Submitted:
29 September 2024
Posted:
30 September 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Ethic Statement
2.2. Laboratory Detection
2.3. Statistical Analysis
3. Results
3.1. The Characteristic of Overall Rate of Pre-Treated Drug Resistance (PDR) Mutations in 80 Samples by SS and NGS
3.2. The analysis of Resistance Mutation Sites Detected by NGS at Different Threshold
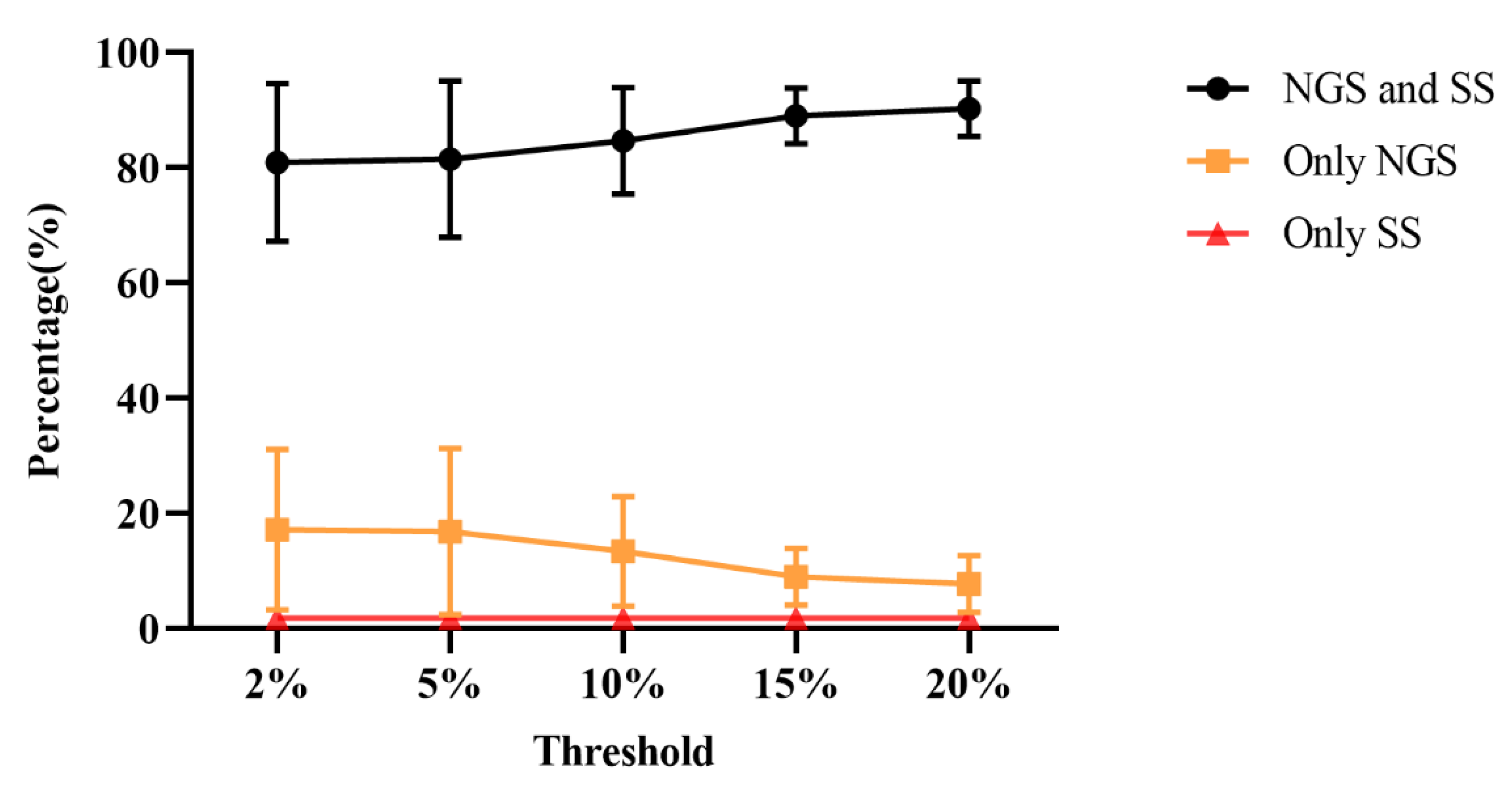
3.3. Comparison of Consistency between SS and NGS
4. Discussion
5. Conclusion
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Ethics Declaration
References
- Perelson, A.S.; Neumann, A.U.; Markowitz, M.; Leonard, J.M.; Ho, D.D. HIV-1 Dynamics in Vivo: Virion Clearance Rate, Infected Cell Life-Span, and Viral Generation Time. Science 1996, 271, 1582–1586. [Google Scholar] [CrossRef] [PubMed]
- Drake, J.W.; Holland, J.J. Mutation rates among RNA viruses. Proceedings of the National Academy of Sciences of the United States of America 1999, 96, 13910–13913. [Google Scholar] [CrossRef] [PubMed]
- Duffy, S.; Shackelton, L.A.; Holmes, E.C. Rates of evolutionary change in viruses: patterns and determinants. Nat. Rev. Genet. 2008, 9, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Jiao, L.Y.; Liu, Y.J.; Guo, D.X.; et al. Comparison of two HIV-1 drug resistance quasispecies analysis methods. Proceedings of Academy of Military Medical Sciences 2010, 34, 261–264. [Google Scholar]
- Cao, W.; Hsieh, E.; Li, T. Optimizing Treatment for Adults with HIV/AIDS in China: Successes over Two Decades and Remaining Challenges. Curr. HIV/AIDS Rep. 2020, 17, 26–34. [Google Scholar] [CrossRef]
- Johnson, J.A.; Li, J.F.; Wei, X.; et al. Minority HIV-1 drug resistance mutations are present in antiretroviral treatment-naïve populations and associate with reduced treatment efficacy. PLoS medicine 2008, 5, e158. [Google Scholar] [CrossRef]
- HIV infection. Nat. Rev. Dis. Prim. 2015, 1, 15060. [CrossRef]
