Submitted:
28 October 2024
Posted:
30 October 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
Sample Collection and Clinical Evaluation
Whole-Exome Sequencing and Data Analyses
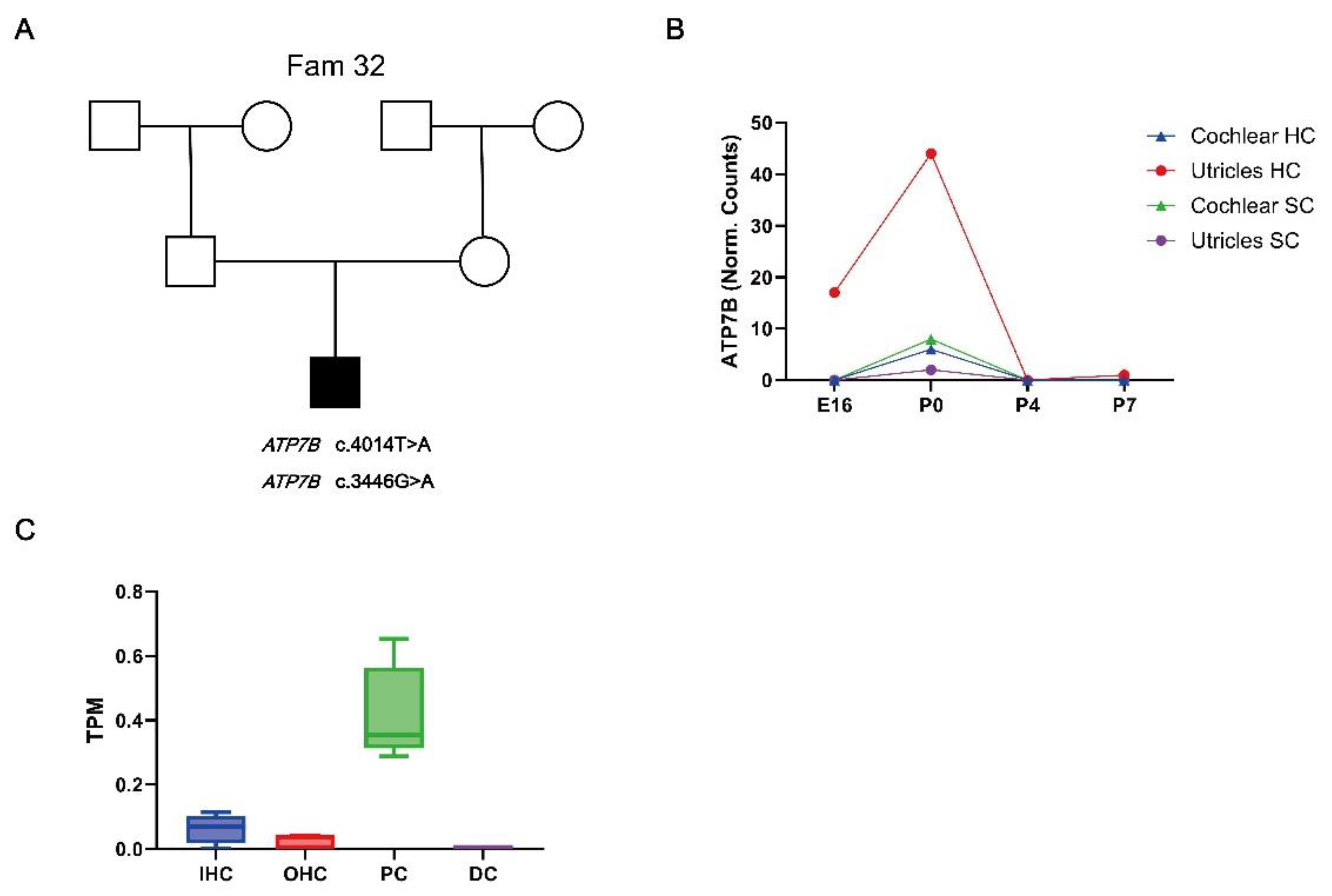
RNA Expression Profiling of ATP7B in Mice Cochlear Tissues
Immunofluorescence of ATP7B Expression in Cochlear Tissue
3. Results
The Pedigree Analysis of the Families in this Cohort
Bioinformatic and Molecular Analysis
