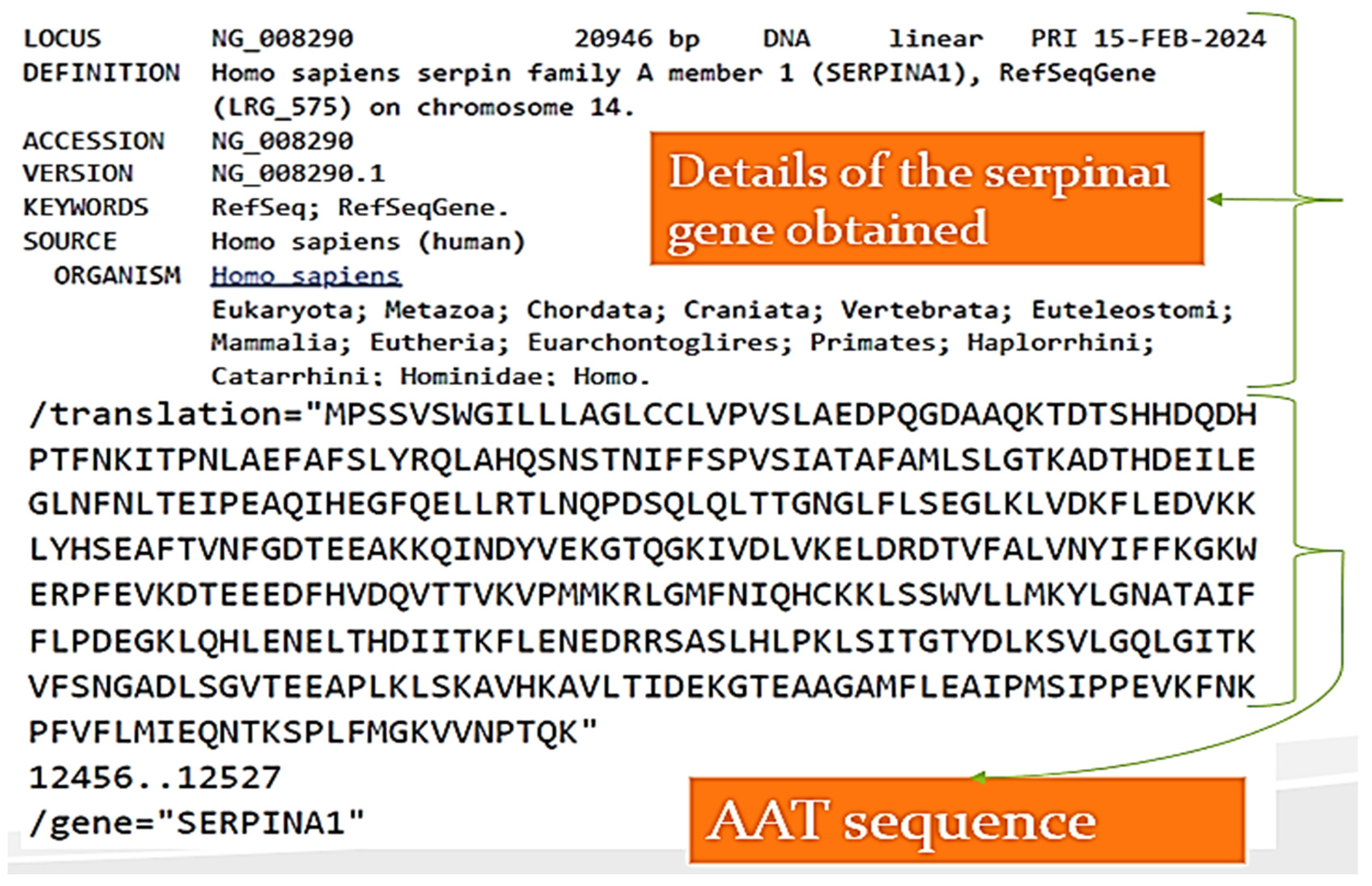
- 1)
- Embryonic and induced pluripotent stem cells and lineage markers pathway.
- 2)
- P73 transcription factor network pathway.
- 3)
- Nuclear receptors meta pathway.
- 4)
- FOXA1 transcription factor network pathway.
- 5)
- Vesicle mediated transport pathway.
- 6)
- Metabolism of proteins pathway.
- 7)
- Lung fibrosis pathway.
- 8)
- Response to elevated platelet cytosolic Ca2+ pathway.
- 9)
- Transport to the golgi and subsequent modification pathway.
- 10)
- Diseases of hemostasis pathway.
- 11)
- Regulation of insulin-like growth factors (IGF) transport and uptake by insulin-like growth factors binding proteins (IGFBPs) pathway.
- 12)
- Innate immune system pathway that included the neutrophil degranulation pathway.
- 13)
- COPII-mediated vesicle transport pathway.
- ❖ Cardioembolic
- ❖ Large artery atherosclerosis
- ❖ Small vessel—this means small vessel occlusion stroke
- ❖ Others—these include those with determined etiology other than cardioembolic, large artery atherosclerosis or small vessel and those with undetermined etiology
- ❖ Variants—in the gene
- ❖ Protein ID—given in form of nucleotide data ID because the gene can be accessed in GenBank and its protein translation is provided on the GenBank file.
- ❖ NA—for “Not Available” represents that the particular data about the gene is not available at the moment of writing.
| Genetic Association and Differential expression with stroke | ||||
| Gene | Regulation |
Stroke subtype (phenotype) | Variants | Protein ID |
| Lung fibrosis pathway | ||||
| SERPINA1 | Up | Cardioembolic, Large artery atherosclerosis & others | intron variant | NM_001002235.2 NM_000295.4 NM_001002235.2 |
| CCR2 | Up | Others Cardioembolic, Large artery atherosclerosis |
synonymous variant, Upstream gene variant | NM_001123396.1 NM_001123041.2 NM_001123041.2 |
| CCR3 | Down | cardioembolic | Intron variant | NM_178328.1 |
| CEBPB | Up & down | Others | Downstream gene variant | NM_005194.3 |
| FAM13A | Up & down | Cardioembolic, Small-vessel occlusion | Intron variant, downstream gene variant | NM_000589.3 NM_172348.2 NM_000589.3 |
| FGF1 | Up | NA | Missense | NM_000800.4 |
| GREM1 | Up | Cardioembolic, large artery atherosclerosis, & others | Intron & upstream variant | NM_013372.6 NM_001191322.1 NM_001191323.1 |
| PDGFB | Down | Large artery atherosclerosis & others | Intron & upstream variant | NM_002608.3 NM_033016.3 |
| PLAU | Up | Cardioembolic, large artery atherosclerosis, small vessel and others | Downstream, intron, splice and 3′ UTR variants | NM_002658.4 NM_001145031.2 NM_001319191.1 |
| TGFB1 | Up | Others | Intron variant | NM_000660.6 |
| TIMP1 | Up | NA | NA | NA |
| Embryonic and induced pluripotent stem cells and linear marker pathway | ||||
| Gene | regulation | Stroke subtype (phenotype) | Variants | Protein ID |
| ATF2 | Up | Cardioembolic, large artery atherosclerosis, small vessel & others | Synonymous, intron, splice-region, & sequence feature variants | NM_005171.4 NM_001256090.1 NM_001256091.1 NM_001256092.1 |
| CXCR4 | Up | Small vessel | Synonymous variant | NM_001008540.1 |
| FGF18 | Up & down | Cardioembolic & large artery atherosclerosis | Intron variant | NM_003862.2 |
| GATA2 | Down | Small vessel, large artery atherosclerosis & others | 5′ UTR, intron, missense, upstream variants | NM_001145661.1 NM_032638.4 |
| HNF1B | Down | Small vessel, Cardioembolic, large artery atherosclerosis & others | 3′ UTR, intron & upstream variants | NM_001165923.3 NM_000458.3 NM_001304286.1 |
| ITGB1 | Up | Small vessel, Cardioembolic, large artery atherosclerosis & others | 5′ UTR, intron, downstream, & upstream gene variants | NM_133376.2 NM_033668.2 NM_002211.3 |
| KDR | Up | Cardioembolic, large artery atherosclerosis & others | Intron, missense, & structural interaction variants | NM_002253.2 |
| KLF5 | Up | Cardioembolic, large artery atherosclerosis, small vessel & others | Upstream, intron & downstream gene variants | NM_001286818.1 NM_001730.4 |
| LGR4 | Down | Cardioembolic, large artery atherosclerosis & others | Intron variant | NM_018490.2 |
| LMO2 | Up | Large artery atherosclerosis & others | Synonymous, intron & upstream gene variant | NM_001142315.1 NM_005574.3 NM_001142316.1 |
| LMO4 | Up & down | Small vessel & others | Intron variant | NM_006769.3 |
| LRIG1 | Down | Cardioembolic, Large artery atherosclerosis & others | Intron & missense variants | NM_015541.2 |
| ONECUT2 | Up & down | Cardioembolic, large artery atherosclerosis & others | 3′ UTR & intron variants | NM_004852.2 |
| PAX4 | Up | NA | Missense & synonymous | NM_006193.2 |
| PROX1 | Down | Cardioembolic, large artery atherosclerosis, small vessel & others | Upstream, downstream, 3′ UTR & intron variant | NM_001270616.1 NM_002763.4 |
| SLC2A2 | Up | Cardioembolic, large artery atherosclerosis & others | Downstream, upstream, intron, synonymous & missense variants | NM_001278658.1 NM_000340.1 |
| SOX9 | Down | Others | Intron variant | NM_000346.3 |
| TCF3 | Down | Cardioembolic, large artery atherosclerosis, small vessel & others | Upstream, downstream, synonymous, splice-region, intron, 3′ UTR & missense | NM_003200.3 NM_001136139.2 |
| THPO | Up | Cardioembolic, small vessel & others | Upstream, downstream & intron variants | NM_001290003.1 NM_000460.3 NM_001177597.2 NM_001290026.1 |
| p73 transcription factor pathway | ||||
| Gene | regulation | Stroke subtype (phenotype) | Variants | Protein ID |
| ADA | Up | Cardioembolic, large artery atherosclerosis, small vessel & others | Downstream, intron, structural interaction, missense, 5′ UTR, upstream, noncoding transcript variant | NM_000022.3 NM_001322050.1 NM_001322051.1 |
| BAK1 | Down | Large artery atherosclerosis | Intron variant | NM_001188.3 |
| BAX | Up | Cardioembolic, large artery atherosclerosis, small vessel | Intron, downstream, upstream gene variant | NM_001291428.1 NM_001291429.1 NM_001291430.1 NM_004324.3 NM_138761.3 |
| BIN1 | Down | NA | Missense & synonymous | NM_139343.2 |
| BUB1 | Up | Cardioembolic, small vessel & others | Intron, sequence feature, upstream, downstream, splice-region variants | NM_001278616.1 NM_004336.4 NM_001278617.1 |
| CCNA2 | Up | Large artery atherosclerosis | Intron & 5′ UTR variant | NM_001237.3 |
| CDK2 | Up | Small vessel | Downstream gene variant | NM_001798.4 NM_001290230.1 |
| CDKN1A | Up | Cardioembolic, large artery atherosclerosis, small vessel & others | Upstream, intron, 5′ UTR, synonymous, missense, 3′ UTR, sequence feature & downstream gene variant | NM_078467.2 NM_000389.4 NM_001220777.1 NM_001291549.1 |
| FBXO45 | Up | Others | Intron variant | NM_00105573.1 |
| GDF15 | Up | NA | Missense, synonymous & conservative in-frame insertion variant | NM_004864.2 |
| HSF1 | Down | Large artery atherosclerosis | Intron variant | NM_005526.2 |
| IL1RAP | Up | NA | Missense, synonymous, & splice region variants | NM_001167931.1NM_001167928.1 |