- Ji, H.; Enns, E.; Brumme, C.J.; Parkin, N.; Howison, M.; Lee, E.R.; Capina, R.; Marinier, E.; Avila-Rios, S.; Sandstrom, P.; et al. Bioinformatic data processing pipelines in support of next-generation sequencing-based HIV drug resistance testing: the Winnipeg Consensus. J. Int. AIDS Soc. 2018, 21, e25193. [Google Scholar] [CrossRef]
- World Health Organization. Global Action Plan. on HIV Drug Resistance 2017-2021; World Health Organization: Geneva, Switzerland, 2017; ISBN 9789241512848. [Google Scholar]
- Dai, L.L.; Chen, R.F.; Chen, Y.K.; et al. Expert consensus on rapid initiation of HIV/AIDS antiviral therapy. China Journal of AIDS and Sexually Transmitted Diseases 2023, 29, 737–744. [Google Scholar]
- Alidjinou, E.K.; Deldalle, J.; Hallaert, C.; Robineau, O.; Ajana, F.; Choisy, P.; Hober, D.; Bocket, L. RNA and DNA Sanger sequencing versus next-generation sequencing for HIV-1 drug resistance testing in treatment-naive patients. J. Antimicrob. Chemother. 2017, 72, 2823–2830. [Google Scholar] [CrossRef]
- McCluskey, S.M.; Siedner, M.J.; Marconi, V.C. Management of Virologic Failure and HIV Drug Resistance. Infect. Dis. Clin. North Am. 2019, 33, 707–742. [Google Scholar] [CrossRef] [PubMed]
- Kyeyune, F.; Gibson, R.M.; Nankya, I.; Venner, C.; Metha, S.; Akao, J.; Ndashimye, E.; Kityo, C.M.; Salata, R.A.; Mugyenyi, P.; et al. Low-Frequency Drug Resistance in HIV-Infected Ugandans on Antiretroviral Treatment Is Associated with Regimen Failure. Antimicrob. Agents Chemother. 2016, 60, 3380–3397. [Google Scholar] [CrossRef]
- Ouyang, F.; Yuan, D.; Zhai, W.; Liu, S.; Zhou, Y.; Yang, H. HIV-1 Drug Resistance Detected by Next-Generation Sequencing among ART-Naïve Individuals: A Systematic Review and Meta-Analysis. Viruses 2024, 16, 239. [Google Scholar] [CrossRef]
- Ávila-Ríos, S.; Parkin, N.; Swanstrom, R.; Paredes, R.; Shafer, R.; Ji, H.; Kantor, R. Next-Generation Sequencing for HIV Drug Resistance Testing: Laboratory, Clinical, and Implementation Considerations. Viruses 2020, 12, 617. [Google Scholar] [CrossRef]
