Submitted:
26 October 2024
Posted:
28 October 2024
You are already at the latest version
Abstract
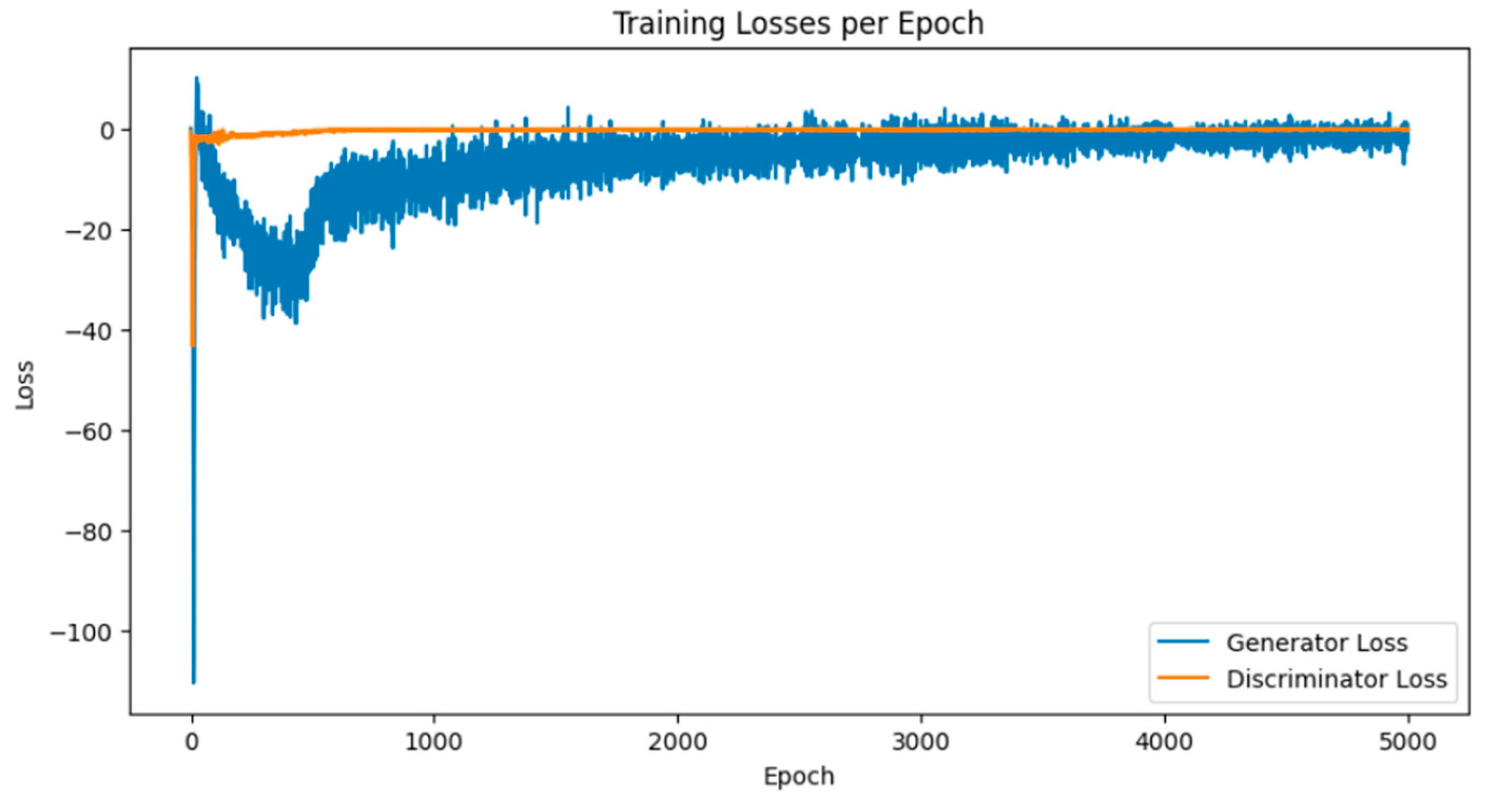
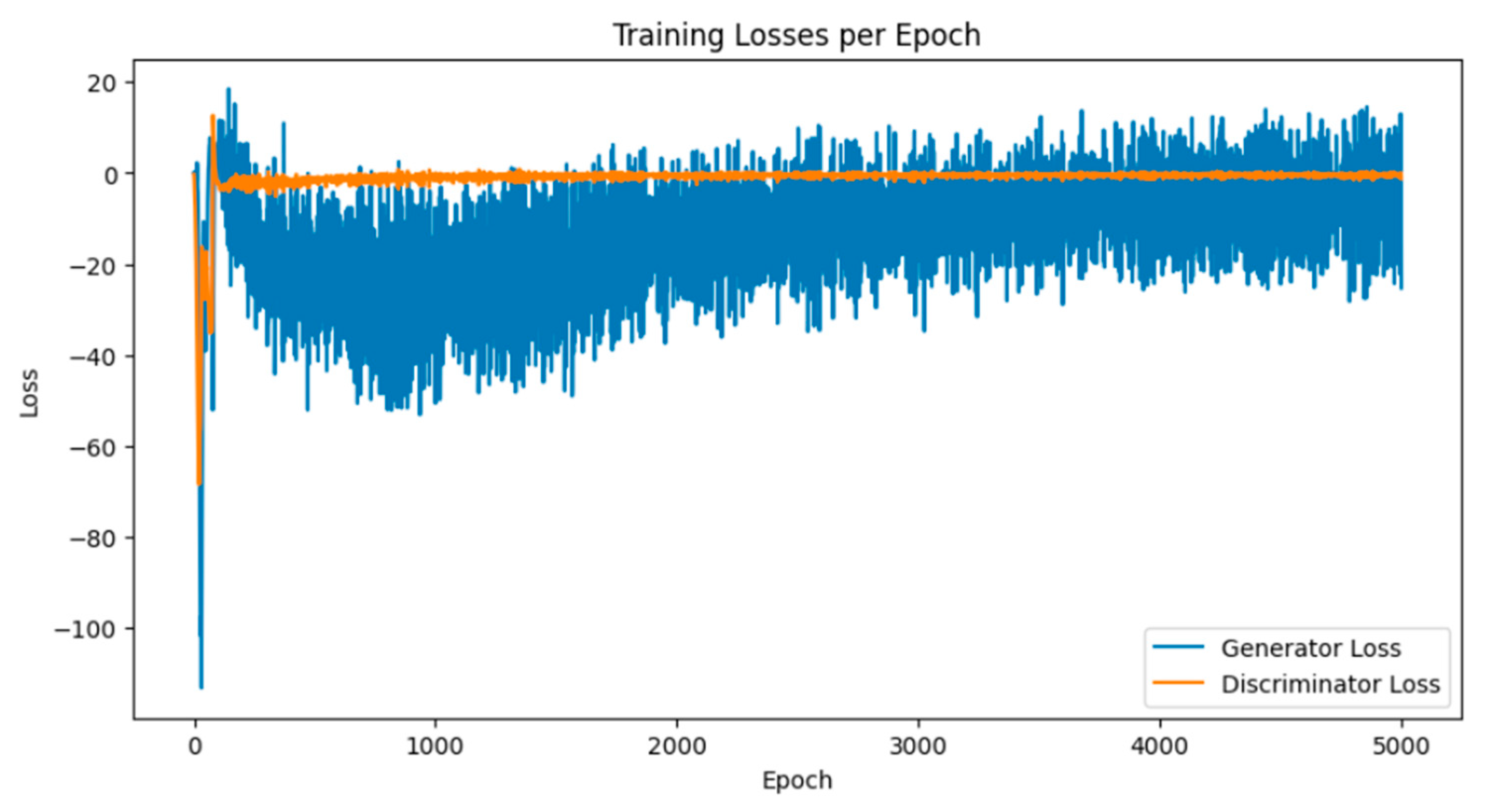
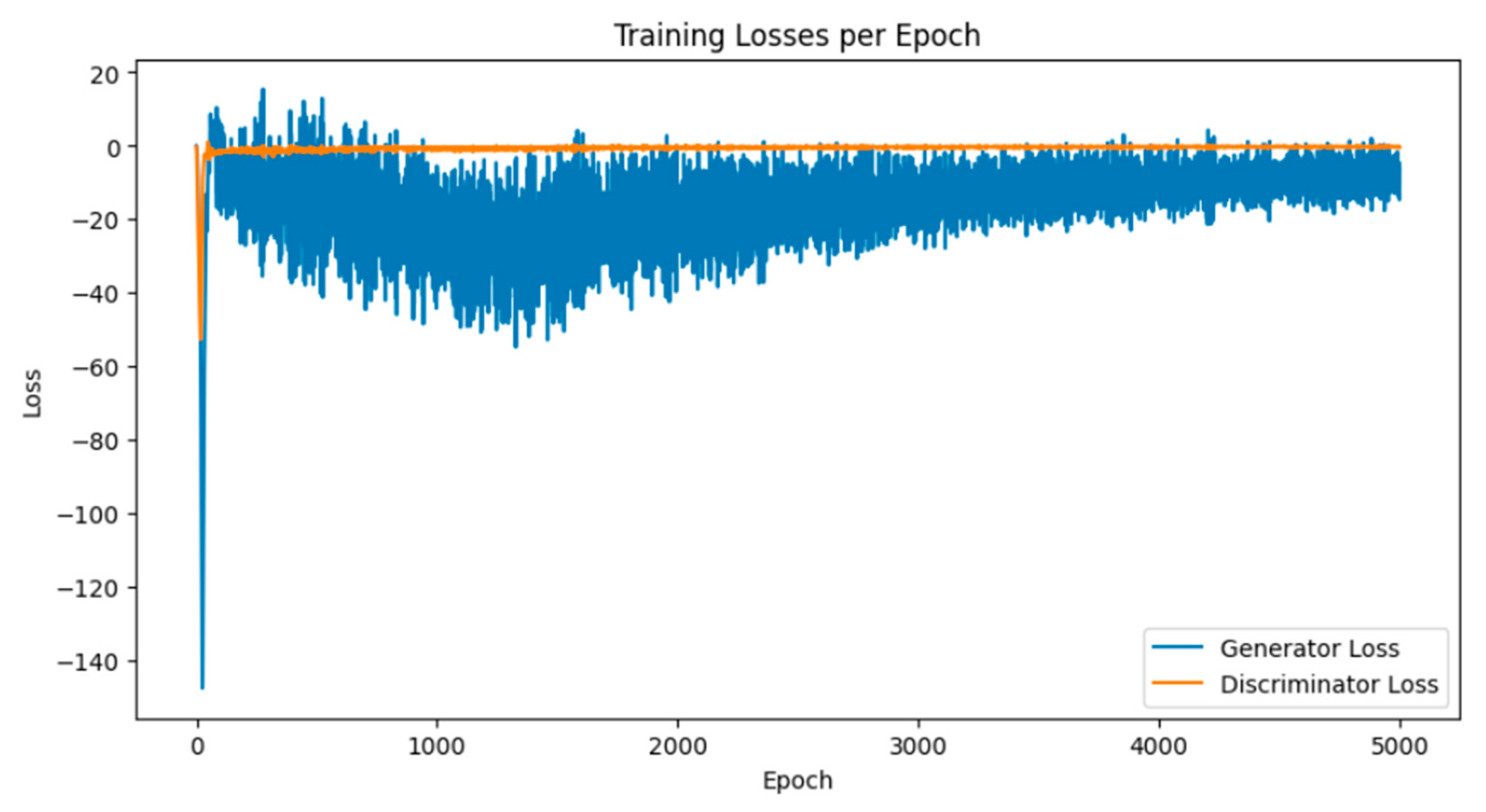
Breast cancer is among the most prevalent cancers in Indonesia. One of the methods for early detection of breast cancer is utilizing ultrasound images to classify breast conditions into normal, benign tumors, or cancer. The advent of deep learning technology facilitates the image analysis process, such as the use of Transfer Learning Convolutional Neural Networks (CNNs). Generally, CNN models require large and balanced datasets to perform well in classification tasks. However, medical datasets like breast ultrasound images tend to be limited and imbalanced. The Wasserstein GAN (WGAN) is a generative data augmentation method capable of producing synthetic images by learning the pattern of the distribution of real image data. The implementation of the Wasserstein distance results in the training process of WGAN demonstrating stability from epochs 3000, 2500, and 3000 out of a total of 5000 epochs. The quality of the synthetic images improves with an increasing number of training iterations. By using WGAN, all of the evaluation metrics of each classifier are increasing, with the best accuracy score is achieved by VGG16 model with 83.33% accuracy.