| IL4R | Up | Cardioembolic, large artery atherosclerosis, small vessel & others | 3′ UTR, downstream, upstream, splice region, sequence feature & intron | NM_000418.3 NM_001257406.1 NM_001257407.1 |
| JAK1 | Up | NA | Missense & synonymous | NM_001320923.1 NM_001321857.1 |
| MAPK14 | Up | Cardioembolic, large artery atherosclerosis, small vessel & others | Upstream, 3′ prime, intron, downstream & sequence feature variants | NM_139013.2 NM_139012.2 NM_001315.2 |
| NSG1 | Down | Small vessel, cardioembolic & others | Intron, upstream & downstream gene variants | NM_001040101.1 NM_001287763.1 NM_001287764.1 |
| PEA15 | Down | Large artery atherosclerosis, cardioembolic & others | Splice-region, intron, 3′ UTR, 5′ UTR, downstream & upstream gene variants | NM_001297576.1 NM_001297577.1 NM_001297578.1 NR_123724.1 |
| PML | Up & down | NA | Missense variants | NM_033239.2 NM_033250.2 NM_033238.2 NM_033249.2 |
| RAD51 | Up | Large artery atherosclerosis & cardioembolic | Intron, upstream downstream, 3′ UTR & 5′ UTR variants | NM_001164269.1 NM_001164270.1 NM_002875.4 NM_133487.3 |
| RB1 | Up | Large artery atherosclerosis, cardioembolic & others | Intron variant | NM_000321.2 |
| RELA | Down | Small vessel, large artery atherosclerosis & others | Downstream, intron & upstream variants | NM_001145138.1 NM_001243984.1 NM_021975.3 |
| SFN | Up | NA | Synonymous variant | NM_006142.3 |
| SP1 | Up | Cardioembolic, large artery atherosclerosis & others | Downstream, intron & upstream variants | NM_138473.2 NM_001251825.1 NM_003109.1 |
| TP63 | Up | NA | Synonymous variant | NM_0061422.4 NM_001329964.1 NM_001114982.1 NM_001329146.1 NM_001329148.1 NM_001114980.1 |
| TUBA1A | Up | Large artery atherosclerosis | Intron variant | NM_001270399.1 NM_001270400.1 |
| Nuclear receptors meta-pathway | ||||
| Gene | regulation | Stroke subtype (phenotype) | Variants | Protein ID |
| ABCB1 | Down | Large artery atherosclerosis, cardioembolic & others | Intron, 5′ UTR & 3′ UTR variants | NM_000927.4 |
| ABCC3 | Down | Cardioembolic, large artery atherosclerosis & small vessel | Upstream & intron variants | NM_003786.3 NM_001144070.1 |
| ABCG8 | Up | Large artery atherosclerosis, small vessel, cardioembolic & others | Upstream, intron, synonymous & missense variants | NM_022437.2 |
| ACKR3 | Down | Large artery atherosclerosis & cardioembolic | Intron & synonymous variants | NM_020311.2 |
| ALAS1 | Up | Cardioembolic | Upstream, intron & downstream gene variants | NM_001304444.1 NM_001304443.1 NM_000688.5 NM_199166.2 |
| APOA2 | Up | NA | Missense & synonymous | NM_001643.1 |
| APOA5 | Up | Cardioembolic, small vessel & others | Downstream & upstream gene variants | NM_001166598.1 |
| ARL5B | Up | NA | Splice region & synonymous | NM-178815.3 |
| ARNT | Up | Cardioembolic & large artery atherosclerosis | Upstream, downstream, intron, 3′ UTR variants | NM_001668.3 NM_001197325.1 NM_001286035.1 NM_001286036.1 |
| BIRC2 | Up | Cardioembolic, small vessel & large artery atherosclerosis | Intron, sequence feature, upstream, 5′ UTR, downstream gene variants | NM_001256163.1 NM_001166.4 NM_001256166.1 |
| BLVRB | Down | NA | Missense & synonymous | NM_000713.2 |
| CAP2 | Down | Large artery atherosclerosis, small vessel & others | Intron variant | NM_006366.2 |
| CBR3 | Down | Cardioembolic | Structural interaction variant | NM_001236.3 |
| CCL20 | Down | Large artery atherosclerosis & cardioembolic | Upstream & downstream variant | NM_004591.2 |
| CPT1A | Up | Cardioembolic, small vessel & others | Upstream, downstream, intron, missense, synonymous, splice region | NM_001876.3 NM_001031847.2 |
| CPT2 | Up | Small vessel & others | Intron & downstream | NM_000098.2 |
| CYP1A2 | Down | Others | Intron & synonymous | NM_000761.4 |
| CYP1B1 | Up | Others | Intron variant | NM_000104.3 |
| CYP2C9 | Up & down | Cardioembolic, large artery atherosclerosis, small vessel & others | Intron & sequence feature variants | NM_000771.3 |
| CYP3A5 | Down | Large artery atherosclerosis, small vessel & others | Intron, splice acceptor, downstream gene variants | NM_000777.4 NR_033807.2 NM_001291829.1 NM_001291830.1 |
| DNAJB1 | Up | Cardioembolic & large artery atherosclerosis | Upstream & 3′ UTR variant | NM_006145.2 NM_001300914.1 |
| EPB41L4B | Up & down | Cardioembolic, large artery atherosclerosis, small vessel & others | Intron, missense, splice region, upstream, | NM_019114.4 |
| EPHA3 | Down | NA | Missense, synonymous & splice region variant | NM_005233.5 |
| ETNK2 | Down | Cardioembolic, large artery atherosclerosis, small vessel & others | Missense, intron, upstream & downstream gene variants | NM_018208.3 NM_001297760.1 NM_001297761.1 NM_001297762.1 |
| FGD4 | Up | NA | Missense & synonymous variants | NM_001304480.1 NM_001304481.1 NM_139241.3 NM_001304484.1 NM_001304483.1 |
| FTH1 | Up | NA | Missense & synonymous | NM_002032.2 |
| GADD45B | Up | NA | Missense & intron variant | NM_015675.3 |
| GCLM | Up | Cardioembolic, large artery atherosclerosis & others | Downstream, upstream & intron variants | NM_002061.3 NM_001308253.1 |
| GPR153 | Up | Others | Downstream, intron, missense & synonymous | NM_207370.2 |
| GSR | Up | Small vessel, cardioembolic & others | Intron, upstream, 3′ UTR, synonymous & missense variants | NM_000637.3 NM_001195102.1 NM_001195103.1 NM_001195104.1 |
| GSTP1 | Down | Small vessel | Missense, synonymous & intron variants | NM_000852.3 |
| GSTT2 | Up | NA | NA | NA |
| IP6K3 | Down | Cardioembolic, large artery atherosclerosis, small vessel & others | Downstream, intron, synonymous, 5′ UTR & upstream gene variants | NM_001142883.1 NM_054111.4 |
| IRS2 | Up | Cardioembolic, large artery atherosclerosis & others | Intron & 3′ UTR variants | NM_003749.2 |
| KTN1 | Up | NA | Missense, synonymous & splice region variants | NM_001079521.1 NM_001079522.1 NM_004986.3 NM_001271014.1 |
| MAFF | Up | Cardioembolic, large artery atherosclerosis, small vessel & others | Upstream, downstream, intron, 5′ UTR & 3′ UTR variants | NM_001161572.1 NM_012323.3 |
| MGST1 | Up | Small vessel & others | Upstream, sequence feature, intron, downstream gene variants | NM_001260511.1 NM_145792.2 NM_001260512.1 NM_020300.4 NR_048545.1 |
| MGST2 | Up | Cardioembolic, large artery atherosclerosis, small vessel & others | Upstream, intron, downstream gene variants | NM_001204366.1 NM_001204368.1 NM_002413.4 |
| MYOF | Up | NA | Missense, synonymous, splice region & frameshift | NM_013451.3 NM_133337.2 |
| NAV3 | Up | NA | Missense, synonymous & splice region variant | NM_001024383.1 NM_014903.5 |
| NCOA2 | Up | NA | Missense & synonymous variant | NM_001321703.1 NM_001321712.1 NM_001321713.1 |
| NR1H3 | Up | Cardioembolic, large artery atherosclerosis, small vessel & others | Upstream, downstream, intron, 5′ UTR & synonymous | NM_001251934.1 NM_001251935.1 NM_005693.3 NM_001130102.2 |
