Submitted:
02 September 2024
Posted:
02 September 2024
You are already at the latest version
Abstract

Keywords:
1. Introduction
1.1. The main protease (Mpro) enzyme of SARS-CoV-2
1.2. The need for novel inhibitors
2. Mpro inhibitors for SARS-CoV-2
2.1. Overview of Mpro Inhibitors
2.2. Computer-aided drug design
2.3. Molecular dynamics (MD) in drug development
2.4. Leveraging quantum mechanics for ADME-optimized drugs
3. Summary and outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Acknowledgment
References
- Kumar, D., G. Chauhan, S. Kalra, B. Kumar, and M.S. Gill, A perspective on potential target proteins of COVID-19: Comparison with SARS-CoV for designing new small molecules. Bioorganic chemistry, 2020. 104: p. 104326. [CrossRef]
- Liu, Y., C. Liang, L. Xin, X. Ren, L. Tian, X. Ju, H. Li, Y. Wang, Q. Zhao, and H. Liu, The development of Coronavirus 3C-Like protease (3CLpro) inhibitors from 2010 to 2020. European journal of medicinal chemistry, 2020. 206: p. 112711. [CrossRef]
- Sharma, P., V. Vijayan, P. Pant, M. Sharma, N. Vikram, P. Kaur, T. Singh, and S. Sharma, Identification of potential drug candidates to combat COVID-19: a structural study using the main protease (mpro) of SARS-CoV-2. Journal of Biomolecular Structure and Dynamics, 2021. 39(17): p. 6649-6659. [CrossRef]
- Krammer, F., SARS-CoV-2 vaccines in development. Nature, 2020. 586(7830): p. 516-527. [CrossRef]
- Sharun, K., R. Tiwari, and K. Dhama, Protease inhibitor GC376 for COVID-19: Lessons learned from feline infectious peritonitis. Annals of medicine and surgery, 2021. 61: p. 122-125. [CrossRef]
- Badavath, V.N., A. Kumar, P.K. Samanta, S. Maji, A. Das, G. Blum, A. Jha, and A. Sen, Determination of potential inhibitors based on isatin derivatives against SARS-CoV-2 main protease (mpro): a molecular docking, molecular dynamics and structure-activity relationship studies. Journal of Biomolecular Structure and Dynamics, 2022. 40(7): p. 3110-3128. [CrossRef]
- Astuti, I., Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2): An overview of viral structure and host response. Diabetes & Metabolic Syndrome: Clinical Research & Reviews, 2020. 14(4): p. 407-412. [CrossRef]
- Mondol, W.C., Exploring the potential of organic molecules in the treatment of covid-19. 2021, Brac University.
- Xu, Y.-S., J.-Z. Chigan, J.-Q. Li, H.-H. Ding, L.-Y. Sun, L. Liu, Z. Hu, and K.-W. Yang, Hydroxamate and thiosemicarbazone: Two highly promising scaffolds for the development of SARS-CoV-2 antivirals. Bioorganic Chemistry, 2022. 124: p. 105799. [CrossRef]
- Dai, W., B. Zhang, X.-M. Jiang, H. Su, J. Li, Y. Zhao, X. Xie, Z. Jin, J. Peng, and F. Liu, Structurebased design, synthesis and biological evaluation of peptidomimetic aldehydes as a novel series of antiviral drug candidates targeting the SARS-CoV-2 main protease. BioRxiv, 2020. [CrossRef]
- Li, X., M. Geng, Y. Peng, L. Meng, and S. Lu, Molecular immune pathogenesis and diagnosis of COVID-19. Journal of pharmaceutical analysis, 2020. 10(2): p. 102-108. [CrossRef]
- Mengist, H.M., T. Dilnessa, and T. Jin, Structural basis of potential inhibitors targeting SARSCoV-2 main protease. Frontiers in Chemistry, 2021. 9: p. 622898. [CrossRef]
- Adelusi, T.I., A.-Q.K. Oyedele, O.E. Monday, I.D. Boyenle, M.O. Idris, A.T. Ogunlana, A.M. Ayoola, J.O. Fatoki, O.E. Kolawole, and K.B. David, Dietary polyphenols mitigate SARS-CoV-2 main protease (Mpro)–Molecular dynamics, molecular mechanics, and density functional theory investigations. Journal of molecular structure, 2022. 1250: p. 131879. [CrossRef]
- Muratov, E.N., R. Amaro, C.H. Andrade, N. Brown, S. Ekins, D. Fourches, O. Isayev, D. Kozakov, J.L. Medina-Franco, and K.M. Merz, A critical overview of computational approaches employed for COVID-19 drug discovery. Chemical Society Reviews, 2021. 50(16): p. 9121-9151. [CrossRef]
- Han, S.H., C.M. Goins, T. Arya, W.-J. Shin, J. Maw, A. Hooper, D.P. Sonawane, M.R. Porter, B.E. Bannister, R.D. Crouch, A.A. Lindsey, G. Lakatos, S.R. Martinez, J. Alvarado, W.S. Akers, N.S.