3.2. Figures, Tables and Schemes
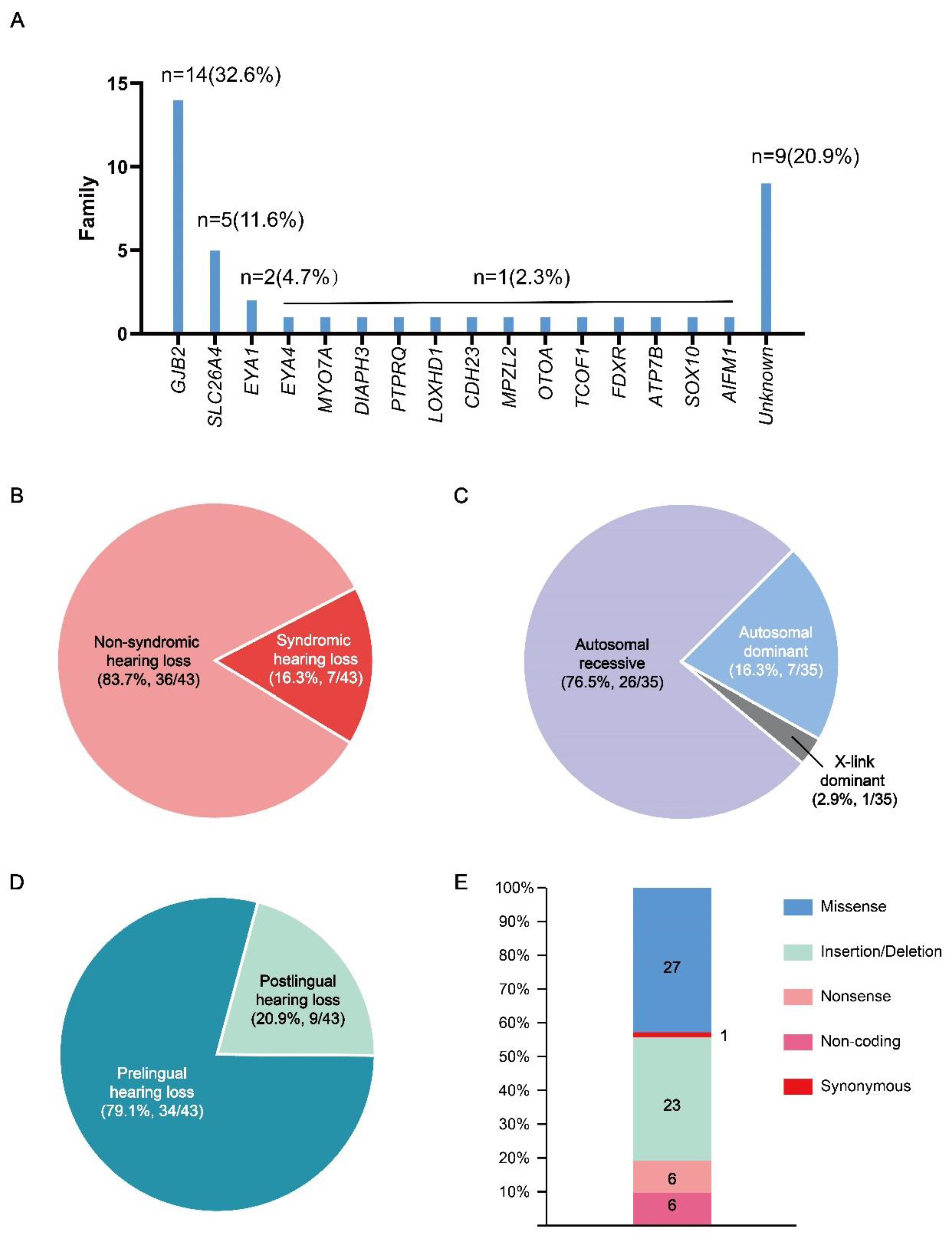
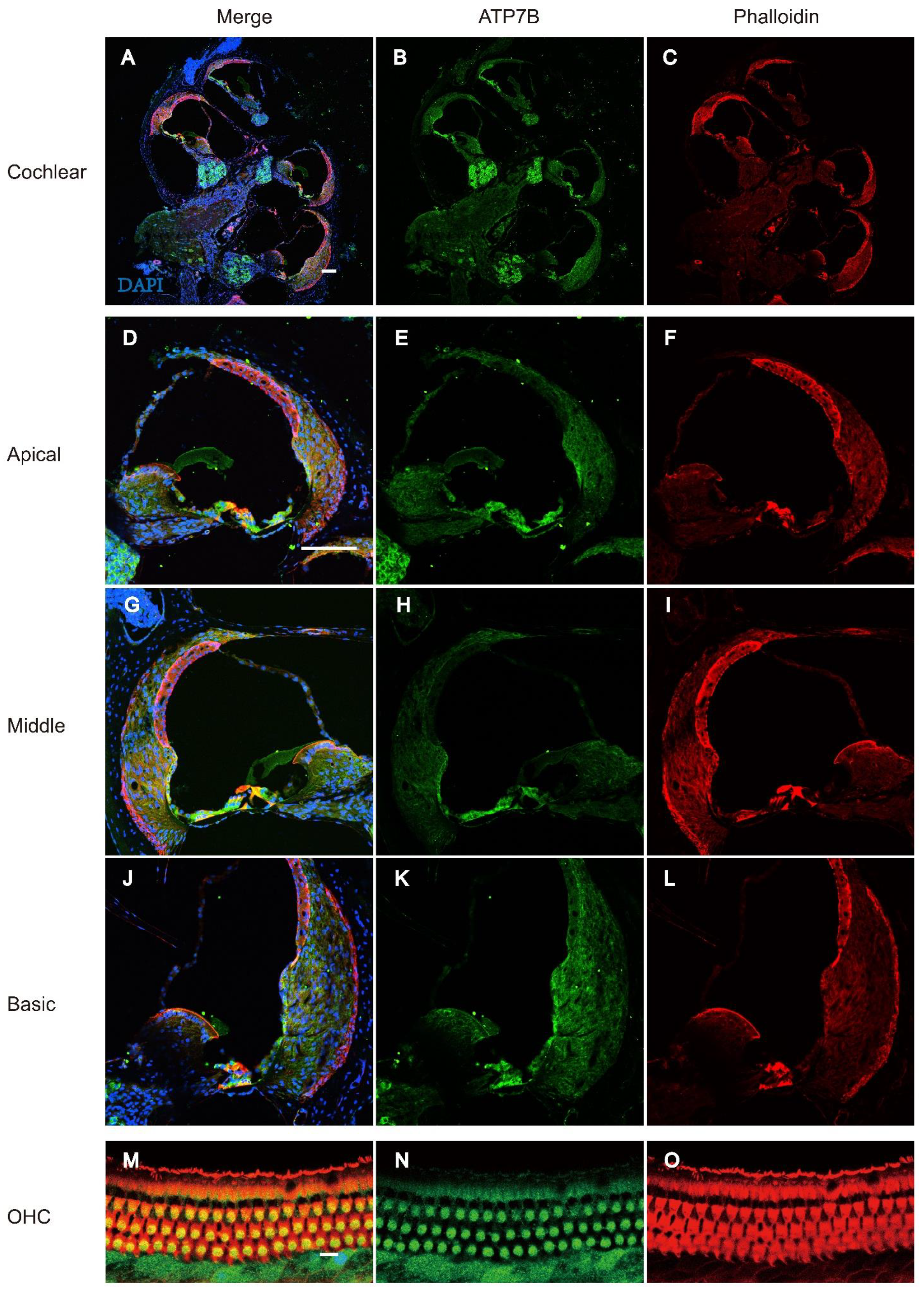
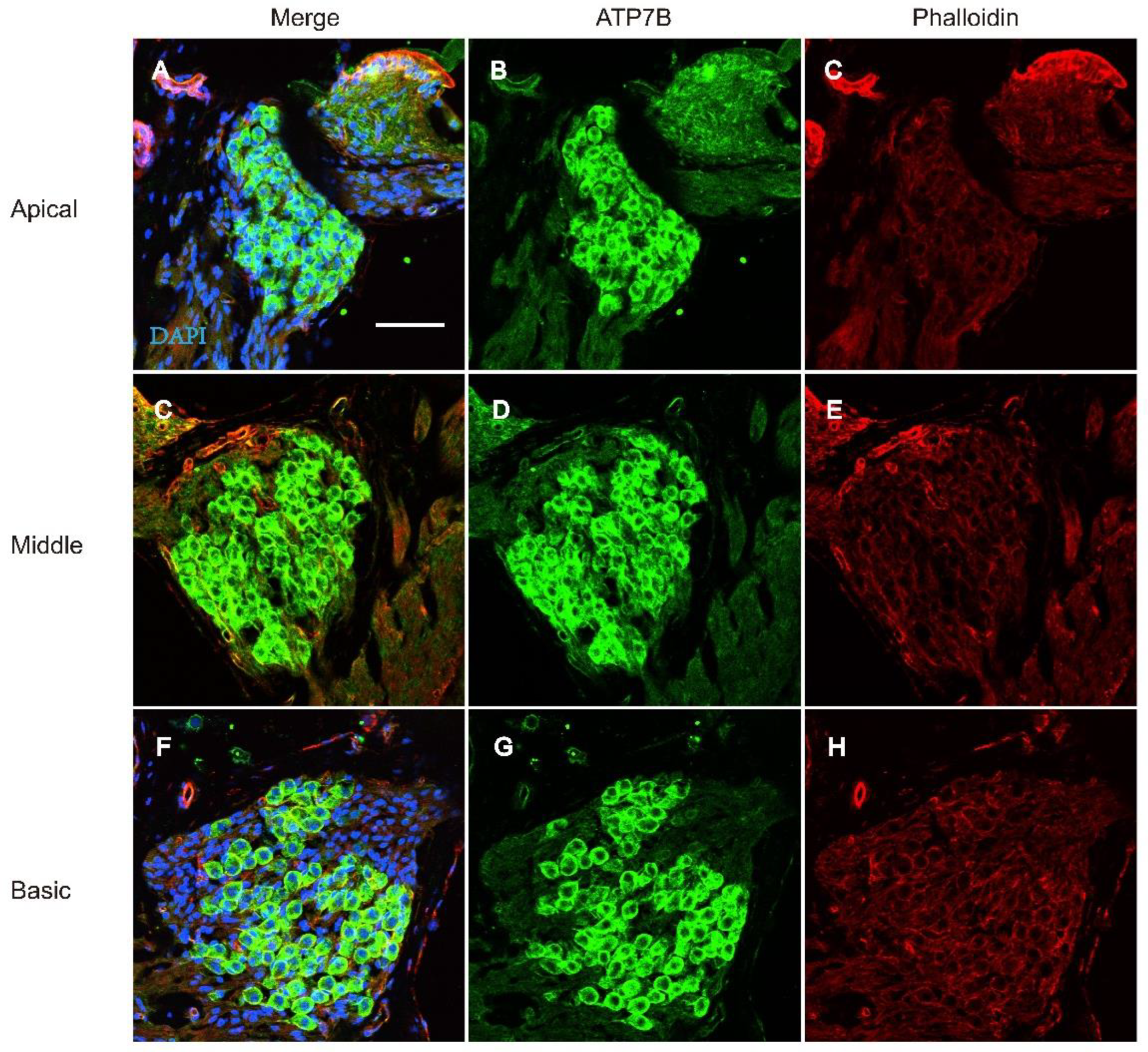
4. Discussion
Author Contributions
Funding
Informed Consent Statement
Conflicts of Interest
References
- Korver, A.M. , et al., Congenital hearing loss. Nat Rev Dis Primers, 2017. 3: p. 16094.
- Cunningham, L.L. and D.L. Tucci, Hearing Loss in Adults. N Engl J Med, 2017. 377(25): p. 2465-2473.
- Wonkam, A.; Adadey, S.M.; Schrauwen, I.; Aboagye, E.T.; Wonkam-Tingang, E.; Esoh, K.; Popel, K.; Manyisa, N.; Jonas, M.; Dekock, C.; et al. Exome sequencing of families from Ghana reveals known and candidate hearing impairment genes. Commun. Biol. 2022, 5, 369. [Google Scholar] [CrossRef] [PubMed]
- Van Camp, G.; Smith, R.J.H. Hereditary Hearing Loss Homepage. 2018. Available online: http://hereditaryhearingloss.org (accessed on 17 December 2021).