- Jingna, X.; Rong, C.; Jun, C.; Hongxia, W.; Ronghui, X.; Zhenyan, W.; Qi, T.; Ren, F.; Li, L.; Hongzhou, L. [Investigation on drug resistance of HIV-infected people before antiviral therapy in three cities in Chinese]. Chinese journal of HIV and STD 2020, 26, 805–808. [Google Scholar] [CrossRef]
- Pyne, M.T.; Simmon, K.E.; Mallory, M.A.; Hymas, W.C.; Stevenson, J.; Barker, A.P.; Hillyard, D.R. HIV-1 Drug Resistance Assay Using Ion Torrent Next Generation Sequencing and On-Instrument End-to-End Analysis Software. J. Clin. Microbiol. 2022, 60, e0025322. [Google Scholar] [CrossRef]
- Lee, E.R.; Parkin, N.; Jennings, C.; Brumme, C.J.; Enns, E.; Casadellà, M.; Howison, M.; Coetzer, M.; Avila-Rios, S.; Capina, R.; et al. Performance comparison of next generation sequencing analysis pipelines for HIV-1 drug resistance testing. Sci. Rep. 2020, 10, 1–10. [Google Scholar] [CrossRef]
- Yuan, D.F. Epidemiological study of HIV-1 subtypes and drug resistance mutations in HIV-1 infected persons under treatment in Jiangsu Province; Southeast University: Jiangsu, 2022. [Google Scholar]
- Teo, C.H.Y.; Norhisham, N.H.B.; Lee, O.F.; Png, S.; Chai, C.N.; Yan, G.; Tang, J.W.-T.; Lee, C.K. Towards Next-Generation Sequencing for HIV-1 Drug Resistance Testing in a Clinical Setting. Viruses 2022, 14, 2208. [Google Scholar] [CrossRef]
- Lataillade, M.; Chiarella, J.; Yang, R.; Schnittman, S.; Wirtz, V.; Uy, J.; Seekins, D.; Krystal, M.; Mancini, M.; McGrath, D.; et al. Prevalence and Clinical Significance of HIV Drug Resistance Mutations by Ultra-Deep Sequencing in Antiretroviral-Naïve Subjects in the CASTLE Study. PLOS ONE 2010, 5, e10952. [Google Scholar] [CrossRef]
- Kojima, S.; Kamada, A.J.; Parrish, N.F. Virus-derived variation in diverse human genomes. PLOS Genet. 2021, 17, e1009324. [Google Scholar] [CrossRef]
- Capina, R.; Li, K.; Kearney, L.; Vandamme, A.-M.; Harrigan, P.R.; Van Laethem, K. Quality Control of Next-Generation Sequencing-Based HIV-1 Drug Resistance Data in Clinical Laboratory Information Systems Framework. Viruses 2020, 12, 645. [Google Scholar] [CrossRef] [PubMed]