- Wang, J.U. Jung, J.D. Macdonald, and S.R. Stauffer, Structure-Based Optimization of ML300-Derived, Noncovalent Inhibitors Targeting the Severe Acute Respiratory Syndrome Coronavirus 3CL Protease (SARS-CoV-2 3CLpro). Journal of Medicinal Chemistry, 2022. 65(4): p. 2880-2904. [CrossRef]
- Fernandes, Q., V.P. Inchakalody, M. Merhi, S. Mestiri, N. Taib, D. Moustafa Abo El-Ella, T. Bedhiafi, A. Raza, L. Al-Zaidan, M.O. Mohsen, M.A. Yousuf Al-Nesf, A.A. Hssain, H.M. Yassine, M.F. Bachmann, S. Uddin, and S. Dermime, Emerging COVID-19 variants and their impact on SARS-CoV-2 diagnosis, therapeutics and vaccines. Annals of Medicine, 2022. 54(1): p. 524-540. [CrossRef]
- Cooper, M.S., L. Zhang, M. Ibrahim, K. Zhang, X. Sun, J. Röske, M. Göhl, M. Brönstrup, J.K. Cowell, and L. Sauerhering, Diastereomeric Resolution Yields Highly Potent Inhibitor of SARSCoV-2 Main Protease. Journal of Medicinal Chemistry, 2022. 65(19): p. 13328-13342. [CrossRef]
- Alexandris, N., G. Lagoumintzis, C.T. Chasapis, D.D. Leonidas, G.E. Papadopoulos, S.J. Tzartos, A. Tsatsakis, E. Eliopoulos, K. Poulas, and K. Farsalinos, Nicotinic cholinergic system and COVID19: In silico evaluation of nicotinic acetylcholine receptor agonists as potential therapeutic interventions. Toxicology reports, 2021. 8: p. 73-83. [CrossRef]
- Yang, F., T.N.-A. Tran, E. Howerton, M.F. Boni, and J.L. Servadio, Benefits of near-universal vaccination and treatment access to manage COVID-19 burden in the United States. BMC Medicine, 2023. 21(1): p. 321. [CrossRef]
- Lockbaum, G.J., A.C. Reyes, J.M. Lee, R. Tilvawala, E.A. Nalivaika, A. Ali, N. Kurt Yilmaz, P.R. Thompson, and C.A. Schiffer, Crystal structure of SARS-CoV-2 main protease in complex with the non-covalent inhibitor ML188. Viruses, 2021. 13(2): p. 174. [CrossRef]
- Li, Q. and C. Kang, Progress in developing inhibitors of SARS-CoV-2 3C-like protease. Microorganisms, 2020. 8(8): p. 1250. [CrossRef]