- Jin, Y.; Liu, X.; Chen, S.; Xiang, J.; Peng, Z.; Sun, Y. Analysis of the Results of Cytomegalovirus Testing Combined with Genetic Testing in Children with Congenital Hearing Loss. J. Clin. Med. 2022, 11, 5335. [Google Scholar] [CrossRef] [PubMed]
- Dai, P.; Yu, F.; Han, B.; Liu, X.; Wang, G.; Li, Q.; Yuan, Y.; Liu, X.; Huang, D.; Kang, D.; et al. GJB2 mutation spectrum in 2063 Chinese patients with nonsyndromic hearing impairment. J. Transl. Med. 2009, 7, 26–26. [Google Scholar] [CrossRef]
- Dai, P.; Yu, F.; Han, B.; Yuan, Y.; Li, Q.; Wang, G.; Liu, X.; He, J.; Huang, D.; Kang, D.; et al. The prevalence of the 235delC GJB2 mutation in a Chinese deaf population. Anesthesia Analg. 2007, 9, 283–289. [Google Scholar] [CrossRef]
- Park, J.H.; Kim, A.R.; Han, J.H.; Kim, S.D.; Kim, S.H.; Koo, J.-W.; Oh, S.H.; Choi, B.Y. Outcome of Cochlear Implantation in Prelingually Deafened Children According to Molecular Genetic Etiology. Ear Hear. 2017, 38, e316–e324. [Google Scholar] [CrossRef]
- Carlson, R.J.; Walsh, T.; Mandell, J.B.; Aburayyan, A.; Lee, M.K.; Gulsuner, S.; Horn, D.L.; Ou, H.C.; Sie, K.C.Y.; Mancl, L.; et al. Association of Genetic Diagnoses for Childhood-Onset Hearing Loss With Cochlear Implant Outcomes. Arch. Otolaryngol. Neck Surg. 2023, 149, 212–222. [Google Scholar] [CrossRef] [PubMed]
- Tropitzsch, A.; Schade-Mann, T.; Gamerdinger, P.; Dofek, S.; Schulte, B.; Schulze, M.; Fehr, S.; Biskup, S.; Haack, T.B.; Stöbe, P.; et al. Variability in Cochlear Implantation Outcomes in a Large German Cohort With a Genetic Etiology of Hearing Loss. Ear Hear. 2023, 44, 1464–1484. [Google Scholar] [CrossRef]
- Vivero, R.J.; Fan, K.; Angeli, S.; Balkany, T.J.; Liu, X.Z. Cochlear implantation in common forms of genetic deafness. Int. J. Pediatr. Otorhinolaryngol. 2010, 74, 1107–1112. [Google Scholar] [CrossRef]
- Lin, P.-H.; Wu, H.-P.; Wu, C.-M.; Chiang, Y.-T.; Hsu, J.S.; Tsai, C.-Y.; Wang, H.; Tseng, L.-H.; Chen, P.-Y.; Yang, T.-H.; et al. Cochlear Implantation Outcomes in Patients with Auditory Neuropathy Spectrum Disorder of Genetic and Non-Genetic Etiologies: A Multicenter Study. Biomedicines 2022, 10, 1523. [Google Scholar] [CrossRef]
- Xing, G.; Yao, J.; Wu, B.; Liu, T.; Wei, Q.; Liu, C.; Lu, Y.; Chen, Z.; Zheng, H.; Yang, X.; et al. Identification of OSBPL2 as a novel candidate gene for progressive nonsyndromic hearing loss by whole-exome sequencing. Anesthesia Analg. 2015, 17, 210–218. [Google Scholar] [CrossRef]
- Morgan, A.; Vuckovic, D.; Krishnamoorthy, N.; Rubinato, E.; Ambrosetti, U.; Castorina, P.; Franzè, A.; Vozzi, D.; La Bianca, M.; Cappellani, S.; et al. Next-generation sequencing identified SPATC1L as a possible candidate gene for both early-onset and age-related hearing loss. Eur. J. Hum. Genet. 2019, 27, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Bharadwaj, T.; Schrauwen, I.; Rehman, S.; Liaqat, K.; Acharya, A.; Giese, A.P.J.; Nouel-Saied, L.M.; Nasir, A.; Everard, J.L.; Pollock, L.M.; et al. ADAMTS1, MPDZ, MVD, and SEZ6: candidate genes for autosomal recessive nonsyndromic hearing impairment. Eur. J. Hum. Genet. 2022, 30, 22–33. [Google Scholar] [CrossRef]
