Submitted:
09 November 2024
Posted:
11 November 2024
You are already at the latest version
Abstract
Keywords:
Introduction
Materials and Methods
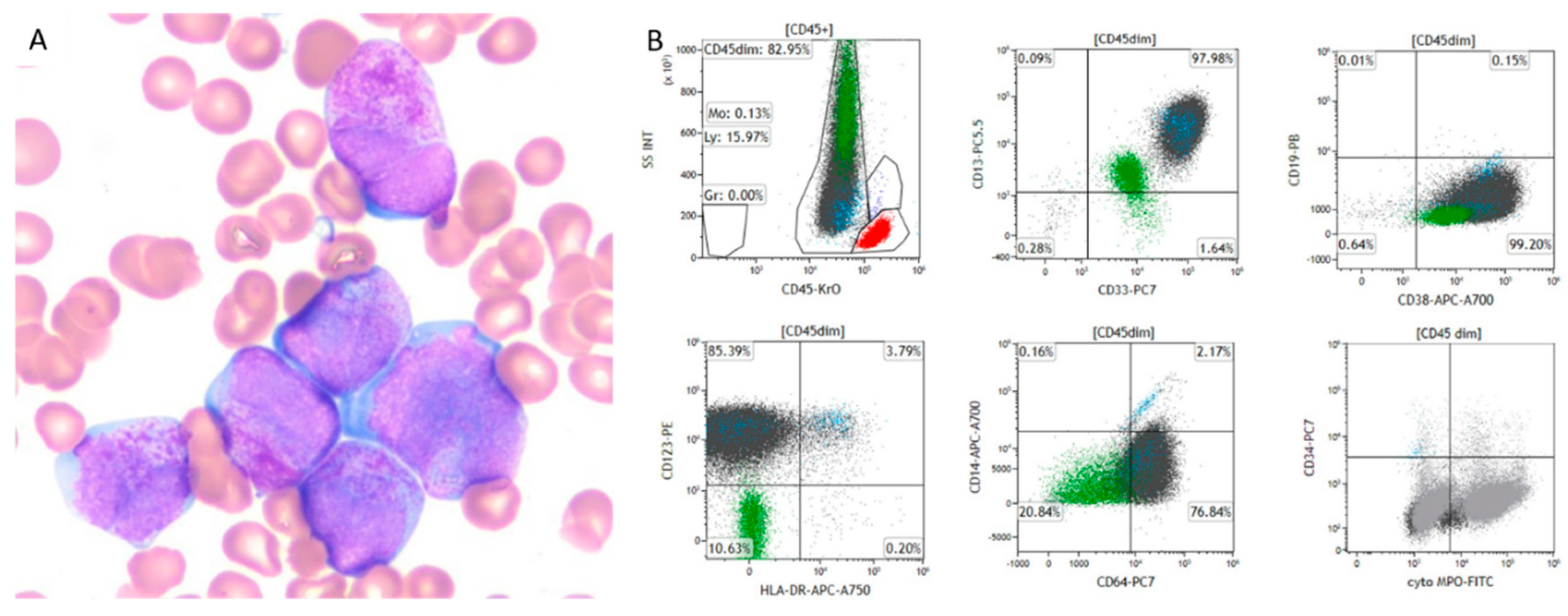
Pathological Examination
Clinical Flow Cytometry Evaluation
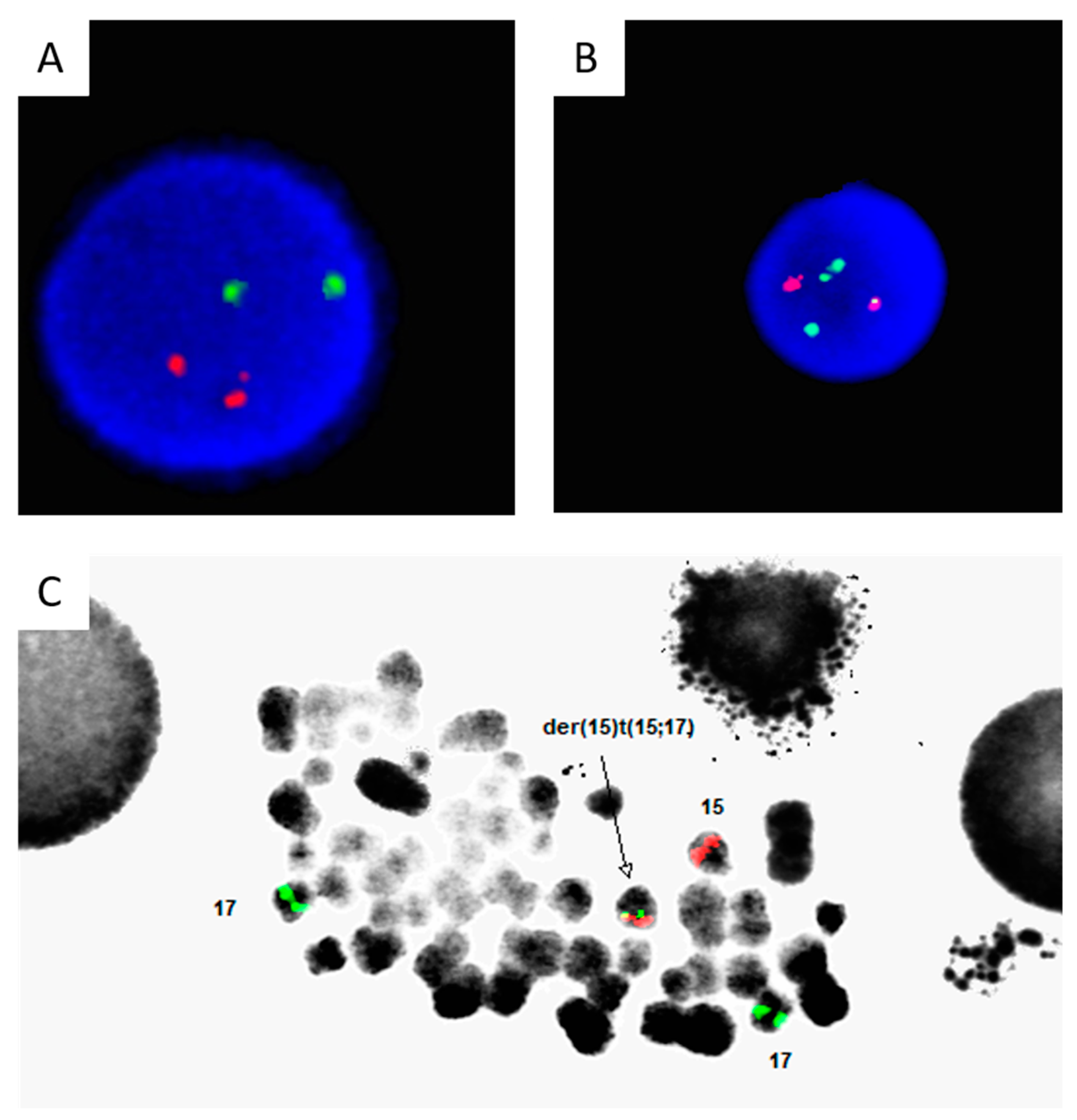
Chromosome G-Banding Karyotype Analysis
Fluorescence In Situ Hybridization
Mutational Analysis
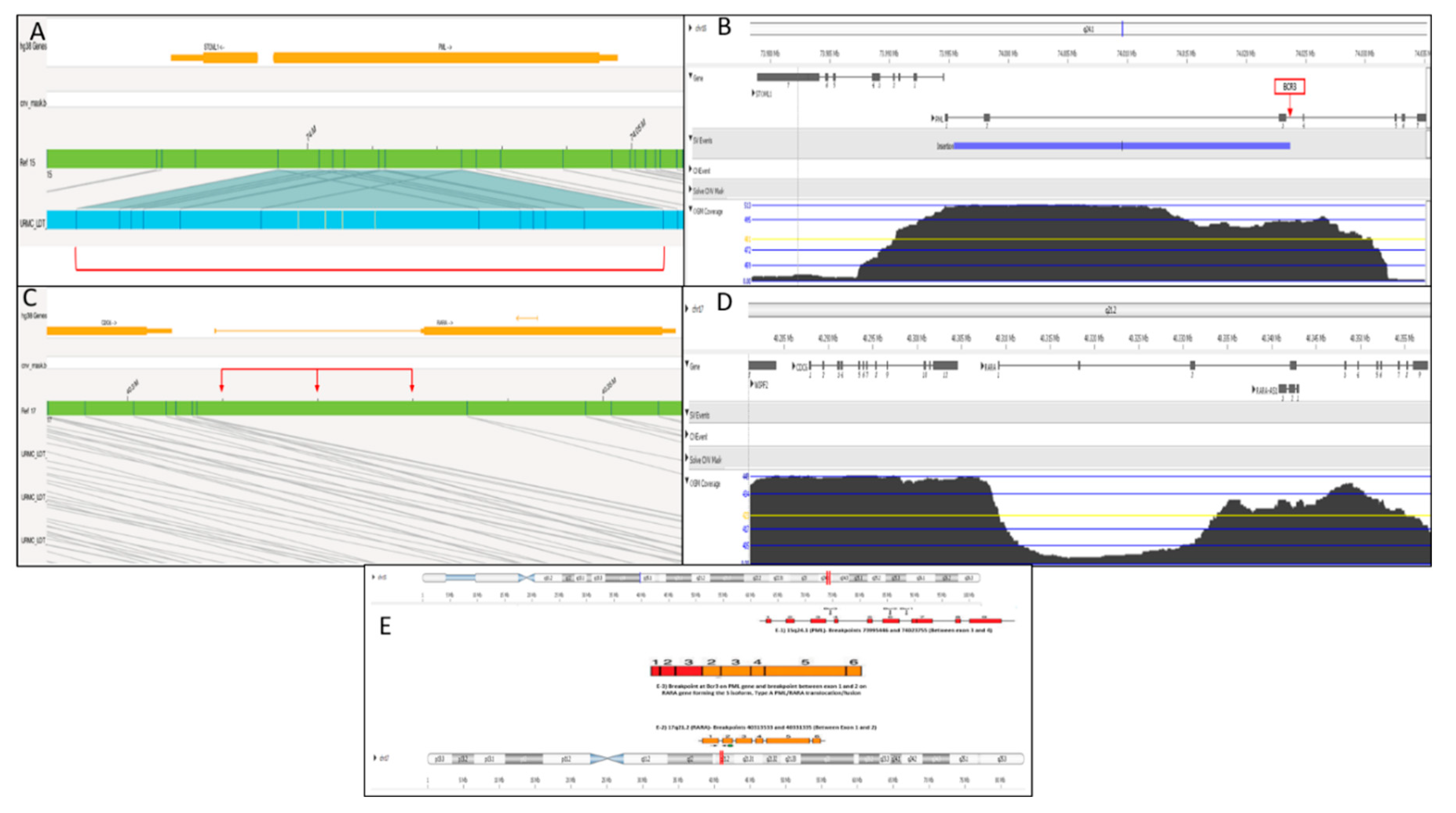
Optical Genome Mapping
Systematic Literature Review
Results
Case Description
Literature Review
Discussion
Author Contributions
