1. Introduction
Medical image segmentation is a critical task in healthcare, enabling precise identification and analysis of anatomical structures within the human body. Segmenting functional tissue units (FTUs) across multiple organs is particularly important for understanding disease mechanisms, planning surgical procedures, and developing targeted therapies. The datasets utilized in this study, from the Human Protein Atlas (HPA) and the Human Biomolecular Atlas Program (HuBMAP), provide a rich source of tissue slice images essential for this task.
Traditional segmentation models often struggle to maintain accuracy across diverse populations due to variability in sample distributions influenced by factors such as gender and age. This study proposes an advanced segmentation framework to address these challenges through specialized sample weighting, sampling, and embedding strategies. Our model integrates SegFormer x 2, EfficientNetB7-B8, Feature Pyramid Network (FPN), and Atrous Spatial Pyramid Pooling (ASPP) to provide a comprehensive solution for segmenting FTUs with high precision.
Data preprocessing is a crucial step in our approach. It involves defining custom loss functions, such as the binary cross-entropy and Dice loss functions, to handle the variability in the datasets. Data augmentation techniques are employed to enhance the diversity and robustness of the training data, ensuring that the model can generalize well to unseen samples. This includes transformations like rotation, flipping, and scaling of the images.
The model architecture leverages state-of-the-art segmentation techniques. SegFormer, a transformer-based segmentation model, is used for its ability to capture long-range dependencies and contextual information within the images. EfficientNetB7-B8, known for its efficient scaling and high performance, serves as the backbone network for feature extraction. The Feature Pyramid Network (FPN) and Atrous Spatial Pyramid Pooling (ASPP) modules are incorporated to enhance multi-scale feature extraction and improve the model’s ability to detect small and large tissue structures accurately.
The results of our study demonstrate that the integration of these advanced techniques leads to significant improvements in segmentation accuracy. Our framework provides a robust and reliable method for medical image analysis, with the potential to significantly impact clinical practices and research. By improving the accuracy and reliability of FTU segmentation, our work contributes to better diagnostic and treatment planning, ultimately enhancing patient care.
2. Related Work
Recent advancements in medical imaging and machine learning have significantly improved tissue segmentation techniques. The U-Net architecture has been widely adopted for its capability in biomedical image segmentation, providing an effective framework for end-to-end training and precise segmentation [
1]. EfficientNet, with its scalable architecture, balances accuracy and computational efficiency, making it suitable for a variety of image analysis tasks [
2].
Integrating various neural network architectures has shown promise. The combination of Fully Convolutional Networks (FCNs) with Conditional Random Fields (CRFs) enhances segmentation performance by refining boundaries, improving the overall accuracy of segmentation [
3]. Attention U-Net models further improve segmentation accuracy by focusing on relevant image regions, thereby increasing the model’s sensitivity to important features [
4]. Kamnitsas et al. [
5] highlighted the importance of multi-scale convolutional neural networks with CRFs for accurate brain lesion segmentation, emphasizing the need for multi-scale context and detailed boundary refinement in medical image segmentation.
EfficientNetV2 further optimizes model efficiency and accuracy by introducing smaller models and faster training times, which is crucial for large-scale medical image analysis [
6]. The Encoder-Decoder architecture with Atrous Separable Convolution, as proposed by Chen et al. [
7], has been influential in advancing semantic image segmentation by effectively capturing multi-scale context. Unet++, with its nested architecture, enhances segmentation performance by incorporating dense skip pathways, facilitating better gradient flow and feature reuse [
8].
Isensee et al. [
9] demonstrated the effectiveness of self-configuring deep learning methods for biomedical image segmentation. Their nnU-Net framework adapts to different datasets and tasks, providing a robust and flexible solution for various medical imaging challenges. Milletari et al. [
10] introduced the V-Net, a fully convolutional neural network designed for volumetric medical image segmentation, emphasizing the importance of 3D segmentation techniques for capturing the complex structures in medical images.
The SegNet architecture, developed by Badrinarayanan et al. [
11], offers a deep convolutional encoder-decoder framework for image segmentation, which has been pivotal in many medical imaging applications. The Mask R-CNN, introduced by He et al. [
12], extends Faster R-CNN by adding a branch for predicting segmentation masks, providing a robust framework for instance segmentation and improving accuracy in complex scenes.