- Liu, M.; Liang, Y.; Huang, B.; Sun, J.; Chen, K. Report of rare and novel mutations in candidate genes in a cohort of hearing-impaired patients. Mol. Genet. Genom. Med. 2022, 10, e1887. [Google Scholar] [CrossRef]
- Liu, X.-Z.; Jin, Y.; Chen, S.; Xu, K.; Xie, L.; Qiu, Y.; Wang, X.-H.; Sun, Y.; Kong, W.-J. F-Actin Dysplasia Involved in Organ of Corti Deformity in Gjb2 Knockdown Mouse Model. Front. Mol. Neurosci. 2022, 14, 808553. [Google Scholar] [CrossRef]
- Wang, Y.; Jin, Y.; Zhang, Q.; Xiong, Y.; Gu, X.; Zeng, S.; Chen, W. Research progress in delineating the pathological mechanisms of GJB2-related hearing loss. Front. Cell. Neurosci. 2023, 17, 1208406. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Ni, W.; Chen, W.-J.; Wan, B.; Zhao, G.-X.; Shi, Z.-Q.; Zhang, Y.; Wang, N.; Yu, L.; Xu, J.-F.; et al. Spectrum and Classification of ATP7B Variants in a Large Cohort of Chinese Patients with Wilson's Disease Guides Genetic Diagnosis. Theranostics 2016, 6, 638–649. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhou, W.; Huang, Y.; Yin, H.; Jin, Y.; Jia, Z.; Zhang, A.; Liu, Z.; Zheng, B. Presumed missense and synonymous mutations in ATP7B gene cause exon skipping in Wilson disease. Liver Int. 2018, 38, 1504–1513. [Google Scholar] [CrossRef] [PubMed]
- Xiang, J.; Jin, Y.; Song, N.; Chen, S.; Shen, J.; Xie, W.; Sun, X.; Peng, Z.; Sun, Y. Comprehensive genetic testing improves the clinical diagnosis and medical management of pediatric patients with isolated hearing loss. BMC Med Genom. 2022, 15, 1–10. [Google Scholar] [CrossRef]
- Ma, J.; Ma, X.; Lin, K.; Huang, R.; Bi, X.; Ming, C.; Li, L.; Li, X.; Li, G.; Zhao, L.; et al. Genetic screening of a Chinese cohort of children with hearing loss using a next-generation sequencing panel. Hum. Genom. 2023, 17, 1–10. [Google Scholar] [CrossRef]
- Wu, J.; Cao, Z.; Su, Y.; Wang, Y.; Cai, R.; Chen, J.; Gao, B.; Han, M.; Li, X.; Zhang, D.; et al. Molecular diagnose of a large hearing loss population from China by targeted genome sequencing. J. Hum. Genet. 2022, 67, 643–649. [Google Scholar] [CrossRef]
- nbsp; Wu, C. -C.; Tsai, C.-Y.; Lin, Y.-H.; Chen, P.-Y.; Lin, P.-H.; Cheng, Y.-F.; Wu, C.-M.; Lin, Y.-H.; Lee, C.-Y.; Erdenechuluun, J.; et al. Genetic Epidemiology and Clinical Features of Hereditary Hearing Impairment in the Taiwanese Population. Genes 2019, 10, 772. [Google Scholar] [CrossRef]