Funding
Data Availability
Conflicts of Interest
Patient Consent
References
- Alaggio R AC, Anagnostopoulos I, Attygalle AD, Araujo IBO, Berti E, Bhagat G, Borges AM, Boyer D, Calaminici M, Chadburn A, Chan JKC, Cheuk W, Chng WJ, Choi JK, Chuang SS, Coupland SE, Czader M, Dave SS, de Jong D, Du MQ, Elenitoba-Johnson KS, Ferry J, Geyer J, Gratzinger D, Guitart J, Gujral S, Harris M, Harrison CJ, Hartmann S, Hochhaus A, Jansen PM, Karube K, Kempf W, Khoury J, Kimura H, Klapper W, Kovach AE, Kumar S, Lazar AJ, Lazzi S, Leoncini L, Leung N, Leventaki V, Li XQ, Lim MS, Liu WP, Louissaint A, Jr., Marcogliese A, Medeiros LJ, Michal M, Miranda RN, Mitteldorf C, Montes-Moreno S, Morice W, Nardi V, Naresh KN, Natkunam Y, Ng SB, Oschlies I, Ott G, Parrens M, Pulitzer M, Rajkumar SV, Rawstron AC, Rech K, Rosenwald A, Said J, Sarkozy C, Sayed S, Saygin C, Schuh A, Sewell W, Siebert R, Sohani AR, Tooze R, Traverse-Glehen A, Vega F, Vergier B, Wechalekar AD, Wood B, Xerri L, Xiao W. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Lymphoid Neoplasms. Leukemia 2022;36:1720-48.