- Novitsky, V.; Nyandiko, W.; Vreeman, R.; DeLong, A.K.; Manne, A.; Scanlon, M.; Ngeresa, A.; Aluoch, J.; Sang, F.; Ashimosi, C.; et al. Added Value of Next Generation over Sanger Sequencing in Kenyan Youth with Extensive HIV-1 Drug Resistance. Microbiol. Spectr. 2022, 10, e0345422. [Google Scholar] [CrossRef]
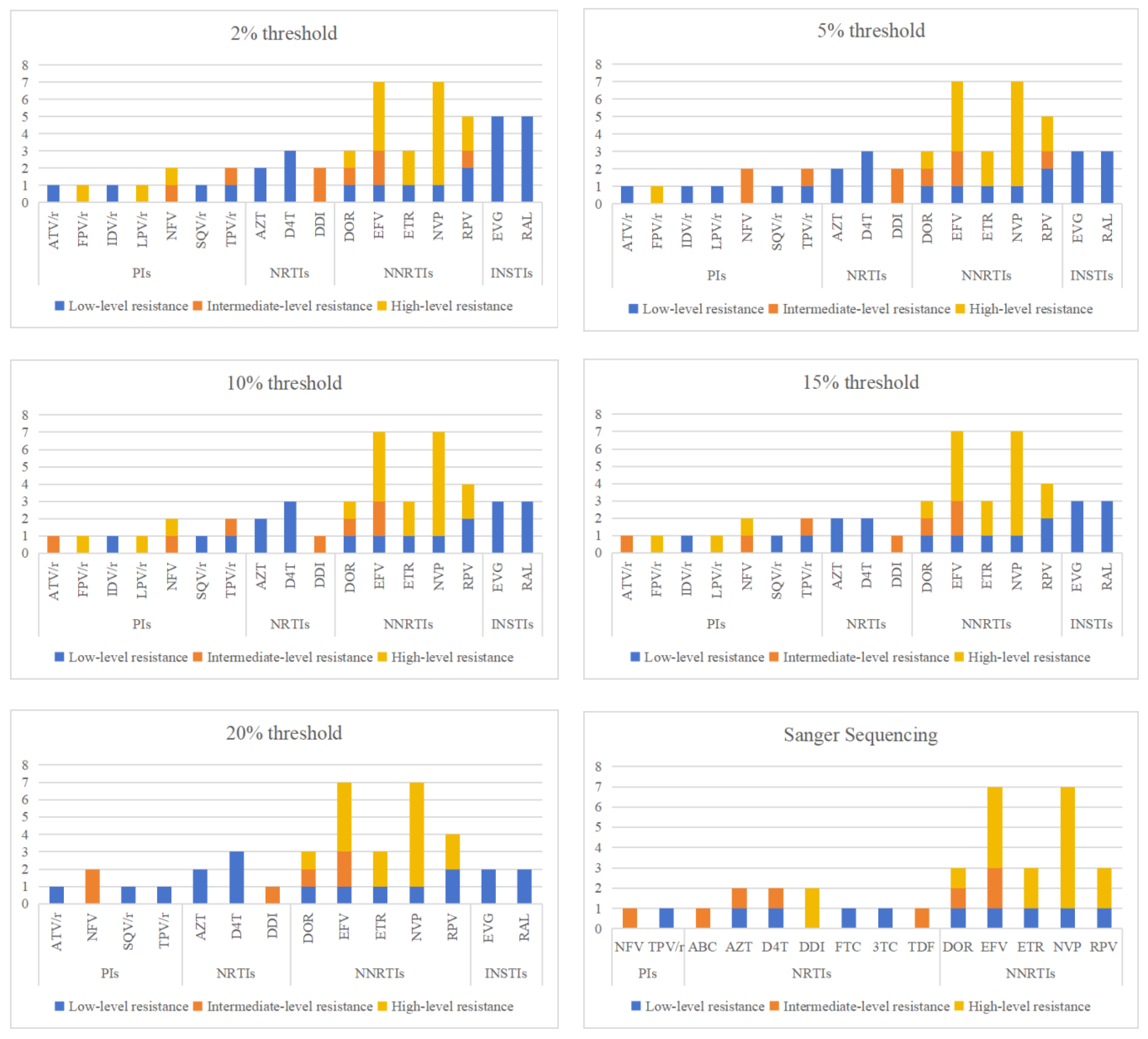
| resistance patterns | Sanger sequencing | Next-generation sequencing | ||||
|---|---|---|---|---|---|---|
| 20% threshold | 15% threshold | 10% threshold | 5% threshold | 2% threshold | ||
| Any drug resistance, n (%) | 11 (13.8) | 14 (17.9) | 16 (20.5) | 17 (21.8) | 19 (23.8) | 20 (25.0) |
| PIs resistance, n (%) | 2 (2.5) | 3 (3.8) | 5 (6.3) | 5 (6.3) | 5 (6.3) | 5 (6.3) |
| NRTIs resistance, n (%) | 2 (2.5) | 2 (2.6) | 2 (2.6) | 3 (3.8) | 4 (5.1) | 4 (5.1) |
| NNRTIs class resistance, n (%) | 8 (10.0) | 8 (10.3) | 8 (10.3) | 8 (10.3) | 8 (10.3) | 8 (10.3) |
| INSTIs resistance, n (%) | 0 | 2 (2.6) | 3 (3.8) | 3 (300.8) | 3 (3.8) | 5 (6.3) |
| drug classes | mutation site | Next-generation sequencing detection threshold | Sanger sequencing |
||||
|---|---|---|---|---|---|---|---|
| 2% | 5% | 10% | 15% | 20% | |||
| PIs | L33F | 3 | 3 | 3 | 3 | 3 | 3 |
| M46I | 1 | 1 | 1 | 1 | 1 | 1 | |
| M46L | 1 | 0 | 1 | 1 | 0 | 0 | |
| M46V | 0 | 0 | 0 | 0 | 1 | 0 | |
| I47AV | 1 | 1 | 1 | 1 | 0 | 0 | |
| F53L | 3 | 3 | 3 | 3 | 1 | 0 | |
| Q58E | 1 | 1 | 1 | 1 | 1 | 1 | |
| L89LMV | 1 | 1 | 1 | 1 | 1 | 0 | |
| total | 11 | 10 | 11 | 11 | 9 | 5 | |
| NRTIs | E40F | 0 | 1 | 1 | 1 | 1 | 0 |
| M41L | 1 | 1 | 1 | 1 | 1 | 1 | |
| E44EDV | 0 | 1 | 0 | 0 | 0 | 0 | |
| S68SG | 27 | 27 | 20 | 12 | 8 | 0 | |
| S68G | 2 | 2 | 2 | 2 | 2 | 3 | |
| D67N | 1 | 1 | 1 | 1 | 1 | 1 | |
| T69D | 1 | 1 | 1 | 1 | 1 | 1 | |
| D67del | 0 | 0 | 0 | 0 | 0 | 1 | |
| T69TADN | 1 | 1 | 0 | 0 | 0 | 0 | |
| F77FL | 1 | 0 | 0 | 0 | 0 | 0 | |
| T215TS | 1 | 1 | 1 | 0 | 0 | 0 | |
| K219KN | 2 | 2 | 2 | 2 | 1 | 0 | |
| K219KQ | 1 | 1 | 0 | 0 | 0 | 0 | |
| K219KR | 1 | 1 | 0 | 0 | 0 | 0 | |
