1. Introduction
Dengue fever is an acute infectious disease caused by dengue virus (DENV). In recent decades, the incidence of dengue fever in the world has increased significantly. According to the World Health Organization (WHO), more than 40% of the world's population (approximately 2.5 billion) is at risk of infection with dengue fever and severe dengue fever (dengue haemorrhagic fever and dengue shock syndrome) [
1]. More than 100 million people are infected each year, approximately 1 to 5 million of whom develop dengue haemorrhagic fever or dengue shock syndrome, causing approximately 25,000 deaths [
2]. Dengue fever is widely prevalent in more than 100 countries in Africa, the Americas, the Eastern Mediterranean, Southeast Asia, and the Western Pacific[
3]. It is currently the most widely distributed, most prevalent and harmful arbovirus disease. DENV belongs to the Flaviviridae Flavivirus genus. The genome of DENV is a single-stranded positive-strand RNA with a total length of approximately 11,000 bp, encoding three structural proteins, C (capsid), prM/M (precursor of membrane), and E (envelope), and seven nonstructural proteins, including NS1, NS2, NS3, NS4A, NS4B and NS5 [
4]. It can be divided into four serotypes (DENV-1, DENV-2, DENV-3, and DENV-4), and each serotype is further subdivided into multiple genotypes according to nucleotide sequence differences[
5,
6].
In China, dengue fever was first discovered in Guangdong Province in 1978. Since then, dengue fever outbreaks have been reported in Hainan, Guangxi, Fujian, Zhejiang and Yunnan Provinces[
7]. However, the coastal city of Jiaxing, which is located in the Yangtze River Delta region, has also experienced cases of imported dengue fever due to the high level of international travel and trade. The first documented case of concurrent human infection with two dengue serotypes was reported in 1982 from Puerto Rico[
8]. It is well established that sequential infections with different dengue serotypes can increase the severity of dengue symptoms[
9]. In addition, cases of coinfection with more than one dengue serotype have been reported in many places, including Thailand and Taiwan[
10,
11,
12,
13]. However, the relationship between coinfection with multiple dengue serotypes and disease severity remains unclear. Although multiple dengue serotypes have been circulating simultaneously since the 1960s, coinfection with two or more dengue serotypes in the same patient has not been previously reported in Jiaxing.
On September 14, 2023, a case of mixed infection with DENV-1 and DENV-2 imported from Xishuangbanna, Yunnan, was discovered in Jiaxing city, Zhejiang Province. An epidemiological investigation was conducted on the patient, and serum DENV antigens, antibodies and nucleic acids were tested. The DENV whole-genome analysis was completed in April 2024, and the pathogen was further verified by tracing the source.
2. Materials and Methods
2.1. Ethical Statement
The Institutional Ethical Committee of the Jiaxing Center for Disease Control and Prevention reviewed and approved this human-participant study. According to national legislation and institutional requirements, no written informed consent was required for participation in this study.
2.2. Sample Collection
Qian Moumou, male, 27 years old, Jiaxing, Zhejiang, a labourer, had been working in Xishuangbanna, Yunnan Province, from June to September 13, 2023. He developed discomfort on September 10 and went to the First People's Hospital of Jinghong City, Xishuangbanna, Yunnan Province. The local doctor considered dengue fever, and the dengue fever NS1 antigen was positive. On the evening of September 13, he returned to Tongxiang. At 14:00 on September 14, the patient needed to visit the fever clinic of Tongxiang First People's Hospital for reexamination. Patients with dengue fever were admitted to the Department of Infectious Diseases of Tongxiang First People's Hospital for mosquito isolation treatment.
2.3. Colloidal Gold Method
Serum samples were tested for dengue NS1 antigen and dengue immunoglobulin M (IgM) and G (IgG) antibodies using the DENG NS1 Ag- IgG/IgM/ Rapid Test Kit (Guangzhou Wanfu, China; Catalog No. W11308002) according to the manufacturer's instructions.