- Stone M, Lilley CM, Tang G, Loghavi S, Mirza KM. Phenotypic clues that predict underlying cytogenetic/genetic abnormalities in myeloid malignancies: A contemporary review. Cytopathology 2023;34(6):530-541. (In eng). [CrossRef]
- El-Hajj Ghaoui R, St Heaps L, Hung D, et al. A Paediatric Acute Promyelocytic Leukaemia Patient Harbouring a Cryptic PML-RARA Insertion due to a Complex Structural Chromosome 17 Rearrangement. Cytogenet Genome Res 2017;153(4):181-189. (In eng). [CrossRef]
- Rashidi A, Fisher SI. FISH-negative, cytogenetically cryptic acute promyelocytic leukemia. Blood Cancer J 2015;5(6):e320. (In eng). [CrossRef]
- Kim MJ, Cho SY, Kim MH, et al. FISH-negative cryptic PML-RARA rearrangement detected by long-distance polymerase chain reaction and sequencing analyses: a case study and review of the literature. Cancer Genet Cytogenet 2010;203(2):278-83. (In eng). [CrossRef]
- Gagnon MF, Berg HE, Meyer RG, et al. Typical, atypical and cryptic t(15;17)(q24;q21) (PML::RARA) observed in acute promyelocytic leukemia: A retrospective review of 831 patients with concurrent chromosome and PML::RARA dual-color dual-fusion FISH studies. Genes Chromosomes Cancer 2022;61(10):629-634. (In eng). [CrossRef]
- Singh MK, Parihar M, Arora N, Mishra DK, Bhave SJ, Chandy M. Diagnosis of variant RARA translocation using standard dual-color dual-fusion PML/RARA FISH probes: An illustrative report. Hematol Oncol Stem Cell Ther 2019;12(1):50-53. (In eng). [CrossRef]
- Catalano A, Dawson MA, Somana K, et al. The PRKAR1A gene is fused to RARA in a new variant acute promyelocytic leukemia. Blood 2007;110(12):4073-6. (In eng). [CrossRef]
- Osumi T, Watanabe A, Okamura K, et al. Acute promyelocytic leukemia with a cryptic insertion of RARA into TBL1XR1. Genes Chromosomes Cancer 2019;58(11):820-823. (In eng). [CrossRef]
- Pardo Gambarte L, Franganillo Suarez A, Cornago Navascues J, et al. ZBTB16-RARalpha-Positive Atypical Promyelocytic Leukemia: A Case Report. Medicina (Kaunas) 2022;58(4) (In eng). [CrossRef]
- Wang Y, Rui Y, Shen Y, et al. Myeloid Sarcoma Type of Acute Promyelocytic Leukemia With a Cryptic Insertion of RARA Into FIP1L1: The Clinical Utility of NGS and Bioinformatic Analyses. Front Oncol 2021;11:688203. (In eng). [CrossRef]
- Peterson JF, He RR, Nayer H, et al. Characterization of a rarely reported STAT5B/RARA gene fusion in a young adult with newly diagnosed acute promyelocytic leukemia with resistance to ATRA therapy. Cancer Genet 2019;237:51-54. (In eng). [CrossRef]
- Yao L, Wen L, Wang N, et al. Identification of novel recurrent STAT3-RARA fusions in acute promyelocytic leukemia lacking t(15;17)(q22;q12)/PML-RARA. Blood 2018;131(8):935-939. (In eng). [CrossRef]
- Seabright M. A rapid banding technique for human chromosomes. Lancet 1971;2(7731):971-2. (In eng). [CrossRef]
- An International System for Human Cytogenomic Nomenclature.: S.Karger AG., 2020.
- Pang AWC, Kosco K, Sahajpal NS, Sridhar A, Hauenstein J, Clifford B, Estabrook J, Chitsazan AD, Sahoo T, Iqbal A, Kolhe R, Raca G, Hastie AR, Chaubey A. Analytic Validation of Optical Genome Mapping in Hematological Malignancies. Biomedicines. 2023 Dec 9;11(12):3263. doi: 10.3390/biomedicines11123263. PMID: 38137484; PMCID: PMC10741484.