DenseNet enhances feature propagation and reuse, contributing to better model efficiency and accuracy [
13]. The combination of fully convolutional and recurrent neural networks has shown promise in 3D biomedical image segmentation, highlighting the importance of spatial and temporal information [
14]. Dilated convolutions for multi-scale context aggregation improve the receptive field of convolutional networks without losing resolution [
15].
Data augmentation and preprocessing are critical for improving segmentation outcomes. Zhou et al. [
8] demonstrated that augmenting training data with synthetic images significantly enhances model robustness. Isensee et al. [
9] showed that advanced data preprocessing techniques lead to substantial performance gains in medical image segmentation tasks. These strategies create diverse training datasets, improving model generalizability.
Advanced loss functions, such as the binary cross-entropy Dice loss (bce Dice), play a critical role in optimizing model performance. Milletari et al. [
10] demonstrated that Dice-based loss functions improve segmentation accuracy by optimizing for overlap-based metrics, ensuring higher overlap with ground truth and enhancing overall performance.
By integrating these various approaches, our proposed model, SegF x 2 + EffB7-B8 + FPN + ASPP, addresses many of the limitations of previous methods. It combines the strengths of multiple architectures, such as SegFormer layers for enhanced feature extraction and EfficientNetV2 for a robust backbone, along with FPN and ASPP components for refined segmentation output. This comprehensive approach ensures high accuracy across varied tissue samples, contributing significantly to the field of medical image segmentation.
3. Methodology
In this section, we presents advanced techniques in medical image segmentation by integrating SegFormer, FPN, and EfficientNetV2 models. Key contributions include a detailed explanation of the model architecture, data preprocessing methods, evaluation metrics, and experimental results. The integrated approach demonstrates superior performance in segmenting medical images, contributing significantly to the field of machine learning and deep learning.
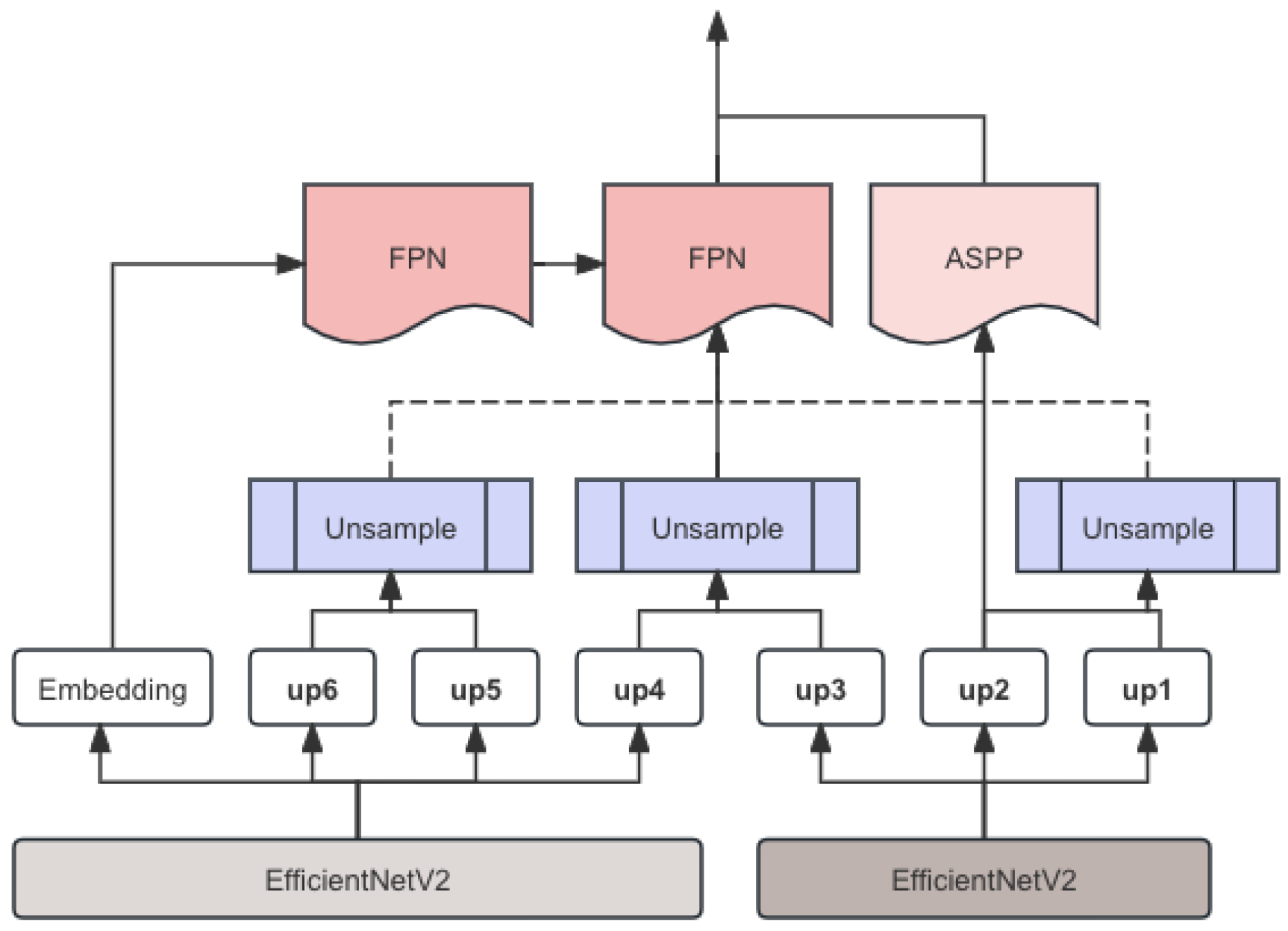