Keywords:
1. Introduction
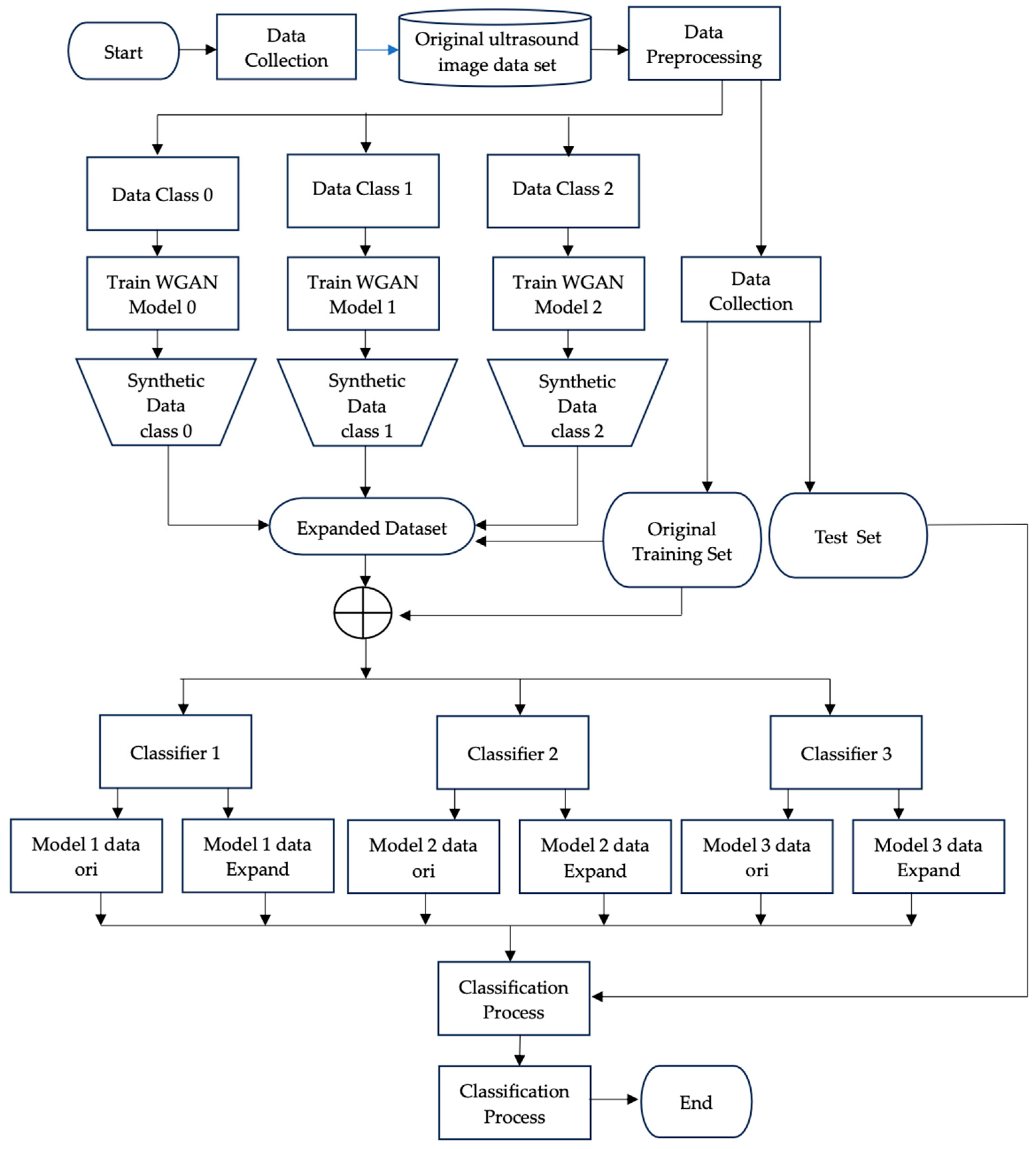
2. Materials and Methods
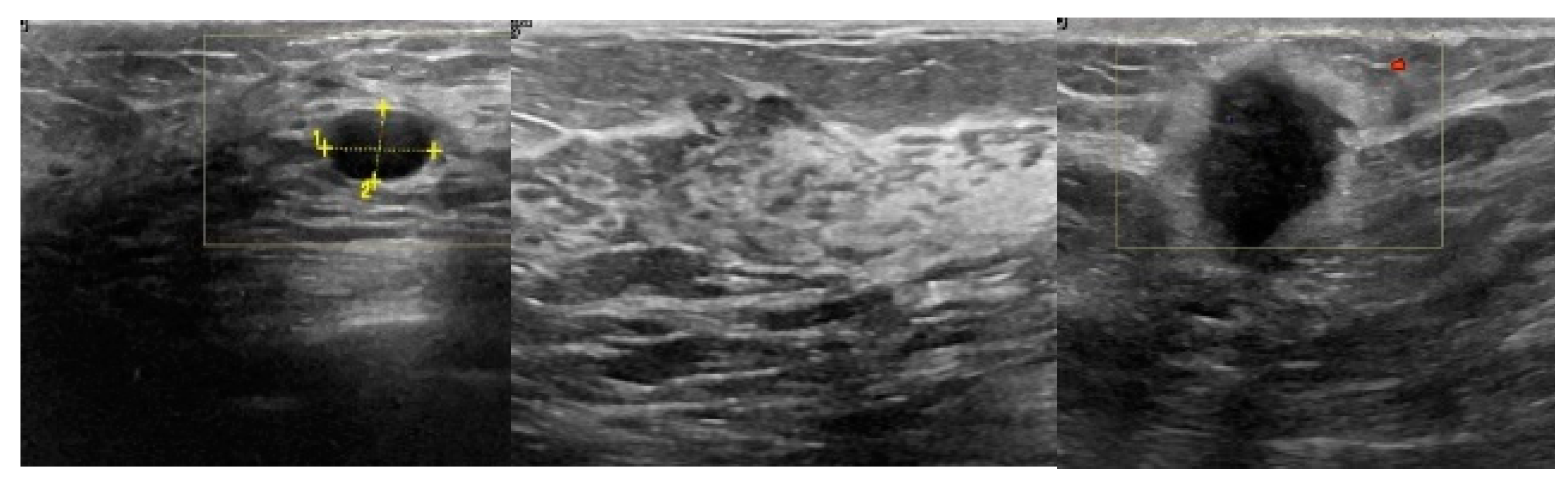
2.1. Breast Ultrasound Image Data Acquisition
2.2. Pre-Processing
-
Normalization is the process of adjusting the pixel intensity range in an image to achieve a uniform distribution. This helps improve the convergence of the optimization algorithm and reduces scale differences that can affect the model's performance [44]. The normalization formula is shown in Equation (1).where:
- Pixel_original refers to the pixel intensity value in the original image.
- The value 127.5 is derived from the maximum pixel intensity in the original image, which is 255, divided by 2.
- Pixel_normalized represents the normalized pixel intensity value.
- Conversion to grayscale is carried out to ensure that breast ultrasound images do not contain any colors originating from external factors such as ultrasound equipment, which are not part of the breast tissue ultrasound. The images should be in grayscale format.
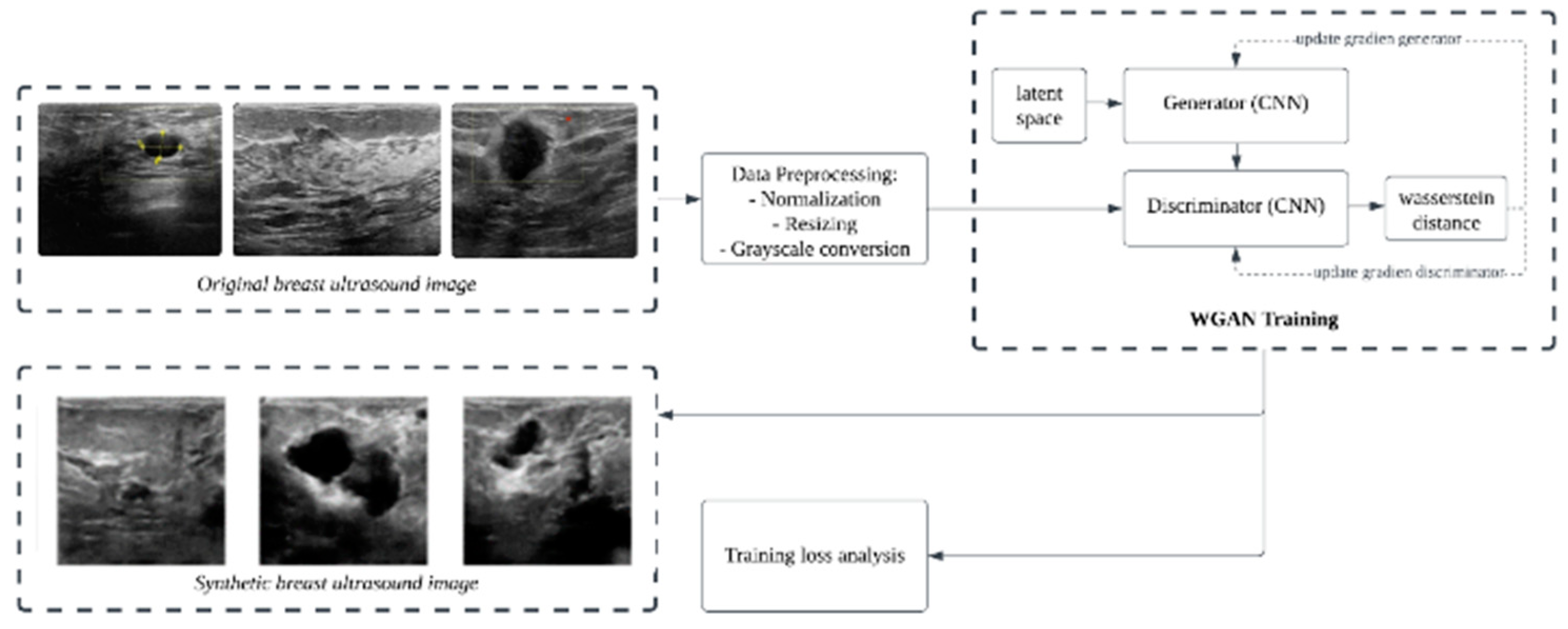
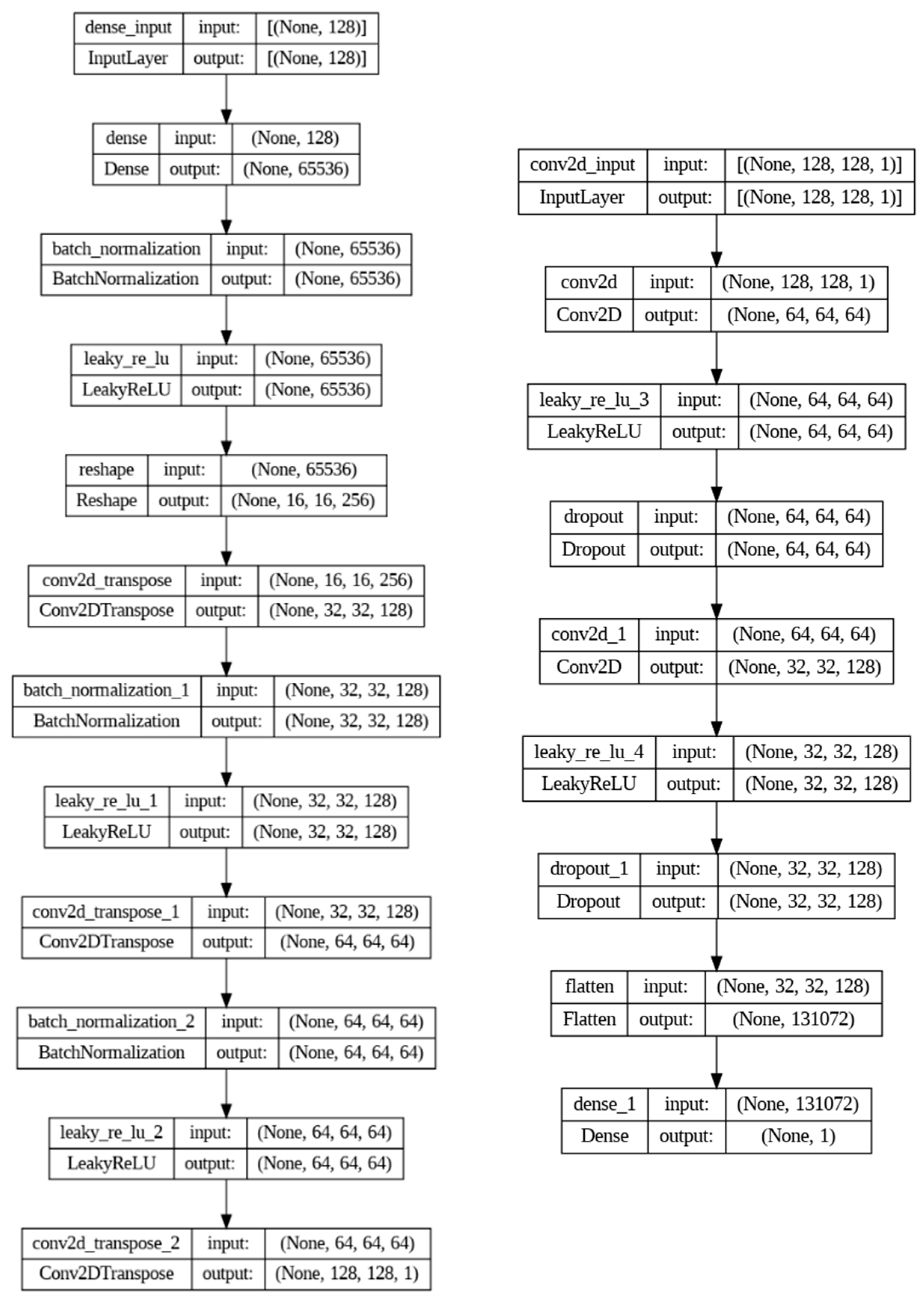
2.3. Wasserstein GAN Training
2.3. Evaluating the Effectiveness of WGAN-Based Augmentation
3. Results and Discussion
3.1. Results of WGAN Training
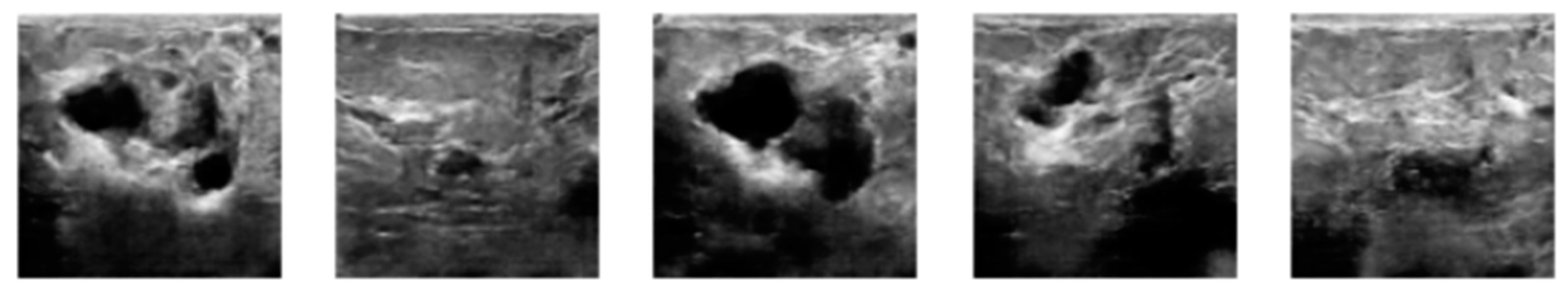
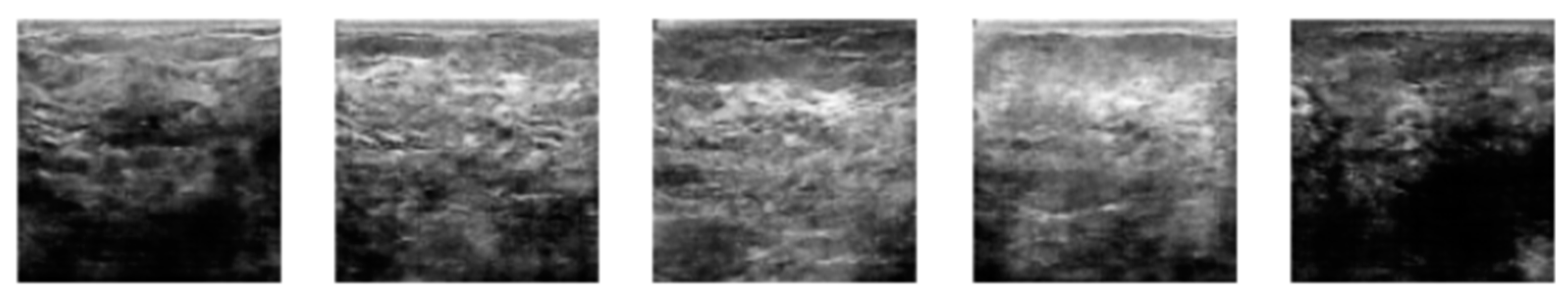
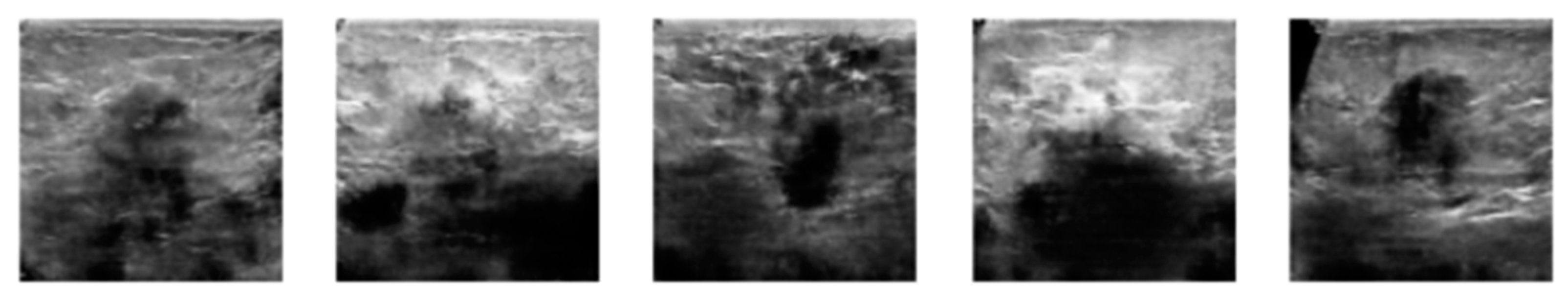
3.2. Image Synthetic Augmented by WGAN
3.3. Prediction Using CNN Classifiers
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hyuna, S.; Jacques, F.; Rebecca, L. S.; Mathieu, L.; Isabelle, S.; Ahmedin, J.; Freddie, B. GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. Global Cancer Statistics 2020 2021, 71, 209–249. [Google Scholar]
- Melina, A.; Eileen, M.; Harriet, R.; Allini, M.; Deependra, S.; Mathieu, L.; Jerome, V.; Julie, R. G.; Fatima, C.; Sabine, S.; Isabelle, S. Current and future burden of breast cancer: Global statistics for 2020 and 2040. The Breast, 2022, 66, 15–23. [Google Scholar]
- Haq, I. U.; Ali, H.; Wang, H. Y.; Cui, L.; Feng, J. BTS-GAN: Computer-aided segmentation system for breast tumor using MRI and conditional adversarial networks. Engineering Science and Technology, an International Journal, 2022, 36, 101154. [Google Scholar] [CrossRef]