- Liu G, Liu L, Bartolo DD, Li KY, Li X. Acute Promyelocytic Leukemia with Rare Genetic Aberrations: A Report of Three Cases. Genes (Basel) 2022;14(1) (In eng). [CrossRef]
- Fasan A, Haferlach C, Perglerova K, Kern W, Haferlach T. Molecular landscape of acute promyelocytic leukemia at diagnosis and relapse. Haematologica 2017;102(6):e222-e224. (In eng). [CrossRef]
- Madan V, Shyamsunder P, Han L, et al. Comprehensive mutational analysis of primary and relapse acute promyelocytic leukemia. Leukemia 2016;30(8):1672-81. (In eng). [CrossRef]
- Blanco EM, Curry CV, Lu XY, et al. Cytogenetically cryptic and FISH-negative PML/RARA rearrangement in acute promyelocytic leukemia detected only by PCR: an exceedingly rare phenomenon. Cancer Genet 2014;207(1-2):48-9. (In eng). [CrossRef]
- Avgerinou G, Katsibardi K, Filippidou M, Tzanoudaki M, Papadhimitriou SI, Kattamis A. Cytogenetically cryptic and fish negative PML/RARA rearrangement in acute promyelocytic leukemia detected by RT-PCR. Leuk Lymphoma 2020;61(14):3526-3528. (In eng). [CrossRef]
- Burns TF, Loo EY, Bengtson EM, Bao L. Cytogenetically cryptic insertion of PML segment into RARA on chromosome 17q resulting PML-RARA fusion in acute promyelocytic leukemia. Ann Hematol 2019;98(1):211-213. (In eng). [CrossRef]
- Goldschmidt N, Yehuda-Gafni O, Abeliovich D, Slyusarevsky E, Rund D. Interstitial insertion of RARalpha gene into PML gene in a patient with acute promyelocytic leukemia (APL) lacking the classic t(15;17). Hematology 2010;15(5):332-7. (In eng). [CrossRef]
- Koshy J, Qian YW, Bhagwath G, Willis M, Kelley TW, Papenhausen P. Microarray, gene sequencing, and reverse transcriptase-polymerase chain reaction analyses of a cryptic PML-RARA translocation. Cancer Genet 2012;205(10):537-40. (In eng). [CrossRef]
- Karlin K, Bryke C, Dias A, Michaels P. Cytogenetically cryptic PML::RARA fusion in acute promyelocytic leukemia: Testing strategies in the modern era. Leuk Res Rep 2022;17:100320. (In eng). [CrossRef]
- Gu S, Zi J, Ma J, Ge Z. Cryptic t(15;17) acute promyelocytic leukemia with a karyotype of add(11)(p15) and t(13,20)- A case report with a literature review. Bosn J Basic Med Sci 2021;21(2):246-251. (In eng). [CrossRef]
- Tang Y, Wang Y, Hu L, et al. Acute promyelocytic leukemia with cryptic t(15;17) on isochromosome 17: a case report and review of literature. Int J Clin Exp Pathol 2015;8(11):15294-300. (In eng) (https://www.ncbi.nlm.nih.gov/pubmed/26823883).