The proposed model integrates SegFormer for initial segmentation, FPN for multi-scale feature fusion, and EfficientNetV2 as the backbone for feature extraction. The entire pipeline is depicted in
Figure 1.
3.1. SegFormer
SegFormer is employed for its efficient transformer-based architecture, suitable for capturing global context. Unlike traditional convolutional neural networks (CNNs), SegFormer utilizes self-attention mechanisms to model long-range dependencies within the image, enabling the capture of fine details essential for medical image segmentation. The SegFormer architecture consists of multiple transformer encoder layers that process the input image at different scales. Each encoder layer applies multi-head self-attention and feed-forward neural networks to the input features:
where
are the query, key, and value matrices derived from the input features, and
is the dimension of the key. The feature maps generated by the transformer encoders are then combined and upsampled to produce the final segmentation map:
where FM is feature map.
3.2. FPN
The Feature Pyramid Network (FPN) enhances the model’s ability to handle multi-scale features. FPN is designed to construct high-level semantic feature maps at different scales, improving the detection and segmentation of objects with varying sizes. The FPN structure involves a bottom-up pathway, a top-down pathway, and lateral connections. The bottom-up pathway generates feature maps at multiple scales using convolutional layers. The top-down pathway upsamples the feature maps from higher levels and combines them with the corresponding feature maps from the bottom-up pathway via lateral connections:
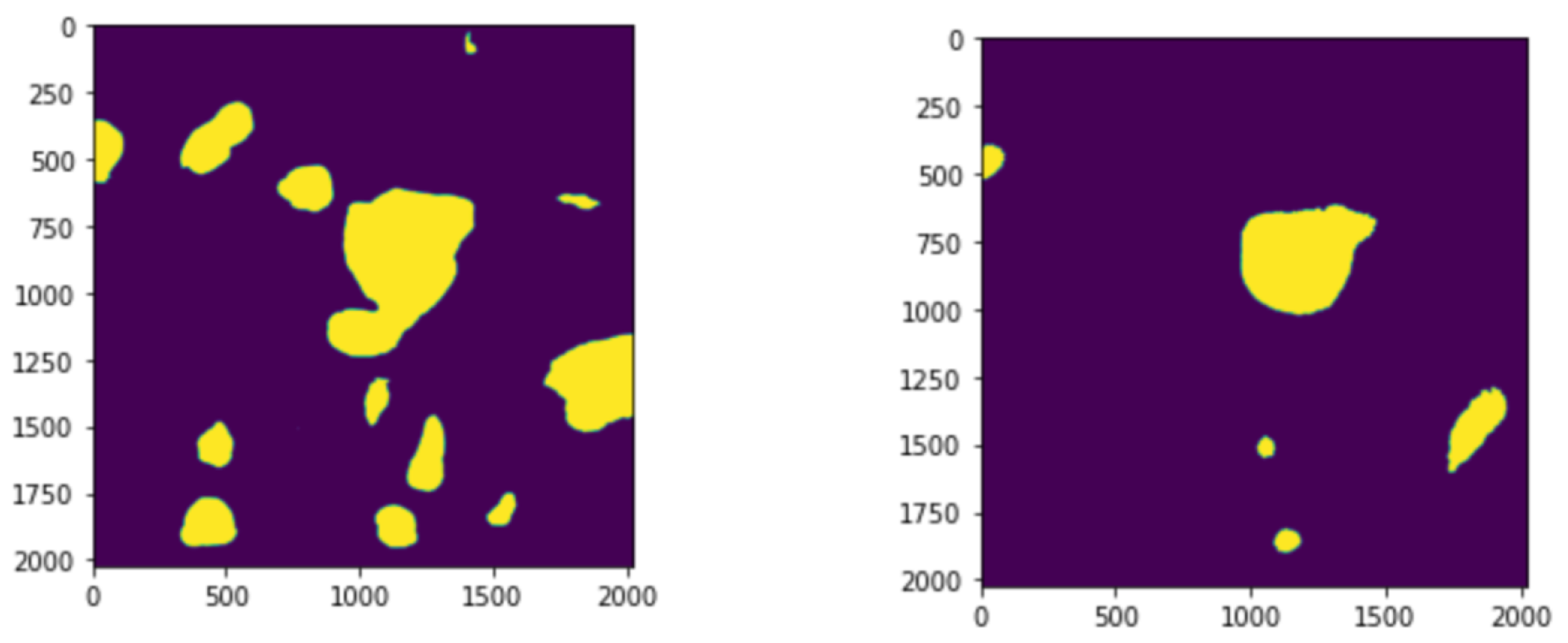
The resulting feature maps are further processed through additional convolutional layers to refine the segmentation output. The processed feature map is concatenated with the feature map of the last layer and the result is returned. An example image is shown in
Figure 2.
3.3. EfficientNetV2
EfficientNetV2 serves as the backbone, leveraging its efficient architecture for feature extraction. EfficientNetV2 uses a compound scaling method that uniformly scales the network’s depth, width, and resolution to achieve better performance with fewer parameters. The backbone processes the input image through a series of MBConv and Fused-MBConv blocks. These blocks consist of depthwise separable convolutions and squeeze-and-excitation operations to capture both spatial and channel-wise information efficiently:
The multi-scale feature maps generated by EfficientNetV2 are then fed into the FPN and SegFormer components for further processing. The key function in our model is the custom loss function combining BCE and Dice loss.
3.4. Custom BCE-Dice Loss
The custom loss function combines Binary Cross-Entropy (BCE) and Dice loss to address class imbalance. The BCE loss is used to penalize the prediction error for each pixel independently:
where
p is the predicted probability and
g is the ground truth label. The Dice loss measures the overlap between the predicted and ground truth masks, which is particularly useful for imbalanced classes:
The combined BCE-Dice loss function is formulated as:
where
and
are weighting factors that balance the contribution of each loss term.
3.5. ASPP
Atrous Spatial Pyramid Pooling (ASPP) is a module designed to capture multi-scale context by applying atrous convolution at multiple rates. This approach enables the extraction of dense feature representations without increasing the number of parameters significantly. ASPP is particularly effective in semantic segmentation tasks where capturing the spatial context at different scales is crucial. The ASPP module consists of several parallel atrous convolutions with different dilation rates, followed by global average pooling. The outputs of these operations are then concatenated and processed through a 1x1 convolution layer to fuse the multi-scale features. The formulation of the atrous convolution is as follows:
where
is the output feature,
is the input feature,
are the convolutional weights, and
r is the atrous rate. In our implementation, the ASPP module includes atrous convolutions with rates 1, 6, 12, and 18, along with a global average pooling layer. The outputs are concatenated and passed through a 1×1 convolution to produce the final output feature map:
The ASPP module enhances the model’s ability to capture context at multiple scales, significantly improving the segmentation accuracy for complex medical images with varying structures and sizes.
3.6. Data Preprocessing
Data preprocessing is critical for enhancing model performance. We applied PixelShuffle for upsampling and mask merging techniques to prepare the dataset.