- Li, J.; Guan, X.; Fan, Z.; Ching, L. M.; Li, Y.; Wang, X.; Cao, W.M.; Liu, D. X. Non-invasive biomarkers for early detection of breast cancer. Cancer MDPI, 2020, 12, 2767. [Google Scholar] [CrossRef] [PubMed]
- Ghulam, G.; Anum, S.; Syeda, N. B.; Aqsa, K.; Hina, S.; Sadia, P.; Akkasha, L.; Sajid, M. and Saeed, M. Digital Image Processing for Ultrasound Images: A Comprehensive Review. International Journal of Innovation, Creativity and Change, 2021, 15, 1335–1354. [Google Scholar]
- Alam, N. A.; Khandaker, M. M. U.; Mahbubur, R.; Manu, M. M. R.; Mostofa, K. N. A Novel Automated System to Detect Breast Cancer From Ultrasound Images Using Deep Fused Features With Super Resolution. intelligence-based Medicine, 2024, 10, 100149. [Google Scholar] [CrossRef]
- Rebeca, T.; David, M.; Carlos, I. I.; Rodrigo, A. G.; Fernando, A. V.; Joaquin, L. H. Recent Advances in Artificial Intelligence-Assisted Ultrasound Scanning. Applied Sciences, 2023, 13, 3693. [Google Scholar]
- Yu, W.; Xinke, G.; He, M.; Shouliang, Q.; Guanjing, Z.; Yudong, Y. Deep Learning in Medical Ultrasound Image Analysis: A Review. IEEE Access, 2016, 4, 1–15. [Google Scholar] [CrossRef]
- Alyaa, R. Effects of Artifacts on the Diagnosis of Ultrasound Image. Medico-legal Update, 2021, 21, 327–336. [Google Scholar] [CrossRef]
- Zeyad, O. M.; Latif, A. E. E. Breast Cancer Detection in Ultrasound Imaging. World Journal of Advanced Research and Reviews 2021, 12, 308–314. [Google Scholar] [CrossRef]
- Randall, J. E.; Ryan, M. S.; Alfonso, L. Twelve key challenges in medical machine learning and solutions. Intelligence-Based Medicine, 2022, 6, 100068. [Google Scholar]
- Imane, A.; Ali, I.; Ikram, C. Cost-sensitive Learning for Imbalanced Medical Data: A Review. Artificial Intelligence Review, 2024, 57, 1–72. [Google Scholar]
- Chiranjib, C.; Manojit, B.; Soumen, P.; Sang-soo, L. From machine learning to deep learning: Advances of the recent data-driven paradigm shift in medicine and healthcare. Current Research in Biotechnology, 2024, 7, 100164. [Google Scholar]
- Yang, Y.; Li, R.; Xiang, Y.; Lin, D.; Yan, A.; Chen, W.; Li, Z.; Lai, W.; Wu, X.; Wan, C.; Bai, W.; Huang, X.; Li, Q.; Deng, W.; Liu, X.; Lin, Y.; Yan, P.; Lin, H. Expert Recommendation on Collection, Storage, Annotation, And Management Of Data Related To Medical Artificial Intelligence. Intelligence Medicine, 2023, 3, 144–149. [Google Scholar] [CrossRef]