- Shepshelovich D, Oniashvili N, Parnes D, et al. Acute promyelocytic leukemia with isochromosome 17q and cryptic PML-RARA successfully treated with all-trans retinoic acid and arsenic trioxide. Cancer Genet 2015;208(11):575-9. (In eng). [CrossRef]
- Cervera J, Montesinos P, Hernandez-Rivas JM, et al. Additional chromosome abnormalities in patients with acute promyelocytic leukemia treated with all-trans retinoic acid and chemotherapy. Haematologica 2010;95(3):424-31. (In eng). [CrossRef]
- Kim M, Lim J, Kim Y, et al. The genetic characterization of acute promyelocytic leukemia with cryptic t(15;17) including a new recurrent additional cytogenetic abnormality i(17)(q10). Leukemia 2008;22(4):881-3. (In eng). [CrossRef]
- Mahmud W, Brown R, Buckingham L, Tira A, Katz DA. Cryptic partial insertion of the RARA gene into the PML gene without reciprocal RARA-PML fusion: a case report and review of literature. Acta Oncol 2020;59(12):1496-1499. (In eng). [CrossRef]
- Greenfield G, Michail O, Merron B, McGimpsey J, Catherwood M, McMullin MF. Acute promyelocytic leukaemia (APML) with cryptic PML-RARA fusion has a clinical course comparable to classical APML with t(15;17)(q24.1;q21.2) translocation. Br J Haematol 2019;186(1):155-157. (In eng). [CrossRef]
- Mohebnasab M, Li P, Hong B, et al. Cytogenetically Cryptic Acute Promyelocytic Leukemia: A Diagnostic Challenge. Int J Mol Sci 2023;24(17) (In eng). [CrossRef]
- Campbell LJ, Oei P, Brookwell R, et al. FISH detection of PML-RARA fusion in ins(15;17) acute promyelocytic leukaemia depends on probe size. Biomed Res Int 2013;2013:164501. (In eng). [CrossRef]
- Schultz MJ, Blackburn PR, Cogbill CH, et al. Characterization of a cryptic PML-RARA fusion by mate-pair sequencing in a case of acute promyelocytic leukemia with a normal karyotype and negative RARA FISH studies. Leuk Lymphoma 2020;61(4):975-978. (In eng). [CrossRef]
- Grimwade D, Biondi A, Mozziconacci MJ, et al. Characterization of acute promyelocytic leukemia cases lacking the classic t(15;17): results of the European Working Party. Groupe Francais de Cytogenetique Hematologique, Groupe de Francais d'Hematologie Cellulaire, UK Cancer Cytogenetics Group and BIOMED 1 European Community-Concerted Action "Molecular Cytogenetic Diagnosis in Haematological Malignancies". Blood 2000;96(4):1297-308. (In eng) (https://www.ncbi.nlm.nih.gov/pubmed/10942371).
- Han Y, Xue Y, Zhang J, Pan J, Wu Y, Bai S. Y-chromosome loss as the sole karyotypic anomaly with 3'RARalpha submicroscopic deletion in a case of M3r subtype of acute promyelocytic leukemia. Leuk Res 2009;33(10):1433-5. (In eng). [CrossRef]
- Singh J, Facey A, O'Malley F, Ryland GL, Blombery P, Gregory GP. Cryptic molecular lesion in acute promyelocytic leukemia with negative initial FISH. Leuk Lymphoma 2021;62(12):3060-3062. (In eng). [CrossRef]
| No. | Authors/ PMID |
Age/Sex | Clinical Presentation | Microscopic Findings | Flow Cytometry Expression | Karyotype Findings |
Interphase FISH | Mutational Analysis |
RT-PCR Results |
Confirmatory Testing |
| 1 | Avgerinou et al/ 32909480 |
12/F | Multiple ecchymoses | Leukocytosis Abnormal promyelocytes with bilobed nuclei and cytoplasmic granules Anemia thrombocytopenia |
MPO, CD34 CD123, CD64, CD33, CD117, CD9, HLA-DR and CD2, while negative for other markers | 46, XX | Abbott Molecular LSI PML/RARA dual-color dual-fusion translocation probe: Negative Cytocell (Cambridge, UK) positive for PML::RARA fusion |
FLT3-ITD | bcr3-PML/RARA transcript | Sanger sequencing: in-frame fusion of PML exon 3 and RARA exon 3 |
| 2 | Blanco et al/ 24561214 |
17/M | Gum bleeding, multiple ecchymoses, abdominal pain, and fever | Leukocytosis Abnormal promyelocytes with bilobed nuclei and cytoplasmic granules |