- Wu, Q., S. Yan, Y. Wang, M. Li, Y. Xiao, and Y. Li, Discovery of 4′-O-methylscutellarein as a potent SARS-CoV-2 main protease inhibitor. Biochemical and biophysical research communications, 2022. 604: p. 76-82.
- Naidu, S.A.G., Y.B. Tripathi, P. Shree, R.A. Clemens, and A.S. Naidu, Phytonutrient Inhibitors of SARS-CoV-2/NSP5-Encoded Main Protease (Mpro) Autocleavage Enzyme Critical for COVID-19 Pathogenesis. Journal of dietary supplements, 2023. 20(2): p. 284-311. [CrossRef]
- Kouam, A.F., F.D. Mabou, L. Fu, R.R. Koagne, Y. Li, B.A. Owona, E.M. Zeuko'o, A.G.K. Fepa, B.R.T. Galani, F. Reyes, F.N. Njayou, P.F. Moundipa, and G.F. Gao, Insights into the inhibition mechanisms of MERS-CoV and SARS-CoV2 papain-like proteases by inhibitors from Crinum distichum: In vitro and in silico analysis. South African Journal of Botany, 2024. 165: p. 290306. [CrossRef]
- Ebenezer, O. and M. Shapi, Promising inhibitors against main protease of SARS CoV-2 from medicinal plants: In silico identification. Acta Pharmaceutica, 2022. 72(2): p. 159-169. [CrossRef]
- Zhu, J., H. Zhang, Q. Lin, J. Lyu, L. Lu, H. Chen, X. Zhang, Y. Zhang, and K. Chen, Progress on SARS-CoV-2 3CLpro Inhibitors: Inspiration from SARS-CoV 3CLpro Peptidomimetics and SmallMolecule Anti-Inflammatory Compounds. Drug Design, Development and Therapy, 2023. 16(null): p. 1067-1082. [CrossRef]
- Song, L., S. Gao, B. Ye, M. Yang, Y. Cheng, D. Kang, F. Yi, J.-P. Sun, L. Menéndez-Arias, and J. Neyts, Medicinal chemistry strategies towards the development of non-covalent SARS-CoV-2 Mpro inhibitors. Acta Pharmaceutica Sinica B, 2023. [CrossRef]
- Singh, E., R.J. Khan, R.K. Jha, G.M. Amera, M. Jain, R.P. Singh, J. Muthukumaran, and A.K. Singh, A comprehensive review on promising anti-viral therapeutic candidates identified against main protease from SARS-CoV-2 through various computational methods. Journal of Genetic Engineering and Biotechnology, 2020. 18(1): p. 69. [CrossRef]