| NR1H4 | Up | NA | Missense, synonymous & splice region variant | NM_001206993.1 NM_001206977.1 NM_005123.3 NM_001206992.1 |
| NR3C1 | Up | NA | Missense & synonymous | NM_001024094.1 |
| PLK2 | Up | Cardioembolic, large artery atherosclerosis, small vessel & others | Downstream, intron, synonymous, 5′ UTR, 3′ UTR, sequence feature & splice region variants | NM_006622.3 NM_001252226.1 |
| PMP2 | Down | NA | Missense & synonymous | NM_002677.3 |
| POLK | Up | Small vessel & large artery atherosclerosis | Intron variant | NM_016218.2 |
| PRDX6 | Up | Cardioembolic | Intron variant | NM_004905.2 |
| PTGS2 | Up | Missense & synonymous | NM_000963.3 | |
| SERPINB2 | Up | Cardioembolic & small vessel | Missense, intron & downstream gene variant | NM_001143818.1 NM_002575.2 |
| SLC2A11 | Down | Cardioembolic, large artery atherosclerosis, small vessel & others | Upstream, intron, missense, non-coding transcript exon, 3′ UTR, downstream, 5′ UTR & synonymous | NM_001282864.1 NR_104248.1 NM_001024938.3 NM_030807.4 |
| SLC2A12 | Up | Large artery atherosclerosis, small vessel & others | Intron variant | NM_145176.2 |
| SLC2A2 | Up | Cardioembolic, large artery atherosclerosis & others | Downstream, intron, missense, synonymous, upstream variants | NM_000340.1 NM_001278658.1 |
| SLC2A3 | Up | Large artery atherosclerosis | Intron, synonymous, sequence feature, | NM_006931.2 |
| SLC2A9 | Up | NA | Missense & synonymous | NM_020041.2 NM_001001290.1 |
| SLC39A1 | Up | Cardioembolic, large artery atherosclerosis & others | 3′ UTR & upstream gene variant | NM_001271957.1 NM_001271959.1 NM_001271958.1 NM_001271961.1 NM_014437.4 |
| SLC39A4 | Up | NA | Missense, synonymous & splice region variants | NM_017767.2 NM_130849.3 NM_001280557.1 |
| SLC39A6 | Up & down | Cardioembolic, large artery atherosclerosis, small vessel & others | Downstream, upstream, intron, synonymous, | NM_012319.3 NM_001099406.1 |
| SLC39A9 | Up | Small vessel & large artery atherosclerosis | Upstream, intron & 3′ UTR variant | NM_018375.4 NM_001252151.1 NM_001252152.1 NM_001252148.1 NM_001252150.1 |
| SLC5A1 | Up | Cardioembolic & large artery atherosclerosis | Downstream, upstream, intron, synonymous, missense & 3′ UTR variants | NM_001256314.1 NM_000343.3 |
| SLC5A8 | Up | Cardioembolic, large artery atherosclerosis & small vessel | Intron variant | NM_145913.3 |
| SLC5A9 | Up | Cardioembolic, large artery atherosclerosis & others | Synonymous, intron, upstream & missense variant | NM_001135181.1 NM_001011547.2 |
| SLC6A1 | Up | Cardioembolic, large artery atherosclerosis, small vessel & others | Intron variant | NM_003042.3 |
| SLC6A2 | Up | Cardioembolic, large artery atherosclerosis & others | Upstream, downstream, 5′ UTR, missense, intron, | NM_001172504.1 NM_001172501.1 NM_001043.3 NM_001172502.1 |
| SLC7A11 | Up | Cardioembolic, large artery atherosclerosis & others | Intron & sequence feature variant | NM_014331.3 |
| SNAI2 | Up | NA | Intron variant | NM_003068.4 |
| SP1 | Up | Cardioembolic, large artery atherosclerosis & others | Upstream, downstream & intron variant | NM_138473.2 NM_001251825.1 NM_003109.1 |
| SQSTM1 | Up & down | Cardioembolic, large artery atherosclerosis & others | Upstream, downstream, 5′ UTR, intron, synonymous & sequence feature variant | NM_001142298.1 NM_003900.4 |
| SRC | Down | Cardioembolic, large artery atherosclerosis, small vessel & others | Intron, 5′ UTR & upstream gene variants | NM_005417.4 NM_198291.2 |
| SREBF1 | Up | Cardioembolic, large artery atherosclerosis & others | 5′ UTR, 3′ UTR, downstream, upstream & intron variant | NM_001005291.2 NM_001321096.2 NM_004176.4 |
| SRGN | Up | Cardioembolic, large artery atherosclerosis, small vessel & others | Downstream, upstream, 3′ UTR, intron variants | NM_001321053.1 NM_001321054.1 NM_002727.3 |
| TGFBR3 | Up | NA | Synonymous, missense & splice region variants | NM_003243.4 NM_001195683.1 |
| TNFAIP3 | Up | Small vessel, large artery atherosclerosis & others | Downstream, upstream & intron variants | NM_001270507.1 NM_001270508.1 NM_006290.3 |
| TSC22D3 | Down | NA | NA | NA |
| TXNRD1 | Up | Cardioembolic, large artery atherosclerosis, small vessel & others | Synonymous, upstream, intron, downstream, 3′ UTR variants | NM_001093771.2 NM_001261445.1 NM_001261446.1 NM_003330.3 NM_182729.2 NM_182742.2 NM_182743.2 |
| VDR | Up | Large artery atherosclerosis, small vessel & others | Downstream & intron variants | NM_001017536.1 NM_001017535.1 NM_000376.2 |
| FOXA1 transcription factor pathway | ||||
| Gene | regulation | Stroke subtype (phenotype) | Variants | Protein ID |
| AR | Down | NA | Missense & synonymous | NM_000044.3 NM_001011645.2 NM_00112707.2 |
| NR2F2 | Up & down | NA | Synonymous variant | NM_021005.3 NM_001145155.1 |
| POU2F1 | Up | NA | Missense & synonymous | NM_002697.3 NM_001198786.1 NM_001198783.1 |
| PRDM15 | Down | NA | Missense, synonymous & splice region variant | NM_022115.4 NM_00104042.2 NM_001282934.1 |
| Disease of hemostasis pathway | ||||
| Gene | regulation | Stroke subtype (phenotype) | Variants | Protein ID |
| C3AR1 | Up | Cardioembolic & large artery atherosclerosis | Downstream gene variant | NM_001326477.1 NM_001326475.1 |
| C8G | Up | NA | Missense, synonymous & splice region variant | NM_000606.2 |
| CD46 | Up | NA | Missense, synonymous & splice region variant | NM_172359.2 NM_153826.2 NM_172350.2 NM_172361.2 |
| CFD | Up | Cardioembolic, small vessel & others | Upstream & 3′ UTR variant | NM_001317335.1 NM_001928.3 |
| CR2 | Down | Cardioembolic & small vessel | 3′ UTR, upstream, downstream & intron variants | NM_001006658.2 NM_001877.4 |
| F11 | Up | Cardioembolic, large artery atherosclerosis & others | Intron & sequence feature variants | NM_000128.3 |
| F12 | Up | NA | Missense, synonymous & splice region variant | NM_000505.3 |
| F5 | Up | Cardioembolic, small vessel, large artery atherosclerosis & others | Intron, synonymous, missense & 3′ UTR variant | NM_000130.4 |
| F8 | Up | NA | Missense & synonymous variant | NM_000132.2 NM_019863.2 |
| FGA | Up | Cardioembolic, large artery atherosclerosis & others | 3′ UTR, missense, intron, downstream, 5′ UTR, upstream, | NM_000508.4 |
| GP5 | Down | Large artery atherosclerosis | 3′ UTR variant | NM_004488.2 |
| GP9 | Down | NA | Missense & synonymous | NM_000174.4 |
| KLK1 | Up | NA | Missense & synonymous | NM_002257.3 |
| LMAN1 | Up | Cardioembolic, large artery atherosclerosis & small vessel | 3′ UTR, synonymous & intron variant | NM_005570.3 |
| MASP1 | Up | Cardioembolic, small vessel, large artery atherosclerosis & others | 3′ UTR, downstream, upstream, sequence feature, non-coding transcript & intron variant | NM_001879.5 NM_001031849.2 NM_139125.3 |
| PLAUR | Up | Cardioembolic, large artery atherosclerosis, small vessel & others | Upstream, 5′ UTR & intron variants | NM_002659.3 NM_001005376.2 NM_001301037.1 |