- Hussain, B. Z.; Andleeb, I.; Ansari, M. S.; Joshi, A. M.; Kanwal, N. Wasserstein GAN based Chest X-Ray Dataset Augmentation for Deep Learning Models: COVID-19 Detection Use-Case. In Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society, EMBS; Institute of Electrical and Electronics Engineers Inc., 2022; pp. 2058–2061. [Google Scholar]
- Haq, D. Z. and Fatichah, C. Ultrasound Image Synthetic Generating Using Deep Convolution Generative Adversarial Network for Breast Cancer Identification. IPTEK The Journal of Technology and Science, 2021, 34, 13–25. [Google Scholar]
- Carriero, A.; Groenhoff, L.; Vologina, E.; Basile, P.; Albera, M. Deep Learning in Breast Cancer Imaging: State of the Art and Recent Advancements in Early 2024. Diagnostics. 2024, 14, 848. [Google Scholar] [CrossRef]
- Nikmah, T. L.; Syafei, R. M.; Anisa, D. N. Inception ResNet v2 for Early Detection of Breast Cancer in Ultrasound Images. Journal of Information System Exploration and Research, 2024, 2, 93–102. [Google Scholar] [CrossRef]
- Gede, I. S. M. D; Eva, Y. P.; Hatta, M.; Ariono, S. New Method For Classification Of Spermatozoa Morphology Abnormalities Based On Macroscopic Video Of Human Semen. 2019 International Seminar on Application for Technology of Information and Communication (iSemantic), 2019; 133–140. [Google Scholar]
- Alessandro, C.; Léon, G.; Elizaveta, V.; Paola, B.; Marco, A. Deep Learning in Breast Cancer Imaging: State of the Art and Recent Advancements in Early 2024. Diagnostics Journal 2024, 14, 848. [Google Scholar]
- Heikal, A.; Amir, E. G.; Samir, E.; Rashad, M. Z. Fine Tuning Deep Learning Models For Breast Tumor Classification. Scientific Reports, 2024, 14, 10753. [Google Scholar] [CrossRef]
- Rguibi, Z.; Hajami, A.; Zitouni, D.; Maleh, Y. and Elqaraoui, A. Medical variational autoencoder and generative adversarial network for medical imaging. Indonesian Journal of Electrical Engineering and Computer Science, 2023, 32, 494–505. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G. E. ImageNet Classification with Deep Convolutional Neural Networks. Communications of the ACM. 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Perez, L.; Wang, L. The Effectiveness of Data Augmentation in Image Classification using Deep Learning. 2017; 1–8. [Google Scholar]