- Reprinted (adapted) with permission from Structure-Based Optimization of ML300-Derived, Noncovalent Inhibitors Targeting the Severe Acute Respiratory Syndrome Coronavirus 3CL Protease (SARS-CoV-2 3CLpro). Sang Hoon Han, Christopher M. Goins, Tarun Arya, Woo-Jin Shin, Joshua Maw, Alice Hooper, Dhiraj P. Sonawane, Matthew R. Porter, Breyanne E. Bannister, Rachel D. Crouch, A. Abigail Lindsey, Gabriella Lakatos, Steven R. Martinez, Joseph Alvarado, Wendell S. Akers, Nancy S. Wang, Jae U. Jung, Jonathan D. Macdonald, and Shaun Journal of Medicinal Chemistry 2022 65 (4), 2880-2904. Copyright 2022 American Chemical Society. [CrossRef]
- Tumskiy, R.S. and A.V. Tumskaia, Multistep rational molecular design and combined docking for discovery of novel classes of inhibitors of SARS-CoV-2 main protease 3CLpro. Chemical physics letters, 2021. 780: p. 138894. [CrossRef]
- Fischer, A., M. Sellner, S. Neranjan, M. Smieško, and M.A. Lill, Potential Inhibitors for Novel Coronavirus Protease Identified by Virtual Screening of 606 Million Compounds. International Journal of Molecular Sciences, 2020. 21(10): p. 3626. [CrossRef]
- Jacobs, J., V. Grum-Tokars, Y. Zhou, M. Turlington, S.A. Saldanha, P. Chase, A. Eggler, E.S. Dawson, Y.M. Baez-Santos, and S. Tomar, Discovery, synthesis, and structure-based optimization of a series of N-(tert-butyl)-2-(N-arylamido)-2-(pyridin-3-yl) acetamides (ML188) as potent noncovalent small molecule inhibitors of the severe acute respiratory syndrome coronavirus (SARS-CoV) 3CL protease. Journal of medicinal chemistry, 2013. 56(2): p. 534-546. [CrossRef]
- Berry, M., B. Fielding, and J. Gamieldien, Human coronavirus OC43 3CL protease and the potential of ML188 as a broad-spectrum lead compound: Homology modelling and molecular dynamic studies. BMC structural biology, 2015. 15: p. 1-10. [CrossRef]
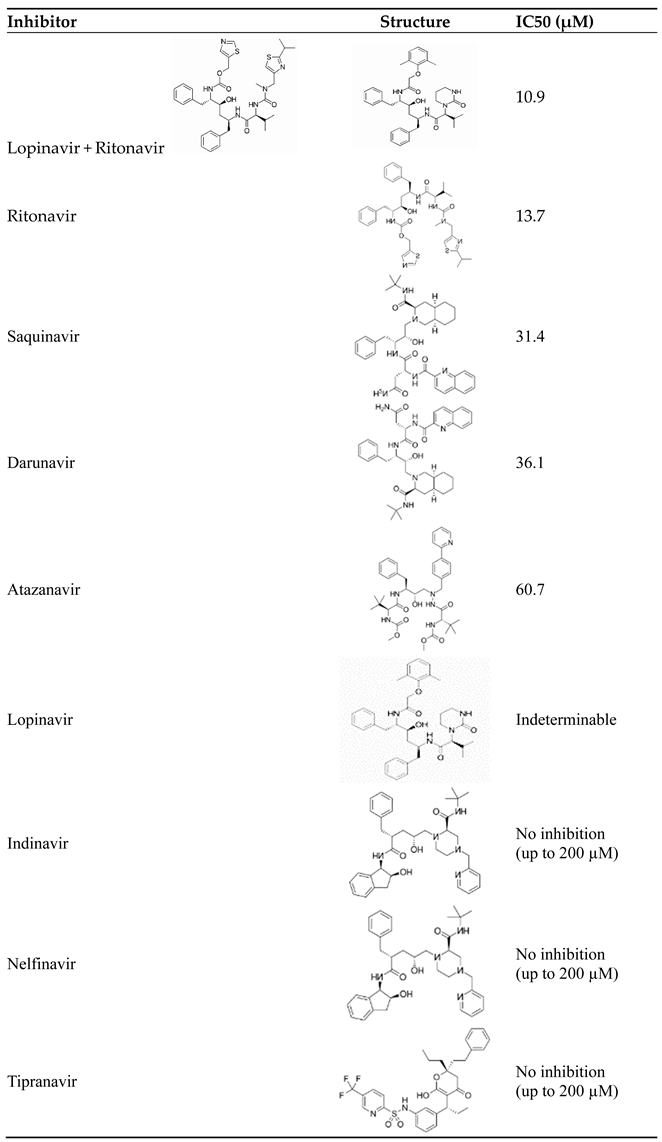
- Mahdi, M., J.A. Mótyán, Z.I. Szojka, M. Golda, M. Miczi, and J. Tőzsér, Analysis of the efficacy of HIV protease inhibitors against SARS-CoV-2′s main protease. Virology Journal, 2020. 17(1): p. 190. [CrossRef]