- Chen, S.; Jin, Y.; Xie, L.; Xie, W.; Xu, K.; Qiu, Y.; Bai, X.; Zhang, H.-M.; Liu, X.-Z.; Wang, X.-H.; et al. A Novel Spontaneous Mutation of the SOX10 Gene Associated with Waardenburg Syndrome Type II. Neural Plast. 2020, 2020, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Liu X, Chen S, Sun Y, Kong W. Lin Chuang Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 2021;35(3):229-237. [CrossRef]
- Shi, X.; Liu, X.; Zong, Y.; Zhao, Z.; Sun, Y. Novel compound heterozygous variants in MARVELD2 causing autosomal recessive hearing loss in two Chinese families. Mol. Genet. Genom. Med. 2024, 12, e2502. [Google Scholar] [CrossRef] [PubMed]
- Liu J, Ding Y, Hu Y. Lin Chuang Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 2022;36(1):27-31. [CrossRef]
- Xia, H.; Hu, P.; Yuan, L.; Xiong, W.; Xu, H.; Yi, J.; Yang, Z.; Deng, X.; Guo, Y.; Deng, H. A homozygous MYO7A mutation associated to Usher syndrome and unilateral auditory neuropathy spectrum disorder. Mol. Med. Rep. 2017, 16, 4241–4246. [Google Scholar] [CrossRef]
- Jin, Y.; Liu, X.-Z.; Xie, L.; Xie, W.; Chen, S.; Sun, Y. Targeted Next-Generation Sequencing Identified Novel Compound Heterozygous Variants in the PTPRQ Gene Causing Autosomal Recessive Hearing Loss in a Chinese Family. Front. Genet. 2022, 13, 884522. [Google Scholar] [CrossRef]
- Liu, X.; Zhao, Z.; Shi, X.; Zong, Y.; Sun, Y. The Effects of Viral Infections on the Molecular and Signaling Pathways Involved in the Development of the PAOs. Viruses 2024, 16, 1342. [Google Scholar] [CrossRef]
- Corriols-Noval, P.; Simón, E.C.L.; Cadiñanos, J.; Diñeiro, M.; Capín, R.; Aguado, R.G.; Marcos, M.C.; Angulo, C.M.; Farpón, R.C. Clinical Impact of Genetic Diagnosis of Sensorineural Hearing Loss in Adults. Otol. Neurotol. 2022, 43, 1125–1136. [Google Scholar] [CrossRef]
- Sloan-Heggen, C.M.; Bierer, A.O.; Shearer, A.E.; Kolbe, D.L.; Nishimura, C.J.; Frees, K.L.; Ephraim, S.S.; Shibata, S.B.; Booth, K.T.; Campbell, C.A.; et al. Comprehensive genetic testing in the clinical evaluation of 1119 patients with hearing loss. Hum. Genet. 2016, 135, 441–450. [Google Scholar] [CrossRef]
- Liu, C.; Huang, Y.; Zhang, Y.; Ding, H.; Yu, L.; Wang, A.; Wang, Y.; Zeng, Y.; Liu, L.; Liu, Y.; et al. Next-generation sequencing facilitates genetic diagnosis and improves the management of patients with hearing loss in clinical practice. Int. J. Pediatr. Otorhinolaryngol. 2022, 161, 111258. [Google Scholar] [CrossRef]
- Ghezzi, D.; Sevrioukova, I.; Invernizzi, F.; Lamperti, C.; Mora, M.; D'Adamo, P.; Novara, F.; Zuffardi, O.; Uziel, G.; Zeviani, M. Severe X-Linked Mitochondrial Encephalomyopathy Associated with a Mutation in Apoptosis-Inducing Factor. Am. J. Hum. Genet. 2010, 86, 639–649. [Google Scholar] [CrossRef]
- Wooton-Kee, C.R. Therapeutic implications of impaired nuclear receptor function and dysregulated metabolism in Wilson's disease. Pharmacol. Ther. 2023, 251, 108529. [Google Scholar] [CrossRef] [PubMed]