- Shangguan, Z.; Zhao, Y.; Fan, W.; Cao, Z. Dog Image Generation using Deep Convolutional Generative Adversarial Networks. International Conference on Universal Village, UV 2020, IEEE Xplor, Institute of Electrical and Electronics Engineers Inc., 2020; 1–6. [Google Scholar]
- Chadebec, C.; Allassonnière, S. and Allassonnière, S. Data Augmentation with Variational Autoencoders and Manifold Sampling. Deep Generative Models, and Data Augmentation, Labelling, and Imperfections, Springer International Publishing, 2021; 184–192. [Google Scholar]
- Strelcenia, E.; Prakoonwit, S. Improving Cancer Detection Classification Performance Using GANs in Breast Cancer Data. IEEE Access, 2017, 11, 1–24. [Google Scholar] [CrossRef]
- Wei, L.; Linchuan, X.; Zhixuan, L.; Senzhang, W.; Jiannong, C.; Chao, M.; Xiaohui, C. Sketch-then-Edit Generative Adversarial Network. Knowledge-Based Systems, 2020, 203, 106102. [Google Scholar]
- Cai, L.; Chen, Y.; Cai, N.; Cheng, W.; Wang, H. Utilizing Amari-Alpha Divergence to Stabilize the Training of Generative Adversarial Networks. Entropy, 22, 410. [CrossRef]
- Zhaoqing, P.; Weijie, Y.; Bosi, W.; Haoran, X.; Victor, S. S.; Jianjun, L.; Sam, K. Loss functions of generative adversarial networks (gans): Opportunities and challenges. IEEE Transactions Emerging Topics in Computational Intelligence 2020, 4, 500–522. [Google Scholar]
- Sharma, P.; Kumar, M.; Sharma, H. K.; Biju, S. M. Generative adversarial networks (GANs): Introduction, Taxonomy, Variants, Limitations, and Applications. Multimedia Tools and Applications, 2024, 1–49. [Google Scholar] [CrossRef]
- Martin, A.; Soumith, C.; Leon, B. Wasserstein Generative Adversarial Networks. Proceedings of Machine Learning Research 2017, 70, 214–223. [Google Scholar]
- Xinyu, G.; Seea, K.W.; Yabin, L.; Bilal, B.; Liang, Z.; Yunpeng, W. A time-series Wasserstein GAN method for state-of-charge estimation of lithium-ion batteries. Journal of Power Sources 2023, 581, 233472. [Google Scholar]
- Emilija, S.; Simant, P. Improving Cancer Detection Classification Performance Using GANs in Breast Cancer Data. IEEE Access 2023, 1, 71594–71615. [Google Scholar]