- Sayed, A.M., H.A. Alhadrami, A.O. El-Gendy, Y.I. Shamikh, L. Belbahri, H.M. Hassan, U.R. Abdelmohsen, and M.E. Rateb, Microbial natural products as potential inhibitors of SARS-CoV2 main protease (Mpro). Microorganisms, 2020. 8(7): p. 970. [CrossRef]
- Thakur, A., G. Sharma, V.N. Badavath, V. Jayaprakash, K.M. Merz, Jr., G. Blum, and O. Acevedo, Primer for Designing Main Protease (Mpro) Inhibitors of SARS-CoV-2. The Journal of Physical Chemistry Letters, 2022. 13(25): p. 5776-5786. [CrossRef]
- Reprinted (adapted) with permission from Primer for Designing Main Protease (Mpro) Inhibitors of SARS-CoV-2. Abhishek Thakur, Gaurav Sharma, Vishnu Nayak Badavath, Venkatesan Jayaprakash, Kenneth M. Merz Jr., Galia Blum, and Orlando Acevedo. The Journal of Physical Chemistry Letters 2022 13 (25), 5776-5786. Copyright 2022 American Chemical Society. [CrossRef]
- Pillaiyar, T., P. Flury, N. Krüger, H. Su, L. Schäkel, E. Barbosa Da Silva, O. Eppler, T. Kronenberger, T. Nie, S. Luedtke, C. Rocha, K. Sylvester, M.R.I. Petry, J.H. McKerrow, A. Poso, Pöhlmann, M. Gütschow, A.J. O’Donoghue, Y. Xu, C.E. Müller, and S.A. Laufer, SmallMolecule Thioesters as SARS-CoV-2 Main Protease Inhibitors: Enzyme Inhibition, Structure– Activity Relationships, Antiviral Activity, and X-ray Structure Determination. Journal of Medicinal Chemistry, 2022. 65(13): p. 9376-9395. [CrossRef]
- Reprinted (adapted) with permission from Small-Molecule Thioesters as SARS-CoV-2 Main Protease Inhibitors: Enzyme Inhibition, Structure–Activity Relationships, Antiviral Activity, and X-ray Structure Determination Thanigaimalai Pillaiyar, Philipp Flury, Nadine Krüger, Haixia Su, Laura Schäkel, Elany Barbosa Da Silva, Olga Eppler, Thales Kronenberger, Tianqing Nie, Stephanie Luedtke, Cheila Rocha, Katharina Sylvester, Marvin R.I. Petry, James H. McKerrow, Antti Poso, Stefan Pöhlmann, Michael Gütschow, Anthony J. O’Donoghue, Yechun Xu, Christa E. Müller, and Stefan A. Laufer. Journal of Medicinal Chemistry 2022 65 (13), 9376-9395. Copyright 2022 American Chemical Society. [CrossRef]