| total | 39 | 40 | 29 | 20 | 15 | 7 | |
| NNRTIs | K101E | 1 | 1 | 1 | 1 | 1 | 1 |
| K103KE | 5 | 5 | 5 | 2 | 1 | 0 | |
| K103N | 2 | 2 | 3 | 3 | 3 | 4 | |
| K103NS | 1 | 1 | 0 | 0 | 0 | 0 | |
| K103S | 1 | 1 | 1 | 1 | 1 | 1 | |
| V106VI | 1 | 0 | 0 | 0 | 0 | 0 | |
| E138EA | 1 | 1 | 1 | 1 | 1 | 0 | |
| E138EG | 1 | 1 | 1 | 1 | 1 | 1 | |
| E138EK | 1 | 1 | 0 | 0 | 0 | 0 | |
| V179D | 3 | 3 | 3 | 3 | 3 | 1 | |
| V179E | 8 | 8 | 8 | 8 | 7 | 5 | |
| Y181C | 1 | 1 | 1 | 1 | 1 | 1 | |
| Y181V | 1 | 1 | 1 | 1 | 1 | 1 | |
| G190S | 1 | 1 | 1 | 1 | 1 | 1 | |
| P225PH | 1 | 1 | 1 | 1 | 1 | 1 | |
| total | 29 | 28 | 27 | 24 | 22 | 17 | |
| INSTIs | T66A | 0 | 0 | 1 | 0 | 0 | 0 |
| E138EA | 5 | 3 | 2 | 3 | 2 | 0 | |
| A128AT | 1 | 0 | 0 | 0 | 0 | 1 | |
| S153A | 4 | 1 | 1 | 1 | 1 | 1 | |
| D232N | 1 | 1 | 1 | 1 | 1 | 0 | |
| total | 11 | 5 | 5 | 5 | 4 | 2 | |
| detection threshold | drug resistance mutation sites | PIs | NRTIs | NNRTIs | INSTIs |
|---|---|---|---|---|---|
| 2% | Only detected by NGS | 6.25% | 36.25% | 18.75% | 7.50% |
| Only detected by SS | 1.25% | 1.25% | 2.50% | 2.50% | |
| Consistency | 92.50% | 62.50% | 78.75% | 90.00% | |
| 5% | Only detected by NGS | 6.25% | 37.50% | 16.25% | 7.50% |
| Only detected by SS | 1.25% | 1.25% | 2.50% | 2.50% | |
| Consistency | 92.50% | 62.50% | 81.25% | 90.00% | |
| 10% | Only detected by NGS | 6.25% | 26.25% | 15.00% | 6.25% |
| Only detected by SS | 1.25% | 1.25% | 2.50% | 2.50% | |
| Consistency | 92.50% | 72.5% | 82.50% | 91.25% | |
| 15% | Only detected by NGS | 5.00% | 15.00% | 11.25% | 5.00% |
| Only detected by SS | 1.25% | 1.25% | 2.50% | 2.50% | |
| Consistency | 93.75% | 83.75% | 86.25% | 92.50% | |
| 20% | Only detected by NGS | 3.75% | 13.75% | 10.00% | 3.75% |
| Only detected by SS | 1.25% | 1.25% | 2.50% | 2.50% | |
| Consistency | 95.00% | 85.00% | 87.50% | 93.75% |
| NGS | SS | Sensitiv-ity | Specifi-city | P | Kappa value | 95%CI | ||
|---|---|---|---|---|---|---|---|---|
| Detected | Not detected | |||||||
| PIs | ||||||||