| PRCP | Up | Cardioembolic, large artery atherosclerosis, small vessel & others | Downstream, upstream & intron variants | NM_199418.3 NM_005040.3 NM_001319214.1 |
| PROCR | Down | Cardioembolic & others | Missense & intron variant | NM_006404.4 |
| TFPI | Up | NA | Missense variant | NM_001032281.3 NM_001329239.1 |
| COPII-mediated vesicle transport pathway | ||||
| Gene | regulation | Stroke subtype (phenotype) | Variants | Protein ID |
| ANKRD28 | Up & down | NA | Missense & synonymous variants | NM_015199.3 NM_00119099.1 |
| AREG | Up | NA | NA | NA |
| CD59 | Up | Large artery atherosclerosis & others | Downstream, intron & 3′ UTR variants | NM_000611.5 NM_001127223.1 NM_001127225.1 NM_001127226.1 NM_001127227.1 NM_203329.2 |
| CNIH1 | Up | Others | Intron & 3′ UTR variants | NM_005776.2 |
| CSNK1D | Up | Large artery atherosclerosis | Intron variants | NM_001893.4 NM_139062.2 NR_110578.1 |
| CTSC | Up | Cardioembolic, large artery atherosclerosis, small vessel & others | Upstream, downstream, 3′ UTR & intron variants | NM_001814.5 NM_001114173.2 NM_148170.4 |
| FOLR1 | Up | Cardioembolic, large artery atherosclerosis, small vessel & others | Upstream, downstream, 5′ UTR & intron variants | NM_000802.3 NM_016729.2 NM_016724.2 NM_016725.2 |
| GOLGA2 | Up | Large artery atherosclerosis & others | Intron variants | NM_004486.4 |
| GOSR2 | Up | Cardioembolic, large artery atherosclerosis, small vessel & others | Upstream, missense, 5′ UTR, 3′ UTR, downstream & intron variant | NM_001321133.1 NM_001012511.2 NM_001321134.1 NM_004287.4 |
| SEC13 | Down | Large artery atherosclerosis, small vessel & others | Sequence feature, downstream, 5′ UTR & intron variants | NM_001136026.2 NM_001136232.2 NM_030673.3 NM_183352.2 |
| SEC22C | Up | Cardioembolic, large artery atherosclerosis & others | Downstream, 3′ UTR, upstream, synonymous, missense & intron variants | NM_032970.3 NM_001201584.1 NM_004206.3 |
| SEC23IP | Up | Cardioembolic, large artery atherosclerosis, small vessel & others | Upstream, 5′ UTR, non-coding transcript, synonymous, 3′ UTR, downstream, missense, sequence feature & intron | NM_007190.3 |
| SEC24A | Up | Cardioembolic, large artery atherosclerosis & others | Downstream, upstream & intron variants | NM_021982.2 NM_001252231.1 |
| SEC24C | Down | Cardioembolic, large artery atherosclerosis, small vessel & others | Upstream, synonymous, downstream, 3′ UTR & intron variants | NM_004922.3 NM_198597.2 |
| SEC16A | Up | Cardioembolic, large artery atherosclerosis, small vessel & others | Downstream, 3′ UTR, missense, synonymous, 5′ UTR, upstream & intron | NM_014866.1 NM_001276418.1 |
| STX17 | Up | Cardioembolic, small vessel & others | Intron & 3′ UTR variants | NM_017919.2 |
| TRAPPC10 | Down | Cardioembolic, small vessel & large artery atherosclerosis | Intron variant | NM_003274.4 |
| TRAPPC2L | Up | Cardioembolic, large artery atherosclerosis & others | Downstream, upstream, 3′ UTR, non-coding transcript & intron variants | NM_001318524.1 NR_134671.1 NM_001318525.1 NM_001318526.1 NM_001318527.1 NM_001318528.1 NM_001318529.1 NM_001318530.1 NM_001318532.1 NM_016209.4 |
| Regulation of insulin-like growth factor (IGF) transport and uptake by insulin-like binding factors binding proteins (IGFBPs) pathway. | ||||
| Gene | regulation | Stroke subtype (phenotype) | Variants | Protein ID |
| AHSG | Up | Large artery atherosclerosis, small vessel & others | Synonymous & intron variants | NM_001622.2 |
| ALB | Up | Cardioembolic & small vessel | Synonymous & intron | NM_000477.6 |
| CALU | Up | Cardioembolic, large artery atherosclerosis, small vessel & others | Upstream, intron, 3′ UTR, non-coding transcript, downstream gene variants | NM_001199671.1 NM_001130674.2 NM_001199672.1 NM_001199673.1 NM_001219.4 NR_074086.1 |
| DNAJC3 | Up | Large artery atherosclerosis, small vessel & others | 3′ UTR & intron variants | NM_006260.4 |
| ENAM | Down | Small vessel & others | Missense, 3′ UTR & intron | NM_031889.2 |
| EVA1A | Up | Cardioembolic, large artery atherosclerosis, small vessel & others | Synonymous, 5′ UTR, upstream & intron variants | NM_001135032.1 NM_032181.2 |
| FAM20A | Up | Cardioembolic, large artery atherosclerosis, small vessel & others | Intron, synonymous, 5′ UTR, upstream, downstream & 3′ UTR gene variants | NR_027751.1 NM_001243746.1 NM_017565.3 |
| FN1 | Up | NA | Synonymous, missense & splice region variants | NM_212482.2 NM_002026.3 NM_212476.2 |
| FSTL3 | Up | NA | Synonymous variant | NM_005860.2 |
| FUCA2 | Up | Cardioembolic | Missense & intron variants | NM_032020.4 |
| MATN3 | Up | Large artery atherosclerosis & small vessel | Intron variant | NM_002381.4 |
| MGAT4A | Up & down | Cardioembolic, large artery atherosclerosis & others | Downstream & intron variants | NM_012214.2 NM_001160154.1 |
| NUCB1 | Down | Large artery atherosclerosis | Intron variant | NM_006184.5 |
| P4HB | Up & down | Cardioembolic, small vessel & others | Synonymous, intron & missense variants | NM_000918.3 |
| PAPPA | Down | Cardioembolic, large artery atherosclerosis, small vessel & others | Intron & 5′ UTR variants | NM_002581.4 |
| PDIA6 | Up | NA | Missense & synonymous variants | NM_005742.3 NM_001282704.1 NM_001282706.1 NM_001282705.1 NM_001282707.1 |
| RCN1 | Down | NA | Synonymous & missense | NM_002901.2 |
| SERPINA10 Down | Cardioembolic & others | Sequence feature, downstream, 3′ UTR, missense & upstream variants | NM_001100607.2 NM_016186.2 | |
| SPARCL1 | Up | NA | Missense & synonymous variants | NM_001128310.2 NM_001291976.1 |
| STC2 | Up | Large artery atherosclerosis & others | Intron variant | NM_003714.2 |
| TGOLN2 | Up | Cardioembolic, large artery atherosclerosis & others | Missense, downstream, 3′ UTR & intron | NM_001206840.1 NM_001206841.1 NM_001206844.1 NM_006464.3 |
| Transport to the Golgi pathway | |||||
| Gene | regulation | Stroke subtype (phenotype) | Variants | Protein ID | |
| ACTR10 | Up | Others | Sequence feature variant | NM_018477.2 | |
| ALG2 | Up | Large artery atherosclerosis | Upstream & downstream | NM_033087.3 | |
| ALG5 | Up | Cardioembolic & large artery atherosclerosis | Intron & upstream variant | NM_001142364.1 NM_013338.4 | |
| ALG6 | Up | Cardioembolic, large artery atherosclerosis & others | Intron variant | NM_013339.3 | |
| ALG8 | Up | Cardioembolic, large artery atherosclerosis & others | Stop lost, stop retained, upstream, intron & downstream variants | NM_024079.4 NM_001007027.2 | |
| AMFR | Up | Cardioembolic & small vessel | Upstream, intron & sequence feature variants | NM_001144.5 NM_001323512.1 NM_001323511.1 | |
| ANK2 | Up | NA | Missense, synonymous & splice region variants | NM_001148.4 NM_001127493.1 NM_02097.3 | |
| ARF3 | Up | Small vessel | Intron variant | NM_001659.2 | |
| ASGR1 | Down | Cardioembolic & others | Downstream, intron & synonymous variants | NM_001671.4 NM_001197216.2 | |
| B4GALT1 | Up | Cardioembolic & large artery atherosclerosis | 3′ UTR & intron variants | NM_001497.3 | |