2.4. Extraction of Viral RNA
Viral RNA was extracted from 140 μl of dengue virus-infected culture supernatant using the QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany; No. 52906) in accordance with the manufacturer’s instructions. The RNA was eluted with 50 μl of RNase- and DNase-free water and stored at -80 °C.
2.5. Reverse Transcription‒Polymerase Chain Reaction (RT‒PCR)
A Dengue virus universal nucleic acid detection kit (Zhuocheng Huisheng, Beijing, China; No. A3801YH-50T) and a DENV I/II/III/IV nucleic acid detection kit (Zhuocheng Huisheng, Beijing, China; No. A5874YH-50T) were used for nucleic acid detection according to the manufacturer’s instructions. RT‒PCR was carried out in one step with the following protocol: initial reverse transcription at 50 °C for 10 min; denaturation at 95 °C for 30 s; and 45 cycles of denaturation at 95 °C for 5 s, annealing and elongation at 60 °C for 30 s.
2.6. DENV Genome Sequencing
Total RNA was extracted from dengue-positive samples as described previously. The virus was amplified using the Dengue Virus Whole Genome Capture Kit (BAIYITECH, Hangzhou, China; No. BK-DENV024). The products were subjected to cycle sequencing on a Nanopore triple sequencer (Oxford Nanopore Technologies, UK) using a third-generation sequencing reagent (BAIYITECH, Hangzhou, China; No. K024) according to the manufacturer’s instructions.
2.7. Whole Genome Sequence Analysis
The whole-genome sequence alignment was performed using BLAST on the NCBI website. The whole-genome sequences of the standard DENV strain and the DENV-1 and DENV-2 genotypes were downloaded from GenBank. GC content analysis was performed using EditSeq in Lasergene 17.2 software. MegAlign was used for nucleotide and amino acid homology comparisons. The phylogenetic analysis was conducted using Molecular Evolutionary Genetics Analysis (MEGA) software version 11 (the neighbour‒joining method, with a bootstrap value of 1 000), and the amino acid sites were analysed using GENEDOC2.7 software. The standard strains used for DENV-1 were the Hawaii strain (GenBank accession no. EU848545) and the New Guinea strain (GenBank accession no. M29095) for DENV-2.
3. Results
3.1. Laboratory Diagnosis
A 27-year-old healthy male patient in Jiaxing was admitted to the fever clinic of the First People's Hospital of Tongxiang due to dengue fever-like symptoms such as fever and headache. Laboratory investigations revealed that the blood parameters, including eosinophil percentage (0.10%), eosinophil count (0.00 × 109/L), and platelet haematocrit (0.13%), were slightly decreased, and the white blood cell count (4.3 × 109/L), platelet count (144.0 × 109/L), and serum amyloid A level were normal. Further analysis revealed that the patient's serum anti-dengue virus IgM antibody was positive, whereas his anti-dengue virus IgG antibody was negative, suggesting acute primary dengue virus infection. Real-time RT‒PCR confirmed that the patient was coinfected with type I and type II viruses, and viral RNA was detected in the patient's plasma.
3.2. Base Sequence Analysis of the JX202301 Sequences
The complete genome sequences of the newly isolated strains (named JX202301) were determined. The complete genome sequence of the newly isolated strain JX202301 DENV-1 was 10,695 nucleotides (nt) in length, with the ORF located between nucleotides 88–10,260, and the length of the other isolated strain, JX202301 DENV-2, was 10,678 nt, with the ORF located between nucleotides 96–10,268. The EditSeq analysis results revealed that the nucleotide composition of the JX202301 DENV-1 strain was 32.18% A, 25.98% G, 21.22% U, and 20.62% C. In comparison, the nucleotide compositions of the JX202301 DENV-2 strain were 32.24% A, 25.94% G, 21.16% U, and 20.66% C. The A + G contents of JX202301 DENV-1 and DENV-2 were 46.77% and 45.70%, respectively. It includes a highly conserved 5'-UTR, a 3'-UTR with a stable stem‒loop structure, and an open reading frame (ORF). The ORF is composed of 10,176 nucleotides and encodes 3392 amino acids. It contains three structural proteins (C, PrM/M, E) and seven nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, NS5).