CD117, CD45 (dim), CD13, CD33, CD15 (weak), and CD64 while negative for HLA-DR, CD34, and other markers | 46, XY | Negative | FLT3-ITD | bcr1-PML/RARA transcript | Sequencing: in-frame fusion of PML exon 6 and RARA exon 3 |
| 3 | Burns et al/ 30030569 |
23/F | Epistaxis and easy bruising | Leukocytosis Abnormal myeloblasts/promyelocytes with ovoid nuclei and cytoplasmic granules (rare Auer rods) Anemia thrombocytopenia |
CD13, CD33 (partial), CD56 (partial, dim), CD64, and MPO, while negative for HLA-DR and CD34 | 46, XX,+8 | Abbott Molecular LSI PML/RARA dual-color dual-fusion translocation probe: Negative | Not mentioned | Cryptic PML::RARA fusion | Metaphase FISH: interstitial insertion of PML into the RARA gene |
| 4 | El-Hajj Ghaoui et al/ 29550828 |
8/M | Bruising and bleeding gums | Leukocytosis Blasts and abnormal promyelocytes with large irregularly folded or bi-lobed nuclei and abnormal granulation (rare Auer rods) Anemia thrombocytopenia |
CD13, CD33, CD117, CD123, and CD45, while negative for other markers | 46,XY,der(17)ins(17; 15) (q21;q24q24)?del(17)(p11.2)add(17)(q21) | MetaSystems, Germany dual colour dual fusion PML-RARA probe: single fusion signal and 2 copies of PML and RARA; second expected reciprocal fusion signal not present, and one each of the PML and RARA signals was of diminished intensity | FLT3-ITD | bcr3-PML/RARA transcript and a faint ∼ 350-bp product of unknown origin | Metaphase FISH (using RARA break-apart probe [Abbott Molecular, USA], 15q11.2 control locus [RP11-160D9 from Australasian Genome Research Facility, Melbourne, Australia], subtelomere clones for chromosome 15q [GS-154P1], 17p [cosmid 2111b1], and 17q [PAC GS-362K4], and NF1 within chromosome band 17q11.2): single fusion with diminished RARA signals on the derivative chromosome 17; i.e., der(17) 850K SNP chromosome microarray: no clinically relevant chromosome copy number abnormality across the tumor genome |
| 5 | Fan et al/ 24673420 |
61/F | Fatigue and easy bruising | Leukocytosis Blasts/promyelocytes 40% Anemia thrombocytopenia |
Dim CD45, CD13, CD33,CD117, variable CD34, and lacking HLA-DR | 46, XX, +8 (17/20 cells) | Peripheral blood: variant abnormal signal pattern with 1fusion (1F1O2G) in 52.5% of the nuclei Bone marrow: variant abnormal signal pattern with 1fusion (1F1O2G) in 42% of the nuclei |
Not mentioned | Cryptic PML::RARA fusion without reciprocal RARA-PML fusion transcripts in either the diagnostic or follow-up samples | Metaphase FISH: Non-reciprocaltranslocation with the fusion signal on chromosome 15 and absence of the fusion signal on chromosome 17 Metaphase FISH using whole chromosome paint: RARA(green) signal on chromosome15, without the corresponding PML (orange) signal on chromosome 17, demonstrating an insertion |
| 6 | Mai et al/ 32366568 |
17/M | Seizure (with recent history of nausea, blood-tinged vomiting, lethargy, and right-sided weakness) | Leukocytosis Blasts/promyelocytes 83% Anemia thrombocytopenia |
CD2 (partial), CD4 (partial), CD13, CD33, CD38, CD45, CD64, CD117 (partial), HLA-DR (small subset), and MPO (bright), while negative for other markers | 46, XY | Negative | Not mentioned | Cryptic PML::RARA fusion | Not performed |
| 7 | Zhang et al/ 31959056 |
66/M | Petechiae and bruises | Leukopenia Blasts/promyelocytes 68% (irregular nuclear shapes, misty nucleoli) Anemia thrombocytopenia |
CD34, CD7, CD13, CD33, CD117, CD38, HLA-DR and MPO, while negative for other markers | 46, XY | Negative | Biallelic CEBPA mutation | bcr2-PML/RARA transcript (nested RT-PCR) | Not performed |
| 8 | Gu et al/ 33052080 |
62/M | Pharyngalgia, fatigue, and gum bleeding | Anemia Leukopenia Hypercellular bone marrow with abnormal promyelocytes |
CD117, CD33, myeloperoxidase (MPO), CD13, CD58, CD38, and CD81 | 46,XY, add(11) (p15), and ?t(13;20)(q12;q11.2) |