- Verma, S., C.N. Patel, and M. Chandra, Identification of novel inhibitors of SARS-CoV-2 main protease (Mpro) from Withania sp. by molecular docking and molecular dynamics simulation. Journal of computational chemistry, 2021. 42(26): p. 1861-1872. [CrossRef]
- Amin, S.A., S. Banerjee, S. Singh, I.A. Qureshi, S. Gayen, and T. Jha, First structure–activity relationship analysis of SARS-CoV-2 virus main protease (Mpro) inhibitors: an endeavor on COVID-19 drug discovery. Molecular diversity, 2021. 25(3): p. 1827-1838. [CrossRef]
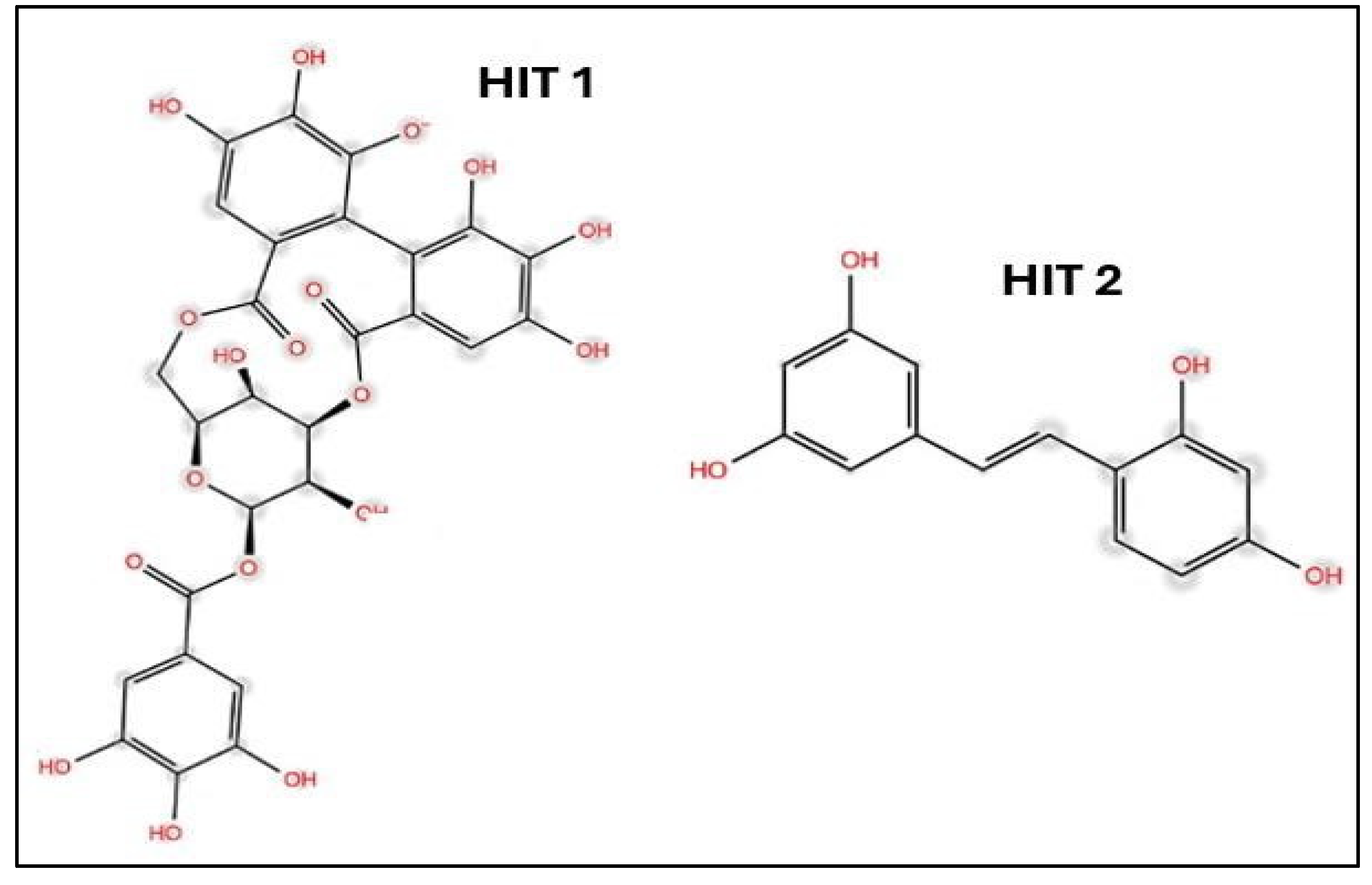
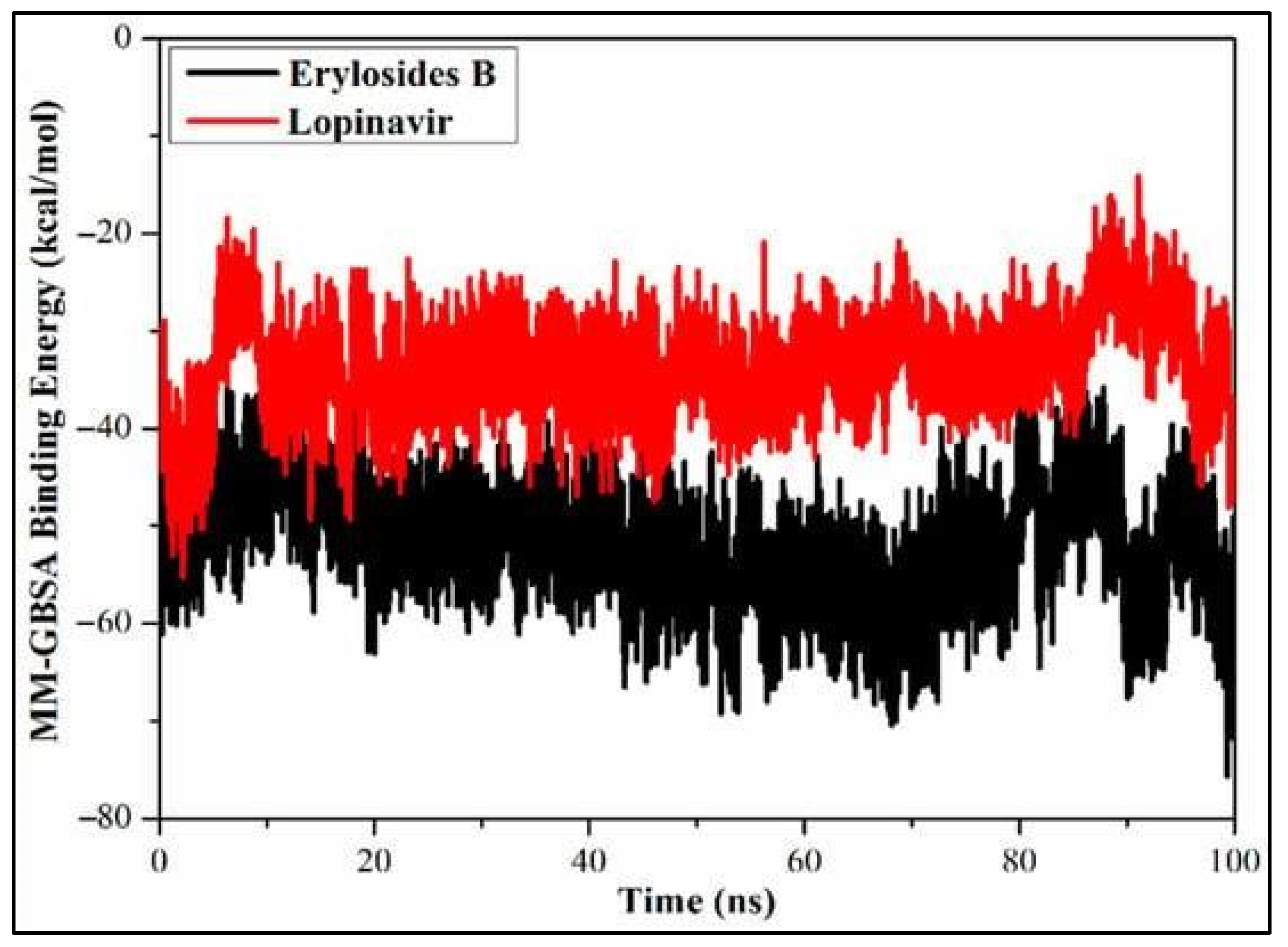
- Ibrahim, M.A.A., A.H.M. Abdelrahman, T.A. Mohamed, M.A.M. Atia, M.A.M. Al-Hammady, K.A.A. Abdeljawaad, E.M. Elkady, M.F. Moustafa, F. Alrumaihi, K.S. Allemailem, H.R. El-Seedi, P.W. Paré, T. Efferth, and M.-E.F. Hegazy, In Silico Mining of Terpenes from Red-Sea Invertebrates for SARS-CoV-2 Main Protease (Mpro) Inhibitors. Molecules, 2021. 26(7): p. 2082. [CrossRef]
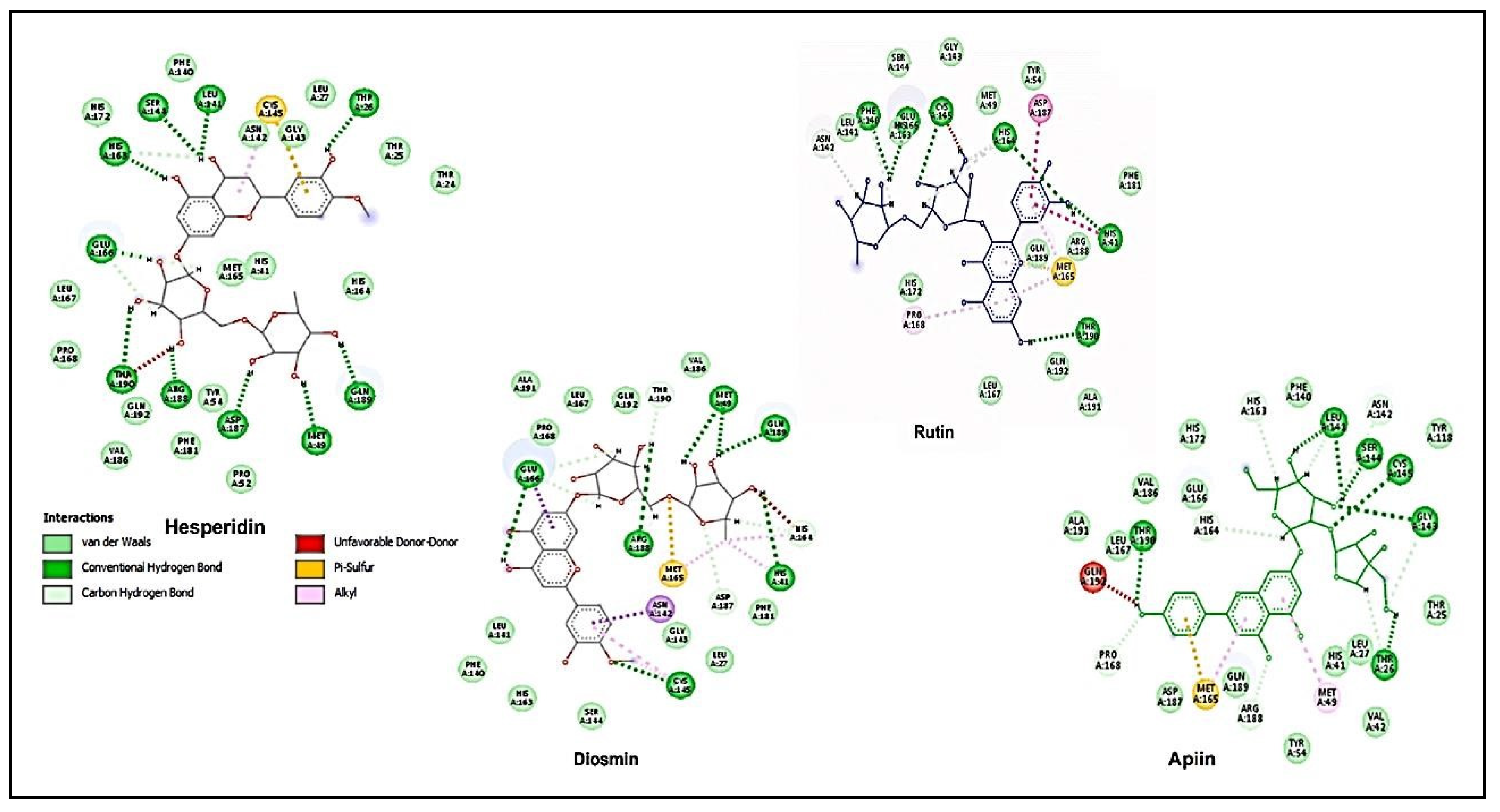
- Adem, Ş., V. Eyupoglu, I.M. Ibrahim, I. Sarfraz, A. Rasul, M. Ali, and A.A. Elfiky, Multidimensional in silico strategy for identification of natural polyphenols-based SARS-CoV-2 main protease (Mpro) inhibitors to unveil a hope against COVID-19. Computers in biology and medicine, 2022. 145: p. 105452-105452. [CrossRef]
- Reprinted from Computers in Biology and Medicine, Vol 145, Adem, Şevki; Eyupoglu, Volkan; Ibrahim, Ibrahim M.; Sarfraz, Iqra; Rasul, Azhar; Ali, Muhammad; Elfiky, Abdo A., Multidimensional in silico strategy for identification of natural polyphenols-based SARS-CoV2 main protease (Mpro) inhibitors to unveil a hope against COVID-19, 105452-105452, Copyright 2022, with permission from Elsevier. [CrossRef]
- Hicks, E.G., S.E. Kandel, and J.N. Lampe, Identification of Aloe-derived natural products as prospective lead scaffolds for SARS-CoV-2 main protease (Mpro) inhibitors. Bioorganic & Medicinal Chemistry Letters, 2022. 66: p. 128732.