3.3. Homology Analysis of the JX202301 Sequences
The complete gene sequences of JX202301 DENV-1 and DENV-2 were compared with those of representative DENV-1 and DENV-2 strains from other geographical sources in GenBank for nucleotide and amino acid homology comparisons. As shown in
Table 1, the results revealed that the JX202301 DENV-1 sequence had the highest homology with the PP563909, PP563875, PP563840 and PP563879 epidemic strains isolated in Guangdong Province in 2023. The nucleotide homology was 99.5–99.8%, and the amino acid homology was 100%. Second, compared with other DENV-1 strains isolated in Guangdong Province in 2023, such as the PP563926, PP563925, PP563914, PP563905, PP563844 and PP563826 epidemic strains isolated in Yunnan in 2023, the nucleotide homology is 99.5–99.8%, and the amino acid homology is 99.9%. The homologies of nucleotides and amino acids with the isolates from Singapore (KX224261), Guangdong (KJ438296) and the United States (EU848545) were 93.7–97.8% and 97.7–99.4%, respectively. The homologies with DENV-1 genotypes II (JN544410), III (GQ357692), IV (EU179861) and V (KY474305) were low, and the nucleotide and amino acid homologies were 90.7–91.6% and 97.2–97.3%, respectively.
As shown in
Table 2, the sequence of JX202301 DENV-2 had the highest homology with the MZ636805, MZ636802, MZ636803, MW512454 and MW512419 epidemic strains isolated from Thailand in 2019. The nucleotide homology was 98.4–99.2%, and the amino acid homology was 99.7%. Second, it had high similarity with DENV-2 strains isolated from Guangdong Province in 2017 (MH827546, MN018344, MH827552, MN018343) and DENV-2 strains isolated from Singapore (MW512473). The nucleotide and amino acid homology were 98.8–98.9% and 99.4–99.6%, respectively. Compared with other DENV-2 strains isolated in 2011, such as Pakistan (KF041234 and KF041232), the nucleotide homology was 98.0%, and the amino acid homology was92.8%. The homology with the reference strain M29095 was low, and the nucleotide and amino acid homology were 92.6% and 97.5%, respectively. The nucleotide and amino acid homology with DENV-2 genotypes I (EU482465), II (AF038403), III (AF119661) and V (AF100465) were 89.3–92.6% and 96.6–97.6%, respectively.
3.4. Phylogenetic Analysis of the JX202301 Sequences
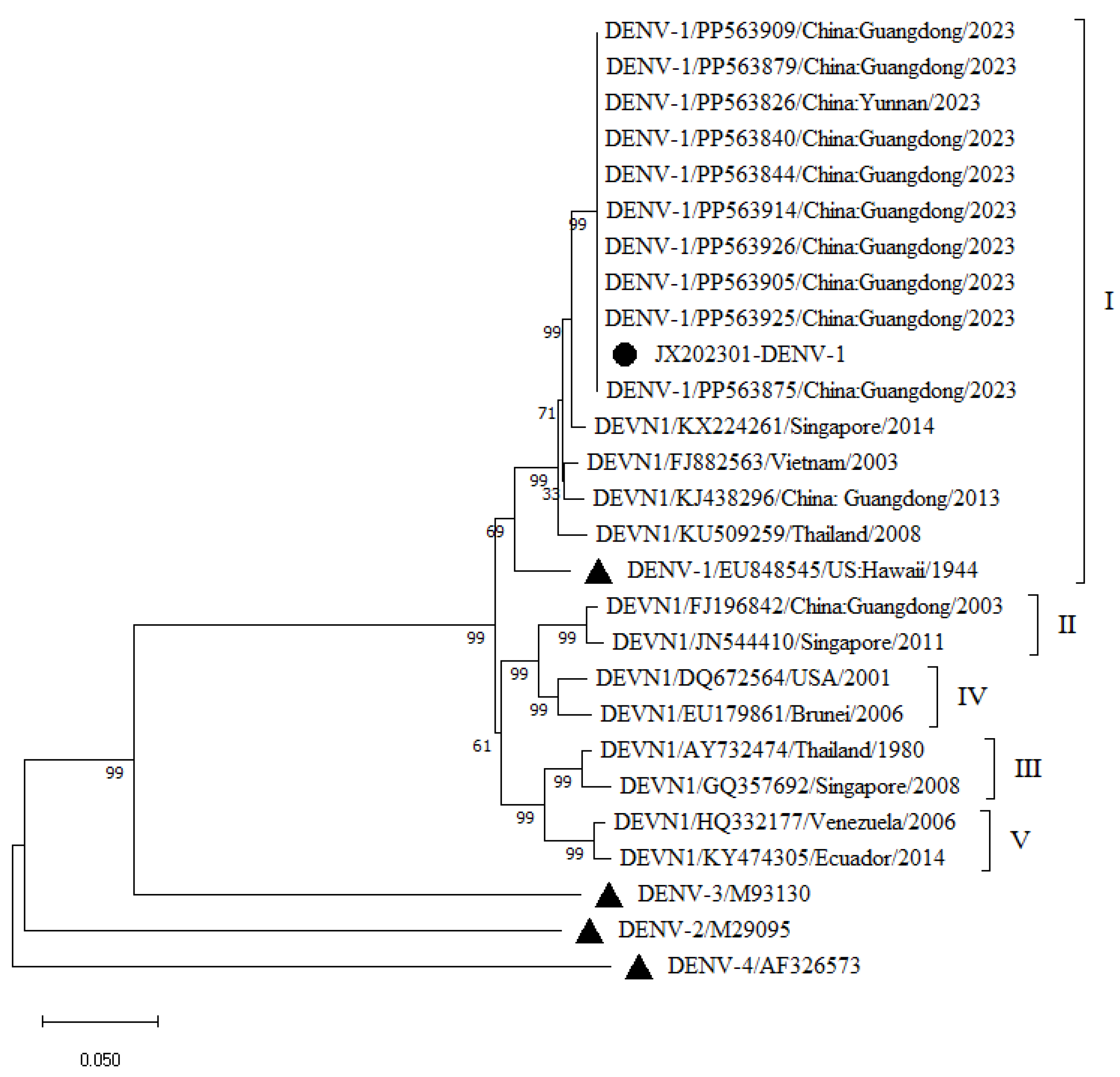
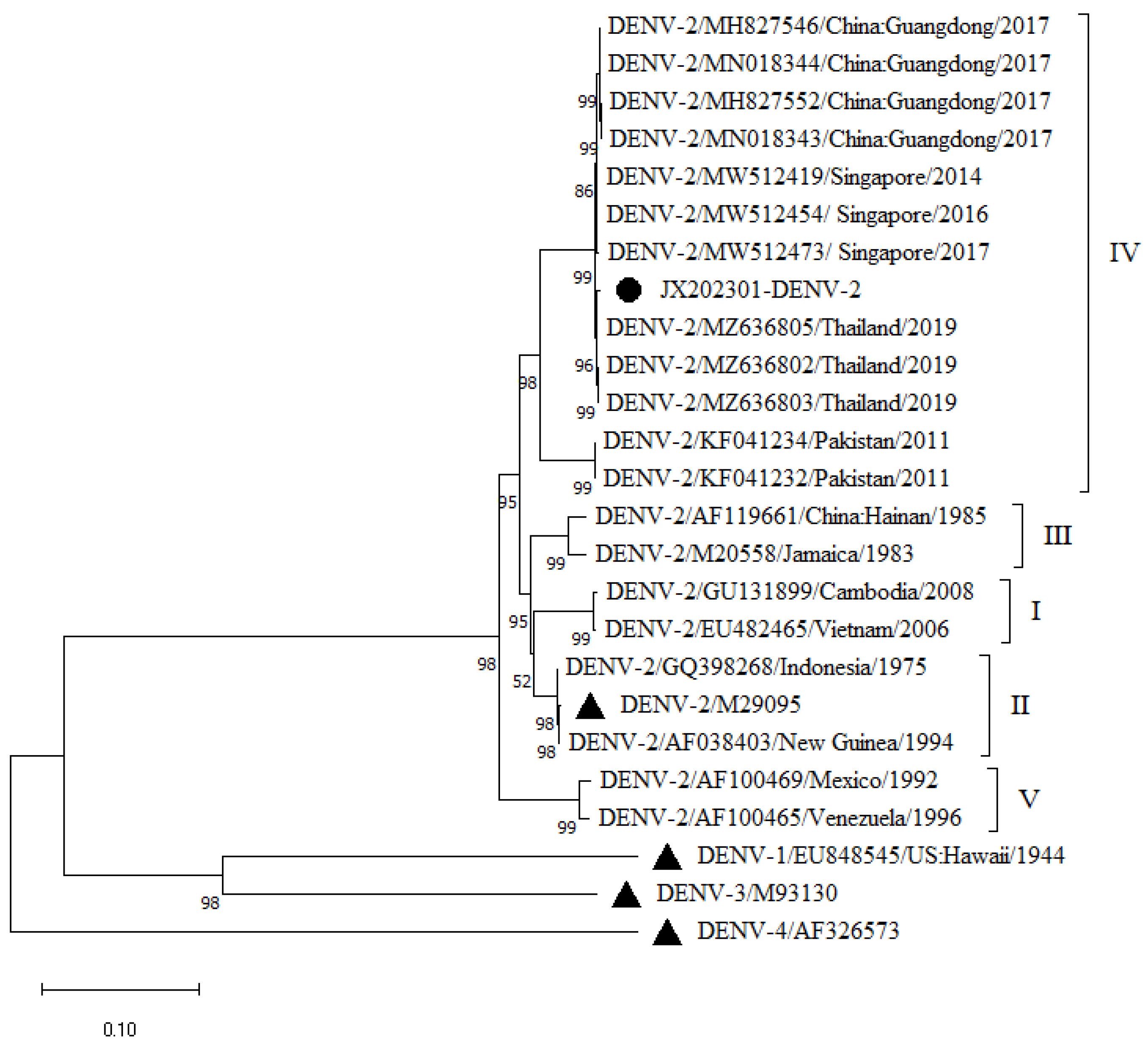
A phylogenetic tree was constructed by comparing the obtained complete JX202301 gene sequence with the complete DENV-1 and DENV-2 gene sequences downloaded from GenBank and the complete gene sequences of the four serotype standard strains of DENV. Phylogenetic tree analysis revealed (
Figure 1 and
Figure 2) that the JX202301 DENV-1 sequence was located in the G-I evolutionary branch of DENV-1. It was closely related to the strains isolated from Guangdong Province in 2023 (PP563926, PP563925, PP563914, PP563909, PP563905, PP563879, PP563875, PP563844, PP563840, and PP563826) and Yunnan Province in 2023 and was in the same evolutionary cluster. It forms a close branch with isolates from Singapore (KX224261), Guangdong (KJ438296), Vietnam (FJ882563), Thailand (KU509259) and the United States (EU848545), all of which belong to genotype I. Other isolates, such as Thailand (AY732474) and Singapore (GQ357692), belong to genotype III; Guangdong (FJ196842) and Singapore (JN544410), belong to genotype II; the United States (DQ672564) and Brunei (EU179861), belong to genotype IV; and Venezuela (HQ332177) and Ecuador (KY474305), belong to genotype V.