- Xiao, Y.; Wu, J.; Lin, Z. Cancer diagnosis using generative adversarial networks based on deep learning from imbalanced data. Computerin Biologi and Medicine, 2021, 135, 104540. [Google Scholar] [CrossRef]
- Kunapinun, A.; Dailey, M. N.; Songsaeng, D.; Parnichkun, M.; Keatmanee, C. and Ekpanyapong, M. Improving GAN Learning Dynamics for Thyroid Nodule Segmentation. Ultrasound in Medicine & Biology, 2023, 49, 416–430. [Google Scholar]
- Abirami, R.N.; Durai Raj Vincent, P. M.; Srinivasan, K.; Tariq, U.; Chang, C. Y. Deep CNN and Deep GAN in Computational Visual Perception-Driven Image Analysis. Complexity, Hindawi, 2021, 2021, 5541134. [Google Scholar] [CrossRef]
- Liu, W.; Duan, l.; Tang, Y.; Yang, J. Data Augmentation Method for Fault Diagnosis of Mechanical Equipment Based on Improved Wasserstein GAN. Proceedings International Conference on Prognostics and System Health Management, PHM-Jinan 2020, IEEE-Xplor, 2020; 103–111. [Google Scholar]
- Jiménez-Gaona, Y.; Carrión-Figueroa, D.; Rodríguez-Álvarez, M. J. Gan-based data augmentation to improve breast ultrasound and mammography mass classification. Biomedical Signal Processing and Control, 2024, 94, 106255. [Google Scholar] [CrossRef]
- Showrov, I.; Tarek, A. MD.; Hadiur, R. N.; Jamin, R. J.; Mridha, M. F.; Mohsin, K.; Nobuyoshi, A.; Jungpil, S. ; Generative Adversarial Networks (GANs) in Medical Imaging: Advancements, Applications, and Challenges. IEEE Access. 2024, 12, 35728–35753. [Google Scholar]
- Alruily, M.; Said, W.; Mostafa, A. M.; Ezz, M.; Elmezain, M. Breast Ultrasound Images Augmentation and Segmentation Using GANwithIdentity Block and Modified U-Net 3+. Sensors, 2023, col. 23, 8599. [Google Scholar] [CrossRef]
- Islam, M. R.; Rahman, M. M.; Ali, M. S.; Nafi, A. A. N.; Alam, M. S.; Godder, T. K.; Miah, M. S.; Islam, M. K. Enhancing Breast Cancer Segmentation and Classification: An Ensemble Deep Convolutional Neural Network And U-Net Approach on Ultrasound Images. Machine Learning with Application, 2024, 16, 100555. [Google Scholar] [CrossRef]
- Sangeeta, P.; Debnath, B. An Enhanced Multi-Scale Deep Convolutional Orchard Capsule Neural Network For Multi-Modal Breast Cancer Detection. Healthcare Analytics, 2024, 5, 100298. [Google Scholar]
- Alhassan, M.; Fuseini, M. Data augmentation: A comprehensive survey of modern approaches. Array. 2022, 16, 100258. [Google Scholar]