| B4GALT3 | Up & down | Cardioembolic & large artery atherosclerosis | Downstream & intron variants | NM_001199874.1 NM_001199873.1 | |
| BET1L | Up | Cardioembolic, large artery atherosclerosis & others | Upstream & downstream variants | NM_001098787.1 | |
| CALR | Up | NA | Missense, intron & synonymous variants | NM_004343.3 NM_145046.4 | |
| COG5 | Up | NA | Missense, splice region & synonymous variants | NM_006348.3 NM_181733.2 | |
| COPA | Up | Cardioembolic, large artery atherosclerosis & others | Intron, upstream & downstream gene variants | NM_001098398.1 NM_004371.3 | |
| COPG1 | Up | Others | Missense & intron | NM_016128.3 | |
| DCTN2 | Up | Cardioembolic & others | Upstream & intron variants | NM_001261412.1 NM_001261413.1 NM_006400.4 | |
| DCTN3 | Up | Cardioembolic & others | Upstream, downstream & intron variants | NM_007234.4 NM_001281425.1 NM_001281426.1 NM_001281427.1 | |
| DCTN5 | Down | Others | Intron variants | NM_032486.3 NM_001199011.1 NM_001199743.1 | |
| DCTN6 | Up | Cardioembolic, small vessel & others | Intron variant | NM_006571.3 | |
| DERL2 | Up | Cardioembolic, large artery atherosclerosis, small vessel & others | Intron, upstream & downstream gene variants | NM_016041.4 NM_001304777.1 NM_001304779.1 NR_130905.1 | |
| DHDDS | Up | Cardioembolic & large artery atherosclerosis | Intron, 3′ UTR, downstream & missense variants | NM_024887.3 NM_001243564.1 NM_001243565.1 NM_205861.2 NM_001319959.1 | |
| DYNLL2 | Up & down | NA | Intron variant | NM_080677.2 | |
| EDEM1 | Down | Large artery atherosclerosis, small vessel & others | Synonymous, intron, 3′ UTR variants | NM_014674.2 | |
| FUOM | Down | Others | Upstream gene variants | NM_001098483.2 NM_001301827.1 NM_001301828.1 | |
| GMPPA | Up | Cardioembolic | Upstream, downstream, 5′ UTR & intron variants | NM_013335.3 NM_205847.2 | |
| GOSR1 | Up & down | Large artery atherosclerosis & others | Intron variants | NM_004871.2 NM_001007024.1 | |
| MAN1C1 | Down | Cardioembolic, large artery atherosclerosis, & others | Intron, upstream, synonymous & 3′ UTR variants | NM_020379.3 NM_001289010.1 | |
| MGAT3 | Down | Cardioembolic, large artery atherosclerosis & small vessel | Downstream, upstream, 3` UTR & intron variants | NM_002409.4 NM_001098270.1 | |
| MPDU1 | Down | Cardioembolic & others | Downstream, upstream, 3′ UTR & intron variants | NM_001330073.1 NM_004870.3 | |
| MPI | Down | Small vessel & others | Intron, synonymous & downstream gene variants | NM_002435.2 NM_001289155.1 NM_001289156.1 NM_001289157.1 | |
| NANS | Up | Large artery atherosclerosis | Intron variant | NM_018946.3 | |
| NEU1 | Up | NA | NA | NA | |
| NEU3 | Up | Large artery atherosclerosis | Intron & missense variants | NM_006656.5 | |
| OS9 | Up | NA | Missense, synonymous & splice region variants | NM_006812.3 NM_001261420.1 | |
| PMM2 | Up | Cardioembolic, large artery atherosclerosis & small vessel | Sequence feature & intron | NM_000303.2 | |
| PSMC1 | Up | Cardioembolic, large artery atherosclerosis & others | Upstream & intron variants | NM_002802.2 | |
| RENBP | Down | NA | Missense variant | NM_002910.3 | |
| RNF103 | Up | Large artery atherosclerosis & small vessel | Upstream, downstream, missense, 5′ UTR & intron | NM_005667.3 NM_001198951.1 | |
| RNF185 | Up | NA | Missense & synonymous variants | NM_152267.3 NM_001135825.1 | |
| RPN1 | Up | Cardioembolic, large artery atherosclerosis, small vessel & others | Synonymous, 3′ UTR & intron variants | NM_002950.3 | |
| RPN2 | Up | NA | Missense & synonymous | NM_001324301.1 NM_0011355771.2 | |
| RPS27A | Up | Others | Intron variants | NM_001135592.2 NM_002954.5 | |
| SPTBN1 | Up & down | NA | Missense & synonymous variants | NM_003128.2 NM_178313.2 | |
| SRP54 | Up | Large artery atherosclerosis, small vessel & others | Upstream, downstream, 5′ UTR, sequence feature, 3′ UTR & intron variants | NM_003136.3 NM_001146282.1 | |
| ST6GALNAC2 | Up | Others | Intron & 5′ UTR variants | NM_006456.2 | |
| ST6GALNAC4 | Down | Cardioembolic, large artery atherosclerosis & others | Upstream & intron variants | NM_175039.3 NM_175040.3 | |
| ST6GALNAC5 Up | NA | Missense & synonymous | NM_030965.2 | ||
| ST6GALNAC6 | Down | Cardioembolic, large artery atherosclerosis, small vessel & others | Upstream, downstream & intron variants | NM_001286999.1 NM_001287001.1 NM_001287003.1 NM_001287000.1 NM_013443.4 NR_104629.1 | |
| ST8SIA2 | Up | Cardioembolic, large artery atherosclerosis, small vessel & others | Synonymous, splice region, 3′ UTR & intron variants | NM_006011.3 | |
| ST8SIA5 | Up | Large artery atherosclerosis, cardioembolic & small vessel | Sequence feature, downstream & intron variants | NM_001307986.1 NM_001307987.1 NM_013305.5 | |
| STT3A | Up | Cardioembolic, large artery atherosclerosis & others | Synonymous, sequence feature, downstream, 3′ UTR, splice region, intron, upstream gene variants | NM_001278503.1 NM_152713.4 NM_001278504.1 | |
| TRIM13 | Up | Cardioembolic, large artery atherosclerosis & small vessel | 5′ UTR, upstream, 3′ UTR, downstream & intron variants | NM_001007278.2 NM_005798.4 NM_052811.3 NM_213590.2 | |
| TUBA1B | Up | NA | Synonymous variant | NM_006082.2 | |
| TUBA4A | Up | Cardioembolic, large artery atherosclerosis, small vessel & others | Downstream, upstream & intron variants | NM_006000.2 NM_001278552.1 | |
| TUBB1 | Down | Others | Missense & synonymous | NM_030773.3 | |
| TUBB2B | Down | NA | |||
| TUBB4A | Up | Cardioembolic, large artery atherosclerosis, small vessel & others | Upstream, 3′ UTR & intron variants | NM_001289123.1 NM_001289130.1 NM_001289131.1 NM_006087.3 NM_001289127.1 NM_001289129.1 | |
| TUBB6 | Up | Others | Upstream, downstream & intron variants | NM_001303524.1 NM_001303526.1 NM_001303527.1 NM_001303528.1 NM_001303529.1 NM_001303530.1 NM_032525.2 | |
| s | ||||||
| Gene | regulation | Stroke subtype (phenotype) | Variants | Protein ID | ||
| A1BG | Up | NA | Missense & synonymous | NM_130786.3 | ||
| AAMP | Down | Small vessel | Intron variant | NM_001302545.1 | ||
| ABHD12 | Up & down | Cardioembolic, small vessel & large artery atherosclerosis | Downstream & intron variants | NM_015600.4 NM_001042472.2 | ||
| ANGPT2 | Down | NA | Missense, synonymous & splice region variants | NM_001147.2 NM_001118887.1 NM_001118888.1 | ||
| ANXA5 | Up | Cardioembolic, small vessel & others | Intron variant | NM_001154.3 | ||
| APBB1IP | Up | Cardioembolic, large artery atherosclerosis, small vessel & others | Intron variant | NM_019043.3 | ||
| ARRB2 | Up | Cardioembolic, large artery atherosclerosis & others | Upstream & downstream gene variants | NM_001257329.1 NM_001257330.1 NM_001257331.1 NM_001330064.1 NM_004313.3 NM_199004.1 | ||
| ATP1B3 | Up | Cardioembolic, large artery atherosclerosis & others | Intron variant | NM_001679.3 | ||
| BRPF3 | Up | Others | Sequence feature & intron | NM_015695.2 | ||