Atypical PML::RARA fusion signal in 91% of nuclei | FLT3, WT1, and KRAS mutations | Major PML/RARα transcript harbored the three type breakpoints | Not performed |
| 9 | Kim et al/ 21156244 |
18/M | Hematuria and hematochezia | Leukocytosis Blasts/promyelocytes 84% Anemia thrombocytopenia |
CD13, CD33, CD45, and CD117 and negative for HLA-DR and CD34 | 46,XY | Negative | Not mentioned | Three PML::RARA fusion transcripts: bcr2, bcr1, and novel transcript (exon 4 of PML and exon 3 of RARA) | Long-distance DNA-PCR: rearrangement between PML (intron 6) and RARA (intron 2) |
| 10 | Goldschmidt et al/ 20863428 |
52/F | Bleeding tendency | Anemia Leukopenia Thrombocytopenia |
Strongly positive for CD33 and CD13 while negative for HLA-DR | 47,XX,zi(5)(p10)[20]/48,idem,z9[2]/46,XX[6] | Negative | Not mentioned | Bcr1-PML/RARa transcript | Metaphase FISH: interstitial insertion of RARa gene into PML gene (low signal retrospectively identified on interphase FISH) |
| 11 | Koshy et al/ 22982005 |
29/M |
Progressive fatigue Bruises |
Pancytopenia 26% abnormal promyelocytes |
Positive for CD117, CD33, and CD13 but negative for HLA-DR and CD34 | 46,XY | Negative with a small PML signal present on another chromosome (20% of cells) | Not mentioned | Positive for PML::RARA transcript | Sanger sequencing across the PML-RARA breakpoint demonstrated a BCR1-type fusion. Whole genome SNP microarray: intragenic duplication of PML on chromosome 15q24.1 (30% of cells). |
| 12 | Karlin et al/ 35572917 |
54/M |
DVT |
Pancytopenia 77% abnormal promyelocytes in the BM |
Positive for MPO and CD117 while negative for CD34 and HLA-DR | 46,XY | Negative | FLT3 p.D835Y variant | Bcr1-PML/RARA transcript | Not performed |
| 13 | Mahmud et al/ 32924730 |
68/F |
Dizziness Fatigue Acute on chronic PE |
Pancytopenia 70% abnormal promyelocytes/blasts |
Not mentioned | 46, XX | Two copies of chromosome 15, but absence of the reciprocal translocation on the two copies of chromosome 17 | Not mentioned | Positive for bcr3-PML::RARA transcript | Metaphase FISH: insertion of a RARA segment into chromosome 15 at the location of PML. Whole-genome sequencing: complex t(15;17) with a possible intrachromosomal rearrangement of chromosome 15. |
| 14 | Schultz et al/ 31809670 |
57/F |
Bruising Gingival bleeding |
Anemia Neutropenia Thrombocytopenia Abnormal promyelocytes/blasts |
Positive for CD13, CD33, CD34 (partial), CD117, MPO, and aberrant partial CD2 expression | 46, XX | Negative | Not mentioned | PML-RARA fusion in 53% of cells | Mate-pair sequencing: cryptic insertional translocation resulting in PML-RARA fusion with breakpoints located within intron 6 of PML and intron 2 of RARA. |
| 15 | Shepshelovich et al/ 26471811 |
53/F | Not mentioned | Not mentioned | Not mentioned | 46XX, iso(17)(q11) | Using the Cytocell probe: negative |
Not mentioned | Major PML−RARA transcript harbored the BCR-1 breakpoint. BCR-2 and BCR-3 were demonstrated as spliced variants. | FISH using the Vysis probe: several clones detected |
| 16 | Tang et al/ 26823883 |
21/M |
Melena Bleeding tendency |
Not mentioned | Not mentioned | Complex karyotype with isochromosome 17q | Two PML/RARA fusion signals | Not mentioned | Not mentioned | Not performed |
| 17 | Venci et al/ 28599418 |
73/F | Diagnosed incidentally (pre-operatively) | Anemia Thrombocytopenia Abnormal promyelocytes/blasts |
Positive for CD13, CD33, MPO, CD2 and CD9, while negative for other markers | 46,XX | Two fusion signals on the two copies of chromosome 15, but absence of the reciprocal on the two copies of chromosome 17 | Not mentioned | Bcr3/short form PML-RARA fusion transcript | Metaphase FISH: two PML/RARA fusion signals (one on each copy of chromosome 15, and two normal RARA signals on the two copies of chromosome 17), raising the possibility of uniparental isodisomy. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