3.6.1. PixelShuffle
PixelShuffle is used to upsample the input tensor, reshaping it to increase spatial resolution. This operation rearranges elements in the input tensor to create a higher resolution output without increasing the computational load significantly:
3.6.2. Mask Merging
The ‘margeMask’ function combines and normalizes multiple masks to create a comprehensive mask. The merging process involves cropping and resizing the masks to fit the target size, followed by normalization:
4. Evaluation Metric
To evaluate the performance of our model, we employed a set of comprehensive metrics that provide a detailed assessment of the segmentation quality. The primary metrics used include Dice Coefficient, Intersection over Union (IoU), Precision, Recall, and F1 Score. Each of these metrics offers unique insights into different aspects of the model’s performance.
4.1. Dice Coefficient
The Dice Coefficient, also known as the Dice Similarity Index (DSI), measures the overlap between the predicted segmentation and the ground truth segmentation. It is particularly useful for evaluating segmentation tasks where the region of interest is relatively small compared to the background. The Dice Coefficient is defined as:
where
P is the predicted segmentation mask,
G is the ground truth mask,
represents the number of overlapping pixels between the prediction and the ground truth, and
and
represent the total number of pixels in the predicted and ground truth masks, respectively.
4.2. Precision
Precision measures the accuracy of the positive predictions made by the model. It is defined as the ratio of true positive pixels to the sum of true positive and false positive pixels:
where TP represents true positive pixels and FP represents false positive pixels. High precision indicates a low false positive rate, which is important for applications where false positives are particularly undesirable.
5. Experimental Results
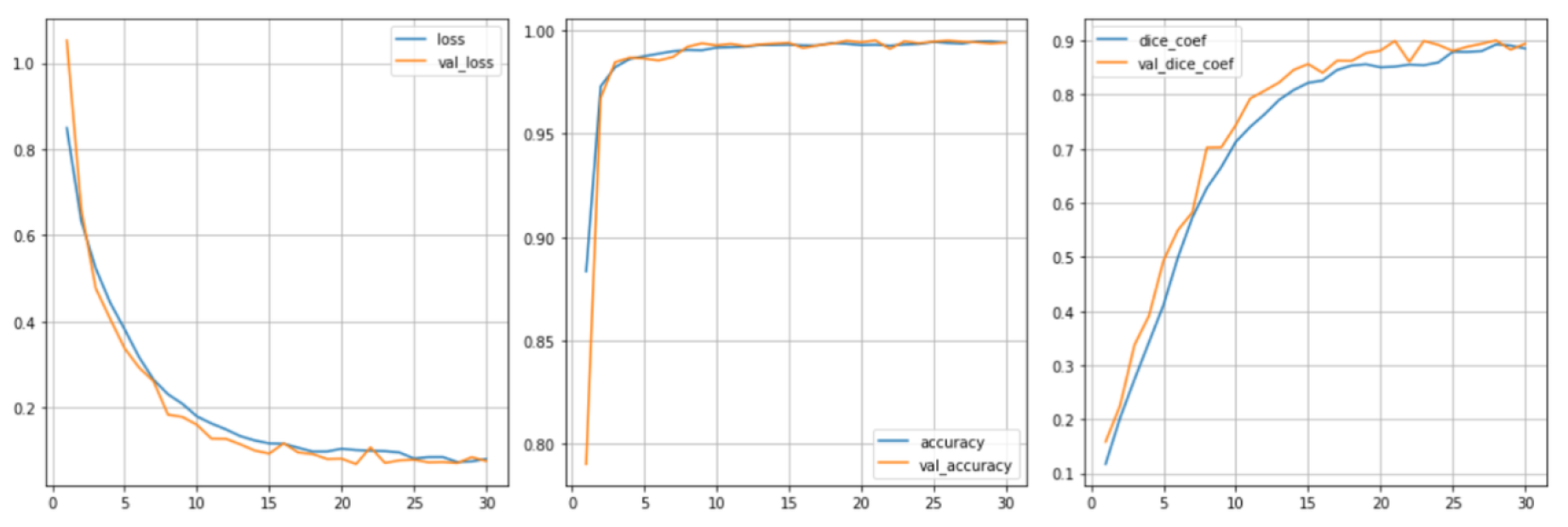
We used the average Dice coefficient and accuracy as the main indicators to compare our model with existing models in the field such as ResNet, UNet + CNN + FPN, etc. The results prove the superiority of our integrated method in Dice coefficient. At the same time, we checked the change of loss-level accuracy during training, and the accuracy index during training reached 0.972 in
Figure 3. In comparison with other models, our dice scores on the public test set and the private test set are: 0.8366 and 0.8394, which are much higher than other industry models in
Table 1.