| CBX5 | Up & down | Cardioembolic, large artery atherosclerosis & others | Downstream, 3′ UTR, 5′ UTR, sequence feature and intron variants | NM_001127322.1 NM_012117.2 NM_001127321.1 | ||
| CD36 | Up | NA | Missense, synonymous & splice region variants | NM_000072.3 NM_001289911.1 NM_001289909.1 NM_001289908.1 | ||
| CD48 | Up | Cardioembolic, small vessel, large artery atherosclerosis & others | Upstream, synonymous & intron variants | NM_001256030.1 NM_001778.3 | ||
| CD58 | Up | Cardioembolic, large artery atherosclerosis & others | Downstream, 3′ UTR & intron variants | NM_001779.2 NM_001144822.1 NR_026665.1 | ||
| CD63 | Up | NA | Missense, synonymous & frameshift variants | NM_001257389.1 NM_001257401.1 NM_001257392.1 | ||
| CDC42 | Up | Large artery atherosclerosis & others | Downstream, upstream & intron variants | NM_001039802.1 NM_044472.2 NM_001791.3 | ||
| CEACAM3 Up | NA | Missense & synonymous variants | NM_00185.4 NM_001277163.2 | |||
| CEACAM6 Up | Cardioembolic & others | Sequence feature & intron | NM_002483.6 | |||
| CFD | Up | Cardioembolic, small vessel & others | Upstream & 3′ UTR variants | NM_001317335.1 NM_001928.3 | ||
| CFL1 | Up & down | NA | Synonymous variant | NM_005507.2 | ||
| DGKB | Up & down | NA | Synonymous, missense & splice region variants | NM_004080.2 | ||
| DGKE | Up | Small vessel | 3′ UTR variant | NM_003647.2 | ||
| DOCK1 | Up | NA | Synonymous, missense & splice region variants | NM_001290223.1 NM_001380.4 | ||
| DOCK2 | Up | NA | Synonymous, missense & splice region variants | NM_004946.2 | ||
| DOCK4 | Up | Cardioembolic, small vessel, large artery atherosclerosis & others | Intron variant | NM_014705.3 | ||
| DOK2 | Down | Cardioembolic & others | Missense, upstream & intron variants | NM_003974.3 NM_001317800.1 NM_201349.2 | ||
| EHD3 | Down | Cardioembolic, large artery atherosclerosis & others | 3′ UTR & intron variants | NM_014600.2 | ||
| ENDOD1 | Up | Cardioembolic, small vessel, large artery atherosclerosis & others | 3′ UTR & intron variants | NM_015036.2 | ||
| FCER1G | Up | NA | Missense variant | NM_004106.1 | ||
| FERMT3 | Up | Small vessel & others | Upstream, downstream, synonymous & intron variant | NM_178443.2 | ||
| FGR | Up | NA | Missense & synonymous | NM_001042729.1 | ||
| GNA13 | Up | Cardioembolic, small vessel & large artery atherosclerosis | Downstream, upstream & intron variants | NM_006572.5 NM_001282425.1 | ||
| GNAI3 | Up | Cardioembolic, small vessel, large artery atherosclerosis & others | Upstream, intron, 3′ UTR & 5′ UTR variants | NM_006496.3 | ||
| GNB5 | Down | Cardioembolic, small vessel, large artery atherosclerosis & others | Downstream, upstream & intron variants | NM_016194.3 | ||
| GNG10 | Up | Small vessel & others | Upstream, downstream, 5′ UTR & intron variant | NM_001017998.3 | ||
| GNG2 | Up | Cardioembolic, small vessel, large artery atherosclerosis & others | 3′ UTR, upstream & intron variants | NM_001243773.1 NM_001243774.1 NM_053064.4 | ||
| GNG4 | Up | Cardioembolic, large artery atherosclerosis & others | Intron variant | NM_001098721.1 NM_001098722.1 | ||
| GNG5 | Up | Small vessel, large artery atherosclerosis & others | 5′ UTR & intron variant | NM_005274.2 | ||
| GNGT2 | Up | Others | Upstream & intron variants | NM_031498.2 NM_001198755.1 NM_001198756.1 NM_001198754.1 | ||
| GRB2 | Up | Cardioembolic, small vessel, large artery atherosclerosis & others | Downstream, upstream, 5′ UTR, 3′ UTR & intron variants | NM_002086.4 | ||
| GTPBP2 | Up & down | Small vessel & others | Intron variant | NM_019096.4 | ||
| GUCY1A2 | Up | NA | Missense & synonymous variants | NM_000855.2 NM_001256424.1 | ||
| GYPB | Up | Large artery atherosclerosis & others | Downstream, upstream & intron variants | NM_002100.5 NM_001304382.1 | ||
| GYPC | Down | Large artery atherosclerosis & others | Downstream & intron variants | NM_002101.4 NM_001256584.1 | ||
| HBE1 | Up | NA | NA | NA | ||
| HRAS | Down | Cardioembolic, small vessel & others | Upstream, downstream & intron variants | NM_001130442.2 NM_001318054.1 NM_005343.3 | ||
| IRF2 | Up | Cardioembolic, small vessel, large artery atherosclerosis & others | Intron variant | NM_002199.3 | ||
| ITGA2 | Up | NA | Synonymous, missense & splice region variants | NM_002203.3 | ||
| ITGA2B | Up | Cardioembolic, small vessel & others | Splice region & intron variant | NM_000419.4 | ||
| ITGA5 | Up | NA | Missense & synonymous | NM_002205.4 | ||
| ITGAX | Up | Large artery atherosclerosis & others | Upstream & intron variants | NM_001286375.1 NM_000887.4 | ||
| ITPR1 | Down | NA | Missense & synonymous variants | NM_002222.5 NM_001099952.2 NM_001168272.1 | ||
| ITPR2 | Up & down | NA | Synonymous, missense, frameshift & splice region | NM_002223.3 | ||
| ITPR3 | Down | Cardioembolic, small vessel, large artery atherosclerosis & others | Splice region, synonymous, downstream & intron | NM_002224.3 | ||
| JMJD1C | Up | NA | Synonymous, missense & disruptive in-frame deletion. | NM_032776.2 NM_001282948.1 NM_001318153.1 NM_001322254.1 | ||
| KIF1C | Up & down | NA | Missense & synonymous variants | NM_006612.5 | ||
| KIF20A | Up | NA | Missense & synonymous | NM_005733.2 | ||
| KIF26A | Down | Cardioembolic, large artery atherosclerosis & others | 3′ UTR, missense, synonymous & intron variant | NM_015656.1 | ||
| KIF26B | Down | NA | Missense & synonymous | NM_018012.3 | ||
| KIF27 | Up | Cardioembolic, large artery atherosclerosis & others | Intron variants | NM_017576.2 NM_001271927.1 NM_001271928.1 | ||
| KIF2C | Up | Cardioembolic, small vessel, large artery atherosclerosis & others | Sequence feature, upstream & intron variants | NM_006845.3 NM_001297655.1 NM_001297656.1 NM_001297657.1 |
||
| KIF3A | Down | Cardioembolic, small vessel & large artery atherosclerosis | 3′ UTR, upstream, downstream & intron variant | NM_001300791.1 NM_001300792.1 NM_007054.6 | ||
| KIF3C | Up | Cardioembolic, large artery atherosclerosis & others | Synonymous & intron variants | NM_007054.6 NM_002254.6 | ||
| KIF5A | Up & down | Cardioembolic, small vessel & others | Intron variant | NM_004984.2 | ||
| KIF6 | Up | NA | Missense & synonymous variants | NM_145027.4 NM_001289024.1 NM_001289020.1 NM_001289021.1 | ||
| KIF9 | Up | Cardioembolic, small vessel & others | Downstream, sequence feature, upstream, synonymous & intron | NM_001134878.1 NM_022342.4 | ||
| KLC1 | Up | Cardioembolic, small vessel, large artery atherosclerosis & others | Intron, downstream & upstream gene variants | NM_001130107.1 NM_005552.4 NM_182923.3 | ||
| KRAS | Up | Cardioembolic, large artery atherosclerosis & others | 3′ UTR & intron variants | NM_033360.3 NM_004985.4 | ||
| LHFPL2 | Up | Cardioembolic, small vessel, large artery atherosclerosis & others | 5′ UTR & intron variant | NM_005779.2 | ||
| MAGED2 | Down | NA | NA | NA | ||
| MANF | Down | NA | Missense variant | NM_006010.5 | ||