The JX202301 DENV-2 sequence is located in the G-IV evolutionary branch of DENV-2, has the closest relationship with the DENV-2 strains (MZ636805, MZ636802 and MZ636803) isolated from Thailand in 2019, and is in the same evolutionary cluster. It clustered on the same evolutionary branch as DENV-2 strains isolated in Guangdong Province in 2017 (MH827546, MN018344, MH827552, MN018343) and DENV-2 strains isolated in Singapore (MW512419, MW512454 and MW512473). Other isolates, Hainan (AF119661) and Jamaica (M20558), belong to genotype III, Cambodia (GU131899) and Vietnam (EU482465) belong to genotype I, and Indonesia (GQ398268), New Guinea (AF038403) and the reference strain (M29095) belong to genotype II. Mexico (AF100469) and Venezuela (AF100465) belong to genotype V.
3.5. Amino Acid Substitutions
The different sites between the amino acids derived from the JX202301DENV-1 sequence and the five DENV-1 isolates reported in the standard strain (EU848545), Guangdong Province in 2023 (PP563840, PP563905) and Guangdong Province in 2013 (KJ438296) were analysed. The results revealed that a total of 52 amino acid sites were mutated. Among them are capsid protein C (6), membrane protein M (3), envelope protein E (7), NS1 protein (11), NS2A protein (3), NS2B protein (1), NS3 protein (7), NS4A protein (1), NS4B protein (3), and NS5 protein (10). The mutation sites were concentrated in nonstructural proteins, mainly in the NS1 and NS5 region.
The amino acid sequence of JX202301 DENV-2 was compared with those of the standard strain (M29095), Thailand isolate (MZ636805), Singapore isolate (MW512454) and Guangdong isolate (MN018344). The results revealed that a total of 51 amino acid sites were mutated. Among them are capsid protein C (2), membrane protein M (5), envelope protein E (9), NS1 protein (6), NS2A protein (4), NS2B protein (2), NS3 protein (6), NS4A protein (0), NS4B protein (4), and NS5 protein (13). The mutation sites were concentrated in nonstructural proteins and were located mainly in the NS5 region. The specific amino acid variation sites and replacement amino acids are shown in
Table 3 and
Table 4.
4. Discussion
Dengue fever has been observed in more than 100 countries and regions, with over 50 million infections occurring annually[
14]. In recent decades, the incidence of dengue fever has increased in China due to various factors, such as urbanization, globalization, climate change, and migration.
Dengue fever is widespread in Southern China. Yunnan Province is located in Southwest China. The diverse geographical environment and climatic conditions make it a hot spot for many infectious diseases, including dengue fever[
15]. Business workers travelling in Zhejiang Province and Yunnan Province have frequent contacts. Dengue fever cases have been reported in each city of the province, and most of them are imported cases. In 2023, Jiaxing city collected a case of imported dengue fever in Xishuangbanna, Yunnan Province. The patient was male, had a history of migrating workers in Xishuangbanna, Yunnan Province, and was admitted to the hospital due to fever. The laboratory dengue antigen and nucleic acid tests were positive[
16]. Combined with the epidemiological and whole-genome sequencing results, the case was confirmed to be a mixed type of DENV-1 and DENV-2.
In this study, the whole-genome sequence of the JX202301 case was determined and analysed. The JX202301 DENV-1 sequence was a DENV-1 genotype I virus containing 10,695 nucleotides, and the DENV-2 sequence was a DENV2 genotype IV virus containing 10,678 nucleotides. The JX202301 DENV-1 sequence was in the same evolutionary cluster as those of the strains from Guangdong and Yunnan in 2023, with the highest homology. The nucleotide homology was 99.5–99.8%, and the amino acid homology was 99.9–100 %, indicating that the DENV-1 genotype I virus was widely prevalent in China in 2023 and that the sequence was highly homologous.