- Reprinted from Bioorganic & Medicinal Chemistry Letters, Vol 66, Hicks,Emily G. Kandel,Sylvie E. Lampe,Jed N., Identification of Aloe-derived natural products as prospective lead scaffolds for SARS-CoV-2 main protease (Mpro) inhibitors, 128732, Copyright 2022, with permission from Elsevier. [CrossRef]
- Vincent, S., S. Arokiyaraj, M. Saravanan, and M. Dhanraj, Molecular docking studies on the anti-viral effects of compounds from Kabasura Kudineer on SARS-CoV-2 3CLpro. Frontiers in molecular biosciences, 2020. 7: p. 613401. [CrossRef]
- Motiwale, M., N.S. Yadav, S. Kumar, T. Kushwaha, G. Choudhir, S. Sharma, and P.K. Singour, Finding potent inhibitors for COVID-19 main protease (Mpro): an in silico approach using SARSCoV-3CL protease inhibitors for combating CORONA. Journal of Biomolecular Structure and Dynamics, 2022. 40(4): p. 1534-1545. [CrossRef]
- Aleissa, M.S., M. Al-Zharani, M.S. Hasnain, and S. Alkahtani, Screening, molecular simulation & in silico kinetics of virtually designed covid-19 main protease inhibitors. Journal of King Saud University - Science, 2022. 34(8): p. 102283. [CrossRef]
- Salo-Ahen, O.M.H., I. Alanko, R. Bhadane, A.M.J.J. Bonvin, R.V. Honorato, S. Hossain, A.H. Juffer, A. Kabedev, M. Lahtela-Kakkonen, A.S. Larsen, E. Lescrinier, P. Marimuthu, M.U. Mirza, G. Mustafa, A. Nunes-Alves, T. Pantsar, A. Saadabadi, K. Singaravelu, and M. Vanmeert, Molecular Dynamics Simulations in Drug Discovery and Pharmaceutical Development. Processes, 2021. 9(1): p. 71. [CrossRef]
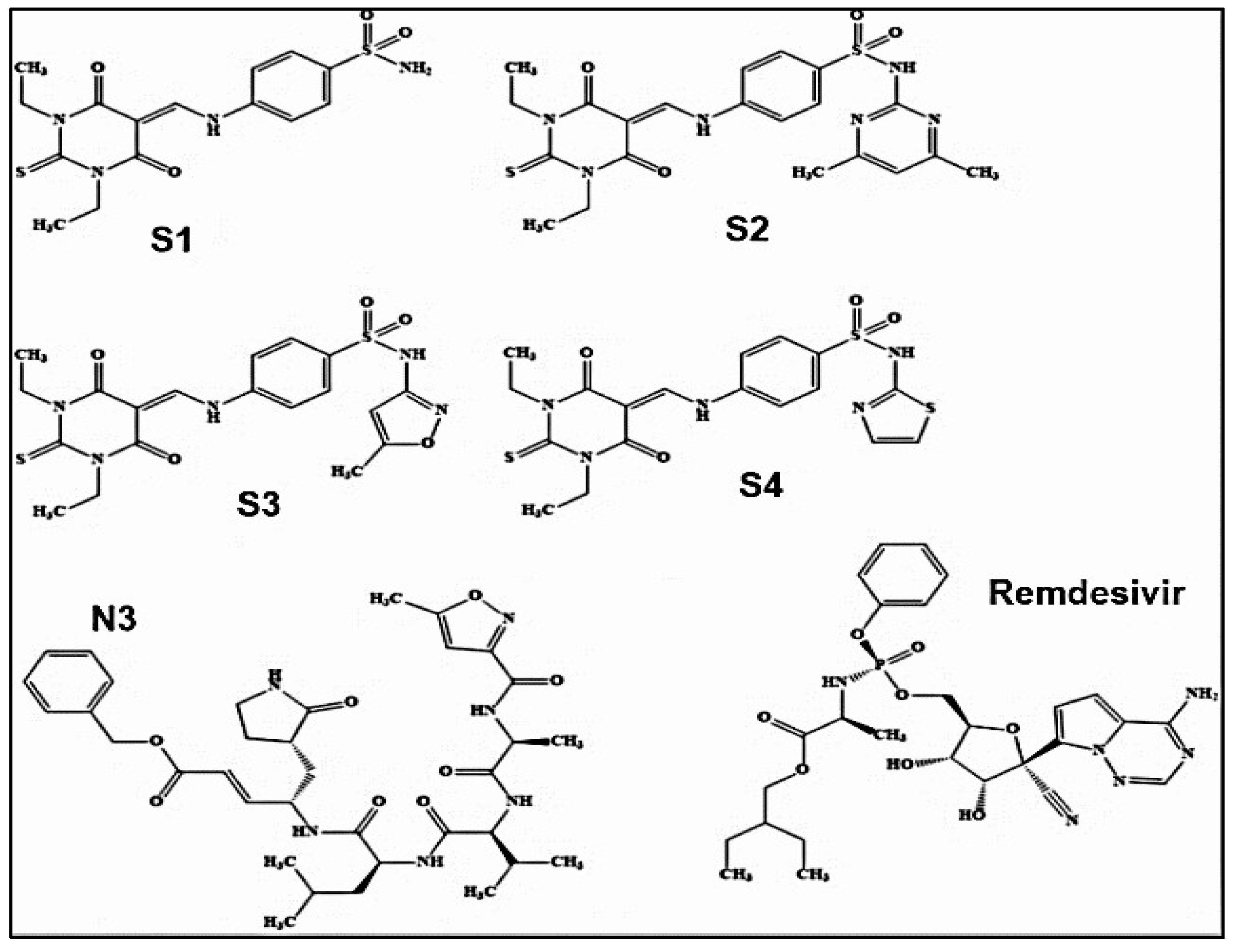
- Sarfraz, M., A. Rauf, P. Keller, and A.M. Qureshi, N,N'-dialkyl-2-thiobarbituric acid based sulfonamides as potential SARS-CoV-2 main protease inhibitors. Canadian Journal of Chemistry, 2021. 99: p. 330+. [CrossRef]
- Reprinted (adapted) with permission from Verma, Surjeet. Patel, Chirag N. Chandra, Muktesh., Identification of novel inhibitors of SARS-CoV-2 main protease (Mpro) from Withania sp. by molecular docking and molecular dynamics simulation, Journal of Computational Chemistry, copyright © 2021 Wiley Periodicals LLC. All rights preserved.