| MGLL | NA | Missense & synonymous variants | NM_007283.6 NM_001256585.1 NM_001003794.2 | |||
| MMRN1 | Up | Cardioembolic, small vessel & others | Intron variant | NM_007351.2 | ||
| MPL | Up | Cardioembolic & others | Sequence feature & intron | NM_005373.2 | ||
| NOS1 | Up & down | NA | Missense & synonymous variants | NM_001204218.1 NM_001204213.1 NM_000620.4 NM_001164757.2 NM_014697.3 | ||
| NOS3 | Down | Cardioembolic, small vessel & others | Upstream, missense, synonymous, 3′ UTR, downstream & intron variant | NM_001160109.1 NM_001160110.1 NM_001160111.1 NM_000603.4 | ||
| NRAS | Down | Large artery atherosclerosis, small vessel & others | Intron variant | NM_002524.4 | ||
| ORAI2 | Up & down | Cardioembolic & others | 3′ UTR variant | NM_001126340.2 NM_001271818.1 NM_001271819.1 NM_032831.3 | ||
| P2RX5 | Down | Large artery atherosclerosis | Downstream, upstream & intron variants | NM_001204519.1 NM_001204520.1 NM_002561.3 | ||
| PCDH7 | Down | NA | Missense & synonymous variants | NM_032457.3 NM_032456.2 NM_001173523.1 | ||
| PDE10A | Up | NA | Missense & synonymous variants | NM_006661.3 NM_001130690.2 | ||
| PDE11A | Up | NA | Missense, synonymous, frameshift, conservative in-frame insertion variants | NM_016953.3 NM_001077197.1 NM_001077196.1 NM_001077358.1 | ||
| PDPN | Up | Cardioembolic, large artery atherosclerosis & others | Intron, upstream, downstream, sequence feature & 3′ UTR variants | NM_006474.4 NM_198389.2 NM_001006624.1 NM_001006625.1 | ||
| PECAM1 | Up | Others | Intron & 3′ UTR variant | NM_000442.4 | ||
| PHF21A | Up | Cardioembolic, large artery atherosclerosis & others | Downstream, 3′ UTR, upstream & intron variants | NM_001101802.1 NM_016621.3 | ||
| PIK3R1 | Up | Cardioembolic, large artery atherosclerosis & others | Upstream, 5′ UTR, 3′ UTR, downstream & intron | NM_181523.2 | ||
| PIK3R5 | Up | Cardioembolic, large artery atherosclerosis & others | Upstream, downstream & intron variants | NM_001142633.2 NM_001251851.1 NM_001251852.1 NM_001251853.1 NM_001251855.1 NM_014308.3 | ||
| PLA2G4A | Up | Cardioembolic, small vessel, large artery atherosclerosis & others | Intron variants | NM_024420.2 NM_001311193.1 | ||
| PLCG1 | Down | Cardioembolic, small vessel, large artery atherosclerosis & others | Upstream, downstream, synonymous & intron variant | NM_002660.2 NM_182811.1 | ||
| PLEK | Up | Cardioembolic, small vessel & others | Sequence feature, 3′ UTR & intron variants | NM_002664.2 | ||
| PPP2CB | Up | Small vessel & others | Intron & 5′ UTR variant | NM_001009552.1 | ||
| PPP2R5B | Down | NA | Missense & synonymous | NM_006244.3 | ||
| PPP2R5D | Up & down | Cardioembolic, small vessel, large artery atherosclerosis & others | Upstream, downstream & intron variants | NM_001270476.1 NM_180976.2 NM_006245.3 | ||
| PPP2R5E | Up | NA | Synonymous variant | NM_001282179.1 NM_001282181.1 | ||
| PRCP | Up | Cardioembolic, small vessel, large artery atherosclerosis & others | Downstream, upstream & intron variants | NM_199418.3 NM_005040.3 NM_001319214.1 | ||
| PRKCB | Up & down | NA | Synonymous & missense variants | NM_002738.6 NM_212535.2 | ||
| PSAP | Up | Cardioembolic, small vessel, large artery atherosclerosis & others | Downstream, 3′ UTR & intron variants | NM_001042465.2 NM_001042466.2 | ||
| PSG11 | Up | Cardioembolic, small vessel, large artery atherosclerosis & others | Upstream, downstream & intron variants | NM_002785.2 NM_001113410.1 NM_203287.1 | ||
| PTGIR | Up & down | Small vessel | 3′ UTR & intron variants | NM_000960.3 | ||
| RBSN | Up | Cardioembolic, large artery atherosclerosis & others | Downstream & intron variants | NM_001302378.1 NM_022340.3 NR_126155.1 | ||
| RHOA | Up & down | NA | Synonymous variant | NM_001313943.1 | ||
| RHOB | Up | NA | Synonymous & missense | NM_004040.3 | ||
| RHOG | Up | Cardioembolic, large atherosclerosis & others | Intron variant | NM_001665.3 | ||
| S100A10 | Up | Cardioembolic & large artery atherosclerosis | Intron variant | NM_002966.2 | ||
| SCCPDH | Up | Cardioembolic & large artery atherosclerosis | 3′ UTR & intron variant | NM_016002.2 | ||
| SELP | Up | Cardioembolic, large artery atherosclerosis & others | Intron variant | NM_003005.3 | ||
| SERPINA3 Up | Cardioembolic, large artery atherosclerosis & others | Missense, synonymous & intron variants | NM_001085.4 | |||
| SERPINB6 | Up | Small vessel & others | Downstream, 5′ UTR, upstream & intron variants | NM_001271823.1 NM_001271825.1 NM_001271822.1 NM_001195291.2 NM_001271824.1 NM_001297699.1 NM_001297700.1 NM_001271823.1 | ||
| SERPINB8 | Up | Cardioembolic, small vessel, large artery atherosclerosis & others | Upstream, downstream, 3′ UTR, missense & intron variants | NM_002640.3 NM_001031848.1 NM_001276490.1 | ||
| SH2B1 | Down | Others | Intron variants | NM_001145795.1 NM_001308293.1 NM_001145796.1 NM_001145797.1 NM_001308294.1 NM_001145812.1 | ||
| SH2B3 | Down | Cardioembolic, small vessel, large artery atherosclerosis & others | Intron, upstream, missense, 3′ UTR & downstream variants | NM_005475.2 NM_001291424.1 | ||
| SRI | Up | Cardioembolic & large artery atherosclerosis | Intron, upstream & downstream gene variants | NM_001256892.1 NM_001256891.1 NM_003130.3 NM_198901.1 | ||
| STX4 | Up | Cardioembolic, small vessel & large artery atherosclerosis | Upstream, 5′ UTR, downstream & intron variants | NM_004604.4 NM_001272096.1 NM_001272095.1 | ||
| STXBP2 | Up | Large artery atherosclerosis, small vessel & others | Sequence feature, downstream, upstream & intron variants | NM_001272034.1 NM_001127396.2 NM_006949.3 | ||
| TFPI | Up | NA | Missense variant | NM_001032281.3 NM_001329239.1 | ||
| TLN1 | Up | Cardioembolic | Intron & 3′ UTR variant | NM_006289.3 | ||
| TNFRSF10D | Up & down | Cardioembolic, large artery atherosclerosis & others | 3′ UTR & intron | NM_003840.4 | ||
| TOR4A | Up | Small vessel | Upstream variant | NM_017723.2 | ||
| TREM1 | Up | Others | Downstream, upstream & intron variant | NM_001242589.2 NM_018643.4 NR_136332.1 NM_001242590.2 | ||
| TRPC3 | Up | Large artery atherosclerosis & others | Downstream & intron variants | NM_001130698.1 NM_003305.2 | ||
| VTI1B | Up | Cardioembolic | Sequence feature & intron | NM_006370.2 | ||
| WDR1 | Up & down | Cardioembolic, small vessel, large artery atherosclerosis & others | Sequence feature, 5′ UTR, upstream, downstream & intron variants | NM_017491.3 NM_005112.4 | ||
| YWHAZ | Up | Cardioembolic & others | Downstream & intron variants | NM_001135699.1 NM_001135702.1 NM_001135700.1 NM_001135701.1 NM_145690.2 | ||
- ➢
- Downstream and Upstream gene variants
- ➢
- 3′ UTR and 5′ UTR variants
- ➢
- Sequence feature variants
- ➢
- Intron variants
- ➢
- Synonymous variant
- ➢
- Missense variants
- ➢
- Frameshift variants
- ➢
- Splice region variants
- ➢
- Conservative in-frame insertions & Disruptive in-frame deletion variants
4.1. Discussion