- Huster, D.; Kühne, A.; Bhattacharjee, A.; Raines, L.; Jantsch, V.; Noe, J.; Schirrmeister, W.; Sommerer, I.; Sabri, O.; Berr, F.; et al. Diverse Functional Properties of Wilson Disease ATP7B Variants. Gastroenterology 2012, 142, 947–956. [Google Scholar] [CrossRef] [PubMed]
- Collins, C.J.; Yi, F.; Dayuha, R.; Duong, P.; Horslen, S.; Camarata, M.; Coskun, A.K.; Houwen, R.H.; Pop, T.L.; Zoller, H.; et al. Direct Measurement of ATP7B Peptides Is Highly Effective in the Diagnosis of Wilson Disease. Gastroenterology 2021, 160, 2367–2382. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Ni, W.; Chen, W.-J.; Wan, B.; Zhao, G.-X.; Shi, Z.-Q.; Zhang, Y.; Wang, N.; Yu, L.; Xu, J.-F.; et al. Spectrum and Classification of ATP7B Variants in a Large Cohort of Chinese Patients with Wilson's Disease Guides Genetic Diagnosis. Theranostics 2016, 6, 638–649. [Google Scholar] [CrossRef] [PubMed]
- Hua, R.; Hua, F.; Jiao, Y.; Pan, Y.; Yang, X.; Peng, S.; Niu, J. Mutational analysis of ATP7B in Chinese Wilson disease patients. . 2016, 8, 2851–61. [Google Scholar]
- Zhang, S.; Yang, W.; Li, X.; Pei, P.; Dong, T.; Yang, Y.; Zhang, J. Clinical and genetic characterization of a large cohort of patients with Wilson’s disease in China. Transl. Neurodegener. 2022, 11, 1–11. [Google Scholar] [CrossRef]
- Bandmann, O.; Weiss, K.H.; Kaler, S.G. Wilson's disease and other neurological copper disorders. Lancet Neurol. 2015, 14, 103–113. [Google Scholar] [CrossRef]
- Xie, J.-J.; Wu, Z.-Y. Wilson’s Disease in China. Neurosci. Bull. 2017, 33, 323–330. [Google Scholar] [CrossRef]
- Wungjiranirun, M.; Sharzehi, K. Wilson's Disease. Semin. Neurol. 2023, 43, 626–633. [Google Scholar] [CrossRef]
- Gouider-Khouja, N. Wilson's disease. Park. Relat. Disord. 2009, 15, S126–S129. [Google Scholar] [CrossRef]
| Family | Gene | Nucleotide change | Protein change | Inh | GT | Mutation type |
|---|---|---|---|---|---|---|
| Fam1 | GJB2 | c.235del | p.Leu79CysfsTer3 | AR | Hom | Deletion |
| Fam2 | GJB2 | c.99del | p.Met34Ter | AR | Het | Deletion |
| c.299_300del | p.His100ArgfsTer14 | AR | Het | Deletion | ||
| Fam3 | GJB2 | c.235del | p.Leu79CysfsTer3 | AR | Hom | Deletion |
| Deletion | ||||||
| Fam4 | GJB2 | c.109G>A | p.Val37Ile | AR | Hom | Missense |
| Fam5 | GJB2 | c.139G>T | p.Glu47Ter | AR | Het | Nonsense |
| c.176_191del | p.Gly59AlafsTer18 | AR | Het | Deletion | ||
| Fam6 | GJB2 | c.109G>A | p.Val37Ile | AR | Hom | Missense |
| Fam7 | GJB2 | c.235del | p.Leu79CysfsTer3 | AR | Hom | Deletion |
| Fam8 | GJB2 | c.235del | p.Leu79CysfsTer3 | AR | Het | Deletion |
| c.176_191del | p.Gly59AlafsTer18 | AR | Het | Deletion | ||
| Fam9 | GJB2 | c.109G>A | p.Val37Ile | AR | Hom | Missense |
| Fam10 | GJB2 | c.109G>A | p.Val37Ile | AR | Hom | Missense |