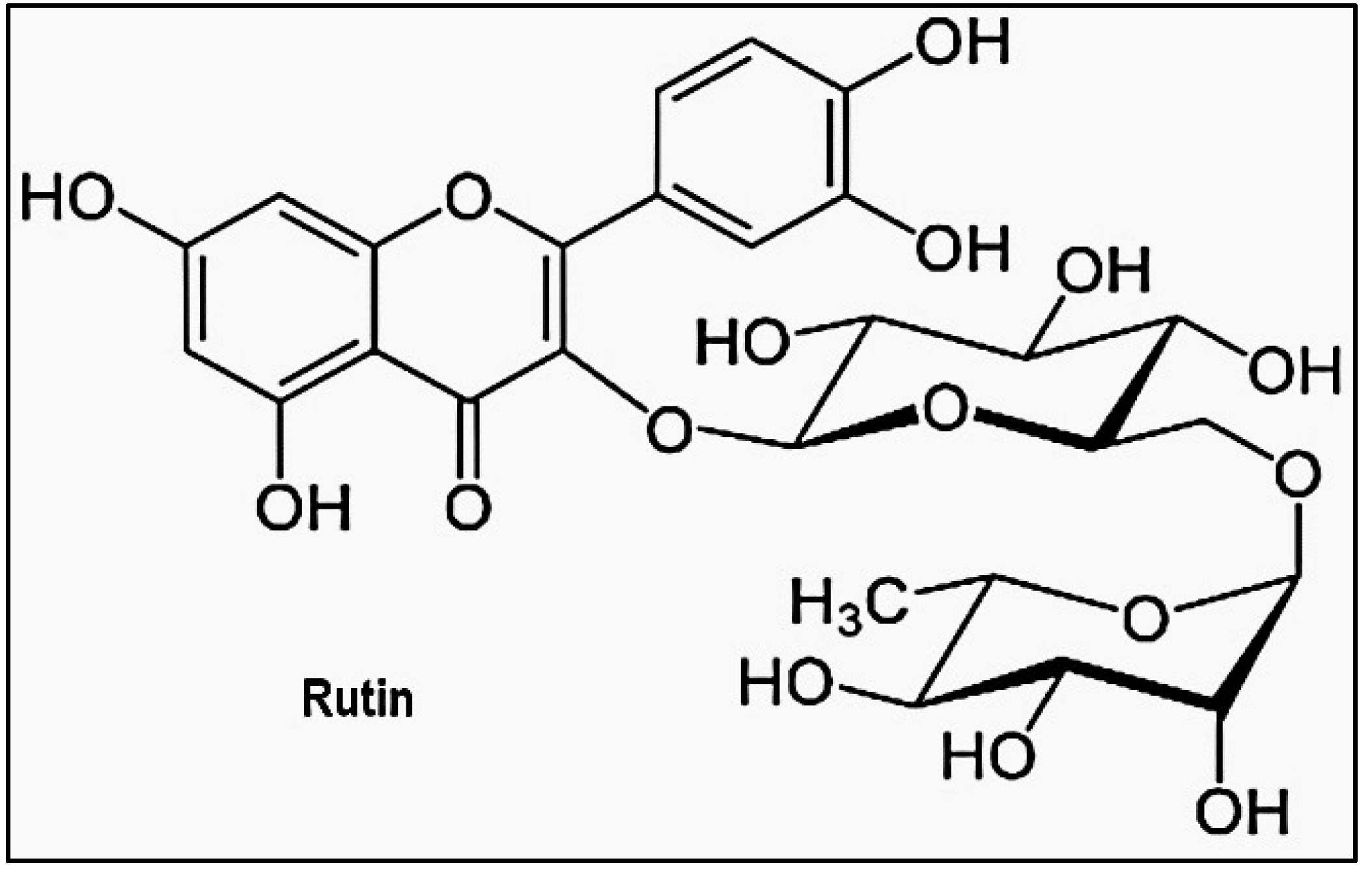
- Agrawal, P.K., C. Agrawal, and G. Blunden, Rutin: A Potential Antiviral for Repurposing as a SARS-CoV-2 Main Protease (Mpro) Inhibitor. Natural product communications, 2021. 16(4): p. 1934578. [CrossRef]
- Adapted from Rutin: A Potential Antiviral for Repurposing as a SARS-CoV-2 Main Protease (Mpro) Inhibitor by Agrawal, Pawan K. Agrawal, Chandan. Blunden, Gerald, used under Creative Commons Attribution 4.0 International License. No changes were made.
- Mohaptra, R.K., K. Dhama, A.A. El-Arabey, A.K. Sarangi, R. Tiwari, T. Bin Emran, M. Azam, M.K. Raval, V. Seidel, and M. Abdalla, Repurposing benzimidazole and benzothiazole derivatives as potential inhibitors of SARS-CoV-2: DFT, QSAR, molecular docking, molecular dynamics simulation, and in-silico pharmacokinetic and toxicity studies. Journal of King Saud UniversityScience, 2021. 33(8). [CrossRef]
- Adapted from Repurposing benzimidazole and benzothiazole