5. Conclusions
6. Scope of Future Work
| S.NO | TITLE | PAGE NO |
| 1 | Serpina1 variants due to substitution mutations | 9 |
| 2 | Alpha-1 Antitrypsin sequence in FASTA format | 12 |
| 3 | Three-dimensional structure of the AAT protein | 12 |
| 4 | Basic information retrieved about serpina1 gene from GenBank | 13 |
| 5 | Graphical statistics data of serpina1 gene expression in bulk tissue | 13 |
| 6 | Serpina1 gene expression in single tissues | 14 |
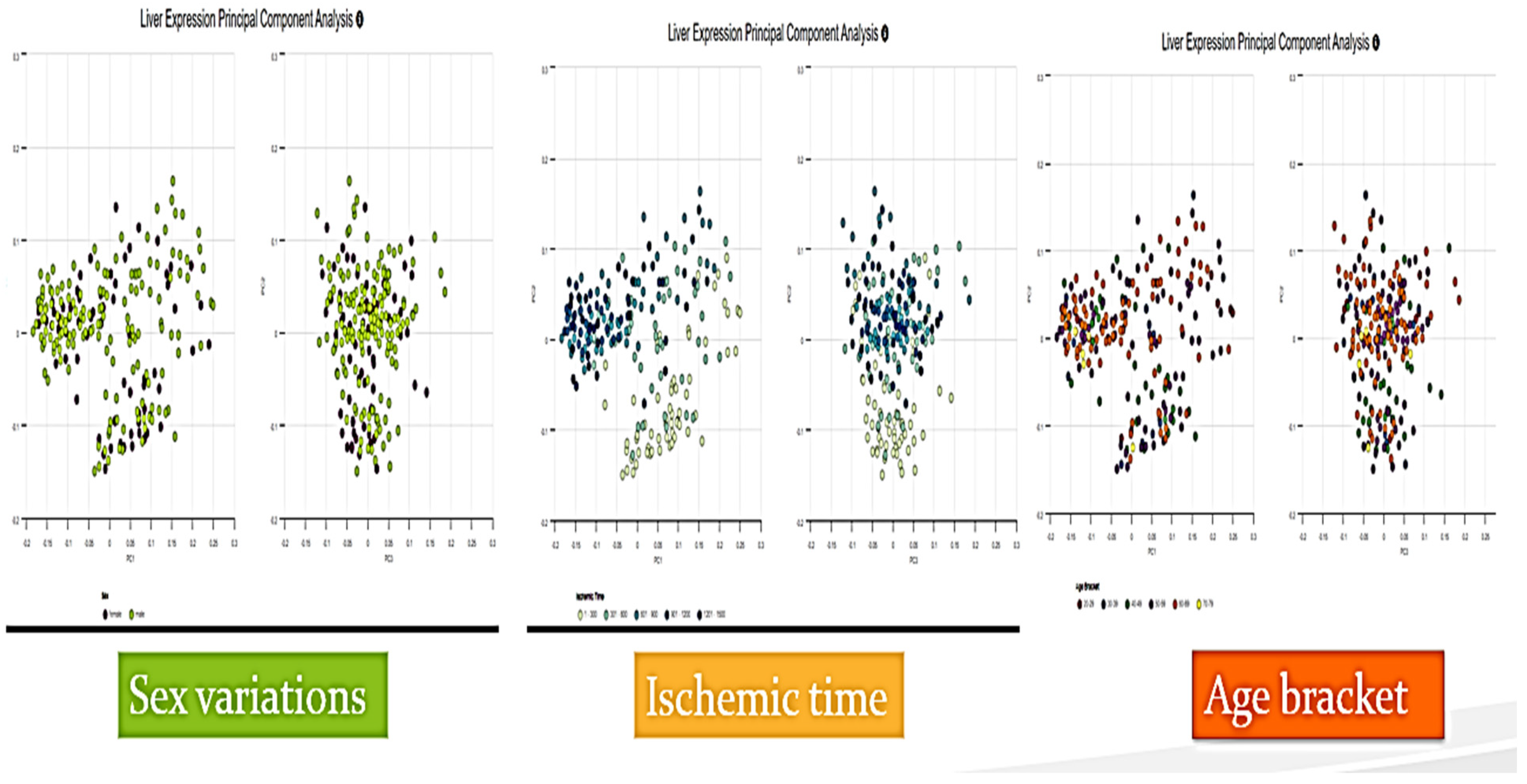
| 7 | Various Serpina1 gene expression in principal component analysis (PCA) | 14 |
| S.NO | TITLE | PAGE NO |
| Genes in various pathways associated with ischemic stroke | ||
| 1 | Lung Fibrosis pathway | 15–16 |
| 2 | Embryonic and induced pluripotent stem cells and lineage markers pathway | 17–18 |
| 3 | P73 transcription factor network pathway | 18–20 |
| 4 | Nuclear receptors meta pathway | 20–24 |
| 5 | FOXA1 transcription factor network pathway | 24–25 |
| 6 | Disease of hemostasis pathway | 25–26 |
| 7 | COPII-mediated vesicle transport pathway | 26–27 |
| 8 | Regulation of insulin-like growth factor (IGF) transport and uptake by insulin-like growth factor binding proteins (IGFBPS) pathway | 27–29 |
| 9 | Transport to the golgi and subsequent modifications pathway | 29–32 |
| 10 | Response to elevated platelet cytosolic Ca2+ pathway | 33–40 |
References
- Cleveland Clinic medical. “Ischemic Stroke (Clots)” ClevelandClinic, 22 Sept. 2022, https://my.clevelandclinic.org/health/diseases/24208-ischemic-stroke-clots.Accessed on 08 Feb. 2024.
- DeLong, Jonathan Howard et al. “Inflammatory Responses After Ischemic Stroke.” Seminars in immunopathology vol. 44,5 (2022): 625-648. [CrossRef]
- Ekkert A, et. al. Ischemic Stroke Genetics: What Is New and How to Apply It in Clinical Practice? Genes (Basel). 2021 Dec 24;13(1):48. [CrossRef] [PubMed]
- Ferrarotti, Ilaria et al. “Rare variants in alpha 1 antitrypsin deficiency: a systematic literature review.” Orphanet journal of rare diseases vol. 19,1 82. 22 Feb. 2024. [CrossRef]
- García-Ramírez, Idoia et al. “Lmo2 expression defines tumor cell identity during T-cell leukemogenesis.” The EMBO journal vol. 37,14 (2018): e98783. [CrossRef]
- Gissen, Paul, and Eamonn R Maher. “Cargos and genes: insights into vesicular transport from inherited human disease.” Journal of medical genetics vol. 44,9 (2007): 545-55. [CrossRef]
- Hart, R G, and M C Kanter. “Hematologic disorders and ischemic stroke. A selective review.” Stroke vol. 21,8 (1990): 1111-21. [CrossRef]
- He, Yi-Fu et al. “HSF1 Alleviates Brain Injury by Inhibiting NLRP3-Induced Pyroptosis in a Sepsis Model.” Mediators of inflammation vol. 2023 2252255. 27 Jan. 2023. [CrossRef]
- Huaux, Francois et al. “Role of Eotaxin-1 (CCL11) and CC chemokine receptor 3 (CCR3) in bleomycin-induced lung injury and fibrosis.” The American journal of pathology vol. 167,6 (2005): 1485-96. [CrossRef]
- Jiyoon Kim, Heon Yung Gee, Min Goo Lee; Unconventional protein secretion—new insights into the pathogenesis and therapeutic targets of human diseases. J Cell Sci 15 June 2018; 131 (12): jcs213686. [CrossRef]
- Ju, Yunfeng et al. “Nuclear receptor 5A (NR5A) family regulates 5-aminolevulinic acid synthase 1 (ALAS1) gene expression in steroidogenic cells.” Endocrinology vol. 153,11 (2012): 5522-34. [CrossRef]
- Khomtchouk et. al., HeartBioPortal 2019, Circulation: Genomic and Precision Medicine e002426124. https://www.ahajournals.org/doi/abs/10.1161/CIRCGEN.118.002426. G. [CrossRef]
- Lacy, P. Mechanisms of Degranulation in Neutrophils. All Asth Clin Immun 2, 98 (2006). [CrossRef]
- Lau, Eric, and Ze’ev A Ronai. “ATF2—at the crossroad of nuclear and cytosolic functions.” Journal of cell science vol. 125, Pt 12 (2012): 2815-24. [CrossRef]
- Lewitt, Moira S, and Gary W Boyd. “The Role of Insulin-Like Growth Factors and Insulin-Like Growth Factor-Binding Proteins in the Nervous System.” Biochemistry insights vol. 12 1178626419842176. 17 Apr. 2019. [CrossRef]
- Lexun Wang et. al., “CCAAT/Enhancer-Binding Proteins in Fibrosis.” Research. 2022;2022. 2022;2022. [CrossRef]
- Luo, Fei et al. “Estrogen lowers triglyceride via regulating hepatic APOA5 expression.” Lipids in health and disease vol. 16,1 72. 4 Apr. 2017. [CrossRef]
- Malik, Rainer et al. “Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes.” Nature genetics vol. 50,4 (2018): 524-537. [CrossRef]
- MedlinePlus [Internet]. Bethesda (MD): National Library of Medicine (US); SERPINA1 gene; [updated Sept 15, 2021; cited 2024 May 3]; Available from: https://medlineplus.gov/genetics/gene/serpina1/#conditions.
- Miller, Richard J et al. “CXCR4 signaling in the regulation of stem cell migration and development.” Journal of neuroimmunology vol. 198,1-2 (2008): 31-8. [CrossRef]
- Okuma, Toshiyuki et al. “C-C chemokine receptor 2 (CCR2) deficiency improves bleomycin-induced pulmonary fibrosis by attenuation of both macrophage infiltration and production of macrophage-derived matrix metalloproteinases.” The Journal of pathology vol. 204,5 (2004): 594-604. [CrossRef]
- Robert, J. (2015). Nuclear Receptor Pathways. In: Textbook of Cell Signaling in Cancer. Springer, Cham. [CrossRef]
- Seixas, S., & Marques, P. I. (2021). Known Mutations at the Cause of Alpha-1 Antitrypsin Deficiency an Updated Overview of SERPINA1 Variation Spectrum. The Application of Clinical Genetics, 14, 173–194. [CrossRef]
- Tsai, Nai-Wen et al. “Dysregulation of Ca2+ movement in platelets from patients with acute ischemic stroke.” Clinical and experimental pharmacology & physiology vol. 36,4 (2009): 380-5. [CrossRef]
- Tucker NR, Middleton RC, Le QP, Shelden EA (2011) HSF1 Is Essential for the Resistance of Zebrafish Eye and Brain Tissues to Hypoxia/Reperfusion Injury. PLoS ONE 6(7): e22268. [CrossRef]
- Visvader, J E et al. “The LIM domain gene LMO4 inhibits differentiation of mammary epithelial cells in vitro and is overexpressed in breast cancer.” Proceedings of the National Academy of Sciences of the United States of America vol. 98,25 (2001): 14452-7. [CrossRef]
- Yu, Dan-Dan et al. “A review on hepatocyte nuclear factor-1beta and tumor.” Cell & bioscience vol. 5 58. 13 Oct. 2015. [CrossRef]
- Yue, Xinping et al. “TGF-β: Titan of Lung Fibrogenesis.” Current enzyme inhibition vol. 6,2 (2010): 10.2174/10067.
- Zhang, Ka et al. “Genetics in Ischemic Stroke: Current Perspectives and Future Directions.” Journal of cardiovascular development and disease vol. 10,12 495. 13 Dec. 2023. [CrossRef]
- Zhou, Ya et al. “Overexpression of GATA2 Enhances Development and Maintenance of Human Embryonic Stem Cell-Derived Hematopoietic Stem Cell-like Progenitors.” Stem cell reports vol. 13,1 (2019): 31-47. [CrossRef]
- The UniProt Consortium, UniProt: the Universal Protein Knowledgebase in 2023, Nucleic Acids Research, Volume 51, Issue D1, 6 January 2023, Pages D523–D531. [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