- Kevin, B.; Anna, M.; Angel, M.; José, R. Automatic generation of artificial images of leukocytes and leukemic cells using generative adversarial networks (syntheticcellgan). Computer Methods and Programs in Biomedicine, 2023, 229, 107314. [Google Scholar]
- Chunxue, W.; Bobo, J.; Yan, W.; Neal, N. X.; Sheng, Z. WGAN-E: AGenerative Adversarial Networks for Facial Feature Security. Electronics, 2020, 9, 486. [Google Scholar]
- Hansoo, L.; Jonggeun, K.; Eun Kyeong, K.; Sungshin, K. Wasserstein Generative Adversarial Networks Based Data Augmentation for Radar Data Analysis. applied sciences, 2020, 10, 1449. [Google Scholar]
- Minsoo, H.; Yoonsik, C. ; De-blurring using Perceptual Similarity Wasserstein Generative Adversarial Network Based De-Blurring Using Perceptual Similarity. Applied Sciences, 2019, 9, 2358. [Google Scholar]
- Luca, T.; Ali, M. A.; Hashim A., H. Novel hybrid integrated Pix2Pix and WGAN model with Gradient Penalty for binary images denoising. Systems and Soft Computing, 2024, 6, 200122. [Google Scholar]

| Hyperparameters | Value |
|---|---|
| Epoch | 5000 |
| Weight clipping value | 0.01 |
| Optimizer | RMSProp |
| Learning rate optimizer | 5e-5 |
| Batch size | 128 |
| Number of discriminator iteration for each generator iteration | 5 |
| Dataset/Class | Class | ||
|---|---|---|---|
| 0 - Benign | 1 - Normal | 2 - Malignant | |
| Original | 437 | 133 | 210 |
| Expand | 437 + 50 | 133 + 354 = 487 | 210 + 277 = 487 |
| Dataset/Metrics | Accuracy | Precision | Recall | F1-score |
|---|---|---|---|---|
| Original data | 0.7628 | 0.7380 | 0.7496 | 0.7433 |
| Expanded data | 0.8333 | 0.8490 | 0.8021 | 0.8219 |
| Dataset/Metrics | Accuracy | Precision | Recall | F1-score |
|---|---|---|---|---|
| Original data | 0.6795 | 0.7000 | 0.6400 | 0.6395 |
| Expanded data | 0.7308 | 0.7100 | 0.6976 | 0.6945 |
| Dataset/Metrics | Accuracy | Precision | Recall | F1-score |
|---|---|---|---|---|
| Original data | 0.8077 | 0.7929 | 0.7951 | 0.7939 |
| Expanded data | 0.8205 | 0.8243 | 0.8151 | 0.8094 |
| Dataset/Metrics | Accuracy | Precision | Recall | F1-score |
|---|---|---|---|---|
| Original data | 0.8030 | 0.8000 | 0.8121 | 0.8005 |
| Expanded data | 0.8133 | 0.8150 | 0.8012 | 0.8175 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