The JX202301 DENV-2 sequence was in the same evolutionary cluster as the strains prevalent in Thailand in 2019, with the highest homology. The nucleotide and amino acid homology were 98.4–99.2% and 99.7%, respectively, suggesting that the DENV-2 genotype IV virus may have originated from Thailand. Amino acid variation analysis of the JX202301DENV1 sequence and 4 DENV-1 epidemic strains revealed that a total of 52 coding region amino acids were mutated, mainly in the nonstructural protein NS1 region and NS5 region, accounting for 21.2% (11/52) and 19.2% (10/52), respectively. Amino acid variation analysis of the JX202301 DENV2 sequence and 4 DENV-2 epidemic strains revealed that a total of 51 coding region amino acids were mutated, mainly in the nonstructural protein NS5 region, accounting for 24.1% (13/54). The variation sites of structural proteins occurred mainly in the outer membrane protein (E) region, accounting for 13.5% (7/52) and 17.6% (9/51) of the total protein content, respectively. Nonstructural proteins play important roles in virus replication, regulation and immune escape[
17,
18]. The outer membrane protein is an important part of the dengue virus and plays a key role in the binding between viruses and cells and the process by which viruses enter cells. Different epidemic strains have mutations in the genes encoding these proteins, which may lead to differences in the functions of these proteins, thus affecting virus replication, the host immune response, virus receptor binding ability and transmission characteristics[
19]. Further functional studies will help to reveal the specific effects of these variations.
Yue et al. reported that the epidemic of dengue fever in Southern China has two modes: local epidemics caused by imported cases and local outbreaks focused on natural epidemics[
20]. In recent years, cases imported from foreign countries and neighbouring provinces have been reported in Zhejiang, but simultaneous infection with more than one serotype of dengue virus is uncommon, and the aetiology of mixed infection cases of dengue virus is poorly understood[
21]. The mixed infection cases mentioned in this paper may be due to their work in Yunnan, which exposed them to both DENV-1 and DENV-2 viral infection sources, and if patients suffer from other diseases, the lowered immunity of the individual may also increase the chances of mixed infections, which needs to be confirmed by further studies. Overall, dengue fever in Jiaxing is still an exotic infectious disease, but the diversity of virus sources is increasing, and the area of infection is expanding. Imported cases, high-density mosquito vectors, and clinical omissions may lead to local outbreaks, which highlights the need to strengthen dengue surveillance and develop prevention and control strategies.
5. Conclusions
In conclusion, we report the first case of mixed infection of DENV-1 and DENV-2 imported from Xishuangbanna, Yunnan, in Jiaxing, Zhejiang Province, and the whole genome sequence analysis was performed to determine the genetic relationship and mutation sites, which provides a scientific basis for dengue surveillance and prevention in Jiaxing City.
Author Contributions
Conceptualization, Y.M.and S.C.; methodology, Y.H.; software, S.Y.;validation, G.L. and J.M.; formal analysis, L.N.; investigation, S.C.; resources, G.Y.; data curation, Y.M.; writing—original draft preparation, Y.M.;writing—review and editing, Y.Y.; visualization, G.L.; supervision, Y.H.; project administration, G.Y.; funding acquisition, G.L and Y.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Jiaxing Science and Technology Bureau (2024AD30124) and Zhejiang Medical Science and Technology Program (2024KY888).
Acknowledgments
The authors are grateful to Ms. Wang Lei, Tang Yi of Hangzhou Baiyi Technology Co., Ltd for the technique support for sequencing services.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Bhadra, S., Dengue guidelines for diagnosis, treatment, prevention and control. 2020.
- Gubler, D.J., Dengue and Dengue Hemorrhagic Fever - ScienceDirect. Tropical Infectious Diseases (Second Edition), 2006. 8(1): p. 813-822.
- Annelies, W.S., Murray, and Q. Mikkel, Epidemiology of dengue: past, present and future prospects. Clinical Epidemiology, 2013. 2013(1): p. 299-309.