| 2%/5%/10% | Detected | 4 | 5 | 80.0% | 93.3% | 0.109 | 0.534 | 0.211-0.857 |
| Not detected | 1 | 70 | ||||||
| 15% | Detected | 4 | 4 | 80.0% | 94.6% | 0.109 | 0.583 | 0.257-0.810 |
| Not detected | 1 | 71 | ||||||
| 20% | Detected | 4 | 3 | 80.0% | 96.0% | 0.109 | 0.640 | 0.315-0.965 |
| Not detected | 1 | 72 | ||||||
| NRTIs | ||||||||
| 2% | Detected | 3 | 29 | 75.0% | 61.8% | <0.001 | 0.085 | -0.042-0.212 |
| Not detected | 1 | 47 | ||||||
| 5% | Detected | 3 | 30 | 75.0% | 60.5% | <0.001 | 0.080 | -0.042-0.202 |
| Not detected | 1 | 46 | ||||||
| 10% | Detected | 3 | 21 | 75.0% | 72.4% | <0.001 | 0.141 | -0.033-0.315 |
| Not detected | 1 | 55 | ||||||
| 15% | Detected | 3 | 12 | 75.0% | 84.2% | 0.003 | 0.257 | -0.004-0.518 |
| Not detected | 1 | 64 | ||||||
| 20% | Detected | 2 | 11 | 50.0% | 86.8% | 0.006 | 0.278 | 0.004-0.552 |
| Not detected | 1 | 66 | ||||||
| NNRTIs | ||||||||
| 2% | Detected | 11 | 10 | 84.6% | 85.1% | 0.019 | 0.558 | 0.342-0.774 |
| Not detected | 2 | 57 | ||||||
| 5% | Detected | 11 | 8 | 84.6% | 88.1% | <0.001 | 0.409 | 0.168-0.650 |
| Not detected | 2 | 59 | ||||||
| 10% | Detected | 11 | 9 | 84.6% | 86.6% | 0.033 | 0.585 | 0.369-0.801 |
| Not detected | 2 | 58 | ||||||
| 15% | detected | 11 | 8 | 84.6% | 88.1% | 0.055 | 0.613 | 0.399-0.823 |
| Not detected | 2 | 59 | ||||||
| 20% | Detected | 11 | 7 | 84.6% | 89.6% | <0.001 | 0.599 | 0.322-0.796 |
| Not detected | 2 | 60 | ||||||
| Any mutation | ||||||||
| 2% | Detected | 20 | 32 | 87.0% | 43.9% | <0.001 | 0.224 | 0.071-0.377 |
| Not detected | 3 | 25 | ||||||
| 5% | Detected | 21 | 30 | 91.3% | 47.4% | <0.001 | 0.284 | 0.131-0.437 |
| Not detected | 2 | 27 | ||||||
| 10% | Detected | 19 | 25 | 82.6% | 56.1% | <0.001 | 0.305 | 0.129-0.481 |
| Not detected | 4 | 32 | ||||||
| 15% | Detected | 19 | 18 | 82.6% | 68.4% | <0.001 | 0.417 | 0.231-0.603 |
| Not detected | 4 | 39 | ||||||
| 20% | Detected | 17 | 19 | 81.0% | 67.8% | <0.001 | 0.396 | 0.206-0.586 |
| Not detected | 4 | 40 | ||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