| Fam11 | GJB2 | c.235del | p.Leu79CysfsTer3 | AR | Het | Deletion |
| c.299_300del | p.His100ArgfsTer14 | AR | Het | Deletion | ||
| Fam12 | GJB2 | c.235del | p.Leu79CysfsTer3 | AR | Hom | Deletion |
| Deletion | ||||||
| Fam13 | GJB2 | c.257C>G | p.Thr86Arg | AR | Het | Missense |
| c.109G>A | p.Val37Ile | AR | Het | Missense | ||
| Fam14 | GJB2 | c.299_300delAT | p.His100ArgfsTer14 | AR | Het | Deletion |
| c.176_191del | p.Gly59AlafsTer18 | AR | Het | Deletion | ||
| Fam15 | SLC26A4 | c.919-2A>G | - | AR | Het | Noncoding |
| c.281C>T | p.Thr94Ile | AR | Het | Missense | ||
| Fam16 | SLC26A4 | c.1594A>C | p.Ser532Arg | AR | Het | Missense |
| c.2168A>G | p.His723Arg | AR | Het | Missense | ||
| Fam17 | SLC26A4 | c.919-2A>G | - | AR | Hom | Noncoding |
| Fam18 | SLC26A4 | c.2009T>C | p.Val670Ala | AR | Hom | Missense |
| Fam19 | SLC26A4 | c.919-2A>G | - | AR | Het | Noncoding |
| c.2168A>G | p.His723Arg | AR | Het | Missense | ||
| Fam20 | MYO7A | c.1183C>T | p.Arg395Cys | AR | Het | Missense |
| c.3696_3706del | p.Arg1232SerTer72 | AR | Het | Deletion | ||
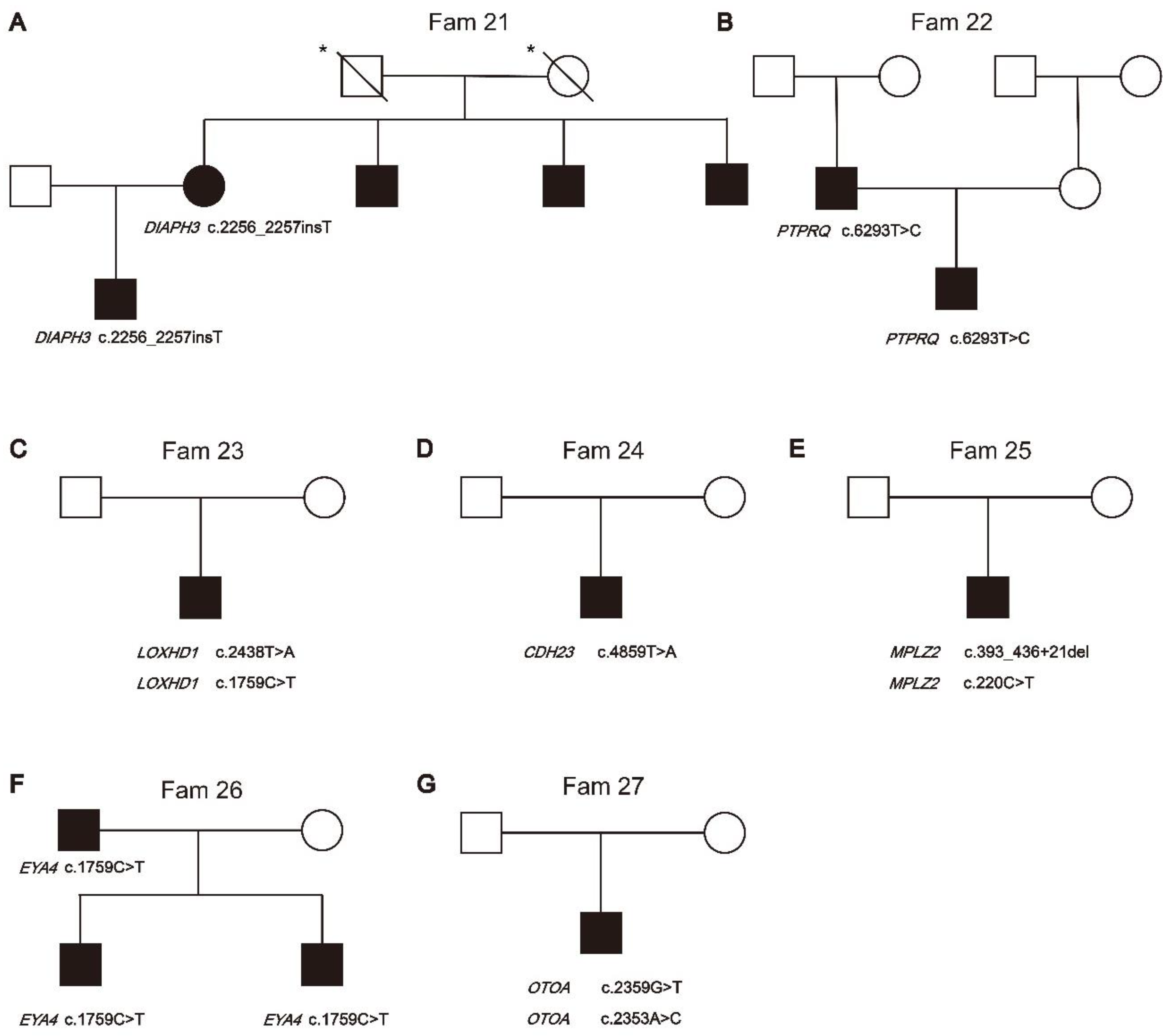
| Fam21 | DIAPH3 | c.2256_2257insT | p.Ser752SerfsTer12 | AD | Het | Deletion |
| Insertion | ||||||
| Fam22 | PTPRQ | c.6293T>C | p.Leu2098Ser | AD | Het | Missense |
| Fam23 | LOXHD1 | c.2438T>A | p.Leu813Ter | AR | Het | Nonsense |
| c.1759C>T | p.Arg587Trp | AR | Het | Missense | ||
| Fam24 | CDH23 | c.4859T>A | p.Val1620Glu | AR | Hom | Missense |
| Fam25 | MPZL2 | c.220C>T | p.Gln74Ter | AR | Het | Nonsense |
| c.393_436+21del | - | AR | Het | Noncoding | ||
| Fam26 | EYA4 | c.1759C>T | p.Arg587Ter | AD | Het | Nonsense |
| Fam27 | OTOA | c.2359G>T | p.Glu787Ter | AR | Het | Nonsense |
| c.2353A>C | p.Thr785Pro | AR | Het | Missense |
| Family | Gene | Nucleotide change | Protein change | Inh | GT | Mutation type |
|---|---|---|---|---|---|---|
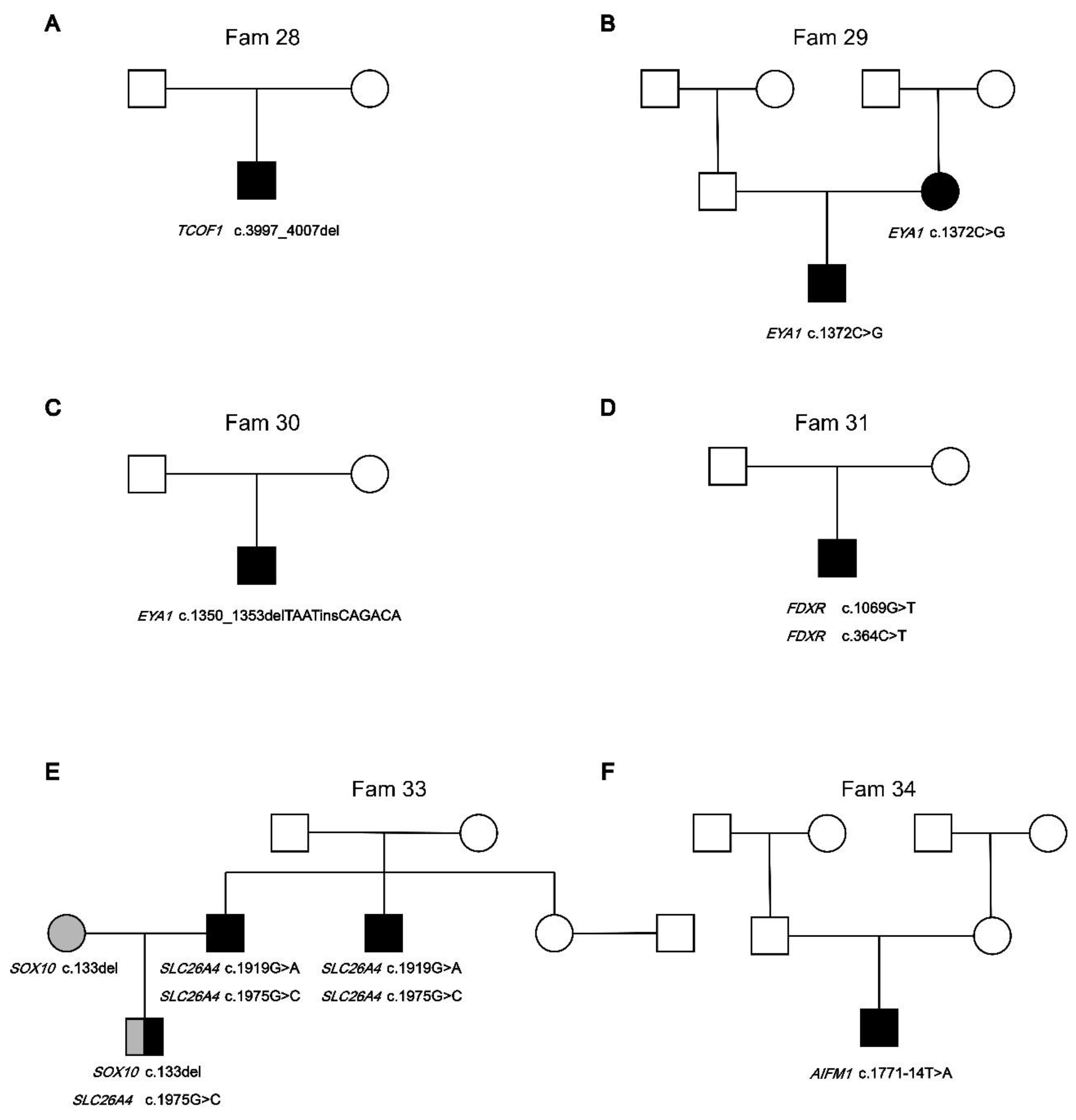
| Fam28 | TCOF1 | c.3997_4007del | p.Ser1333GlnfsTer16 | AD | Het | Deletion |
| Fam29 | EYA1 | c.1372C>G | p.Pro458Ala | AD | Het | Missense |
| Fam30 | EYA1 | c.1350_1353delTAATinsCAGACA | p.Asn451ArgfsTer18 | AD | Het | Deletion/Insertion |
| Fam31 | FDXR | c.1069G>T | p.Val357Leu | AR | Het | Missense |
| FDXR | c.364C>T | p.Arg122Cys | AR | Het | Missense | |
| Fam33 | SOX10 | C.133del | p.Gly38AlafsTer71 | AD | Het | Deletion |
| SLC26A4 | c.1975G>C | p.Val659Leu | AR | Het | Missense | |
| SLC26A4 | c.1919G>A | p.Trp640Ter | AR | Het | Nonsense | |
| Fam34 | AIFM1 | c.1771-14T>A | - | X-link | Het | Noncoding |
| Family | Gene | Nucleotide change | Protein change | Inh | GT | Mutation type |
|---|---|---|---|---|---|---|
| Fam32 | ATP7B | c.4014T>A | p.Ile1338Ile | AR | Het | Synonymous |
| ATP7B | c.3446G>A | p.Gly1149Glu | AR | Het | Missense |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