- Calisher, C.H., N. Karabatsos, J.M. Dalrymple, et al., Antigenic Relationships between Flaviviruses as Determined by Cross-neutralization Tests with Polyclonal Antisera. Journal of General Virology, 1989. 30(1): p. 37-43. [CrossRef]
- Gubler, D.J., Dengue and dengue hemorrhagic fever: its history and resurgence as a global public health problem. Dengue & Dengue Hemorrhagic Fever, 1997.
- Roy, S.K. and S. Bhattacharjee, Dengue Virus: Epidemiology, Biology and Disease Aetiology. Canadian Journal of Microbiology, 2021.
- Wang, Q., Z. Xu, F.-M. Dou, et al., Current situation and surveillance on dengue fever in China, 2005-2007. Zhonghua liu xing bing xue za zhi= Zhonghua liuxingbingxue zazhi, 2009. 30(8): p. 802-806.
- Gubler, D.J., G. Kuno, G.E. Sather, et al., A case of natural concurrent human infection with two dengue viruses. American Journal of Tropical Medicine & Hygiene, 1985. 34(1): p. 170-3. [CrossRef]
- Hammon, W.M., Dengue hemorrhagic fever-do we know its cause? 1973.
- Cnops, L., C. Domingo, V.D.B. Dorien, et al., First dengue co-infection in a Belgian traveler returning from Thailand, July 2013. Journal of Clinical Virology, 2014. 61(4): p. 597-599. [CrossRef]
- Senaratne, U., K. Murugananthan, P. Sirisena, et al., Dengue virus co-infections with multiple serotypes do not result in a different clinical outcome compared to mono-infections. Epidemiology & Infection, 2020. 148: p. e119. [CrossRef]
- Wang, W.-K., D.-Y. Chao, S.-R. Lin, et al., Concurrent infections by two dengue virus serotypes among dengue patients in Taiwan. Journal of microbiology, immunology, and infection= Wei mian yu gan ran za zhi, 2003. 36(2): p. 89-95.
- Mamani, E., D. Figueroa, M.P. García, et al., Concurrent infections by two dengue virus serotypes during an outbreak in northwestern Peru, 2008. Revista peruana de medicina experimental y salud publica, 2010. 27(1): p. 16-21.
- Katherine, B., Poole-Smith, R. Ryan, et al., Comparison of vector competence of Aedes mediovittatus and Aedes aegypti for dengue virus: implications for dengue control in the Caribbean. PLoS neglected tropical diseases, 2015.
- Wei, C., X.-L. Guo, R. Yang, et al., Epidemiological and cluster characteristics of dengue fever in Yunnan province, China, 2013-2020. 2021.
- Cassidy-Seyoum, S., M. Vongsouvath, O. Sengvilaipaseuth, et al., Rapid Diagnostic Tests as a Source of Dengue Virus RNA for Envelope Gene Amplification: A Proof of Concept. The American journal of tropical medicine and hygiene, 2019. 101(2). [CrossRef]
- Fang, E., M. Li, X. Liu, et al., NS1 Protein N-Linked Glycosylation Site Affects the Virulence and Pathogenesis of Dengue Virus. Vaccines, 2023. 11(5): p. 959. [CrossRef]
- Julianna, Z., F.S. Lorena, B. Glauce, et al., Non-Canonical Roles of Dengue Virus Non-Structural Proteins. Viruses, 2017. 9(3): p. 42.
- Modis, Y., S. Ogata, D. Clements, et al., Structure of the dengue virus envelope protein after membrane fusion. Nature, 2004. 427(6972): p. 313-9. [CrossRef]
- Yue, Y., X. Liu, D. Ren, et al., Spatial Dynamics of Dengue Fever in Mainland China, 2019. Int J Environ Res Public Health, 2021. 18(6). [CrossRef]
- Yan, H., Z. Ding, J. Yan, et al., Epidemiological Characterization of the 2017 Dengue Outbreak in Zhejiang, China and Molecular Characterization of the Viruses. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).