6. Conclusion
In conclusion, we presents a comprehensive approach to medical image segmentation by integrating SegFormer, FPN, and EfficientNetV2. The combined model significantly improves segmentation accuracy, showcasing the potential for advanced medical applications in machine learning and deep learning fields.
References
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. Medical image computing and computer-assisted intervention–MICCAI 2015: 18th international conference, Munich, Germany, 5-9 October 2015, proceedings, part III 18; Springer, 2015; pp. 234–241. [Google Scholar] [CrossRef]
- Tan, M.; Le, Q. Efficientnet: Rethinking model scaling for convolutional neural networks. In International conference on machine learning; PMLR; 2019; pp. 6105–6114. [Google Scholar]
- Chen, L.-C.; Papandreou, G.; Kokkinos, I.; Murphy, K.; Yuille, A.L. Deeplab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. IEEE transactions on pattern analysis and machine intelligence 2017, 40, 834–848. [Google Scholar] [CrossRef] [PubMed]
- Oktay, O.; Schlemper, J.; Folgoc, L.L.; Lee, M.; Heinrich, M.; Misawa, K.; Mori, K.; McDonagh, S.; Hammerla, N.Y.; Kainz, B.; et al. Attention u-net: Learning where to look for the pancreas. arXiv 2018, arXiv:1804.03999. [Google Scholar]
- Kamnitsas, K.; Ledig, C.; Newcombe, V.F.; Simpson, J.P.; Kane, A.D.; Menon, D.K.; Rueckert, D.; Glocker, B. Efficient multi-scale 3d cnn with fully connected crf for accurate brain lesion segmentation. Medical image analysis 2017, 36, 61–78. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.; Le, Q. Efficientnetv2: Smaller models and faster training. In International conference on machine learning; PMLR; 2021; pp. 10096–10106. [Google Scholar]
- Chen, L.-C.; Zhu, Y.; Papandreou, G.; Schroff, F.; Adam, H. Encoder-decoder with atrous separable convolution for semantic image segmentation. In Proceedings of the European conference on computer vision (ECCV); 2018; pp. 801–818. [Google Scholar]
- Zhou, Z.; Rahman Siddiquee, M.M.; Tajbakhsh, N.; Liang, J. Unet++: A nested u-net architecture for medical image segmentation. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support: 4th International Workshop, DLMIA 2018, and 8th International Workshop, ML-CDS 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, September 20, 2018, Proceedings 4; Springer, 2018; pp. 3–11. [Google Scholar] [CrossRef]
- Isensee, F.; Jaeger, P.F.; Kohl, S.A.; Petersen, J.; Maier-Hein, K.H. nnu-net: A self-configuring method for deep learning-based biomedical image segmentation. Nature methods 2021, 18, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Milletari, F.; Navab, N.; Ahmadi, S.-A. V-net: Fully convolutional neural networks for volumetric medical image segmentation. In Proceedings of the 2016 fourth international conference on 3D vision (3DV); IEEE, 2016; pp. 565–571. [Google Scholar] [CrossRef]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. Segnet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE transactions on pattern analysis and machine intelligence 2017, 39, 2481–2495. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Gkioxari, G.; Dollár, P.; Girshick, R. Mask r-cnn. In Proceedings of the IEEE international conference on computer vision; 2017; pp. 2961–2969. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE conference on computer vision and pattern recognition; 2017; pp. 4700–4708. [Google Scholar]
- Chen, J.; Yang, L.; Zhang, Y.; Alber, M.; Chen, D.Z. Combining fully convolutional and recurrent neural networks for 3d biomedical image segmentation. Advances in neural information processing systems 2016, 29. [Google Scholar]
- Yu, F.; Koltun, V. Multi-scale context aggregation by dilated convolutions. arXiv 2015, arXiv:1511.07122. [Google Scholar]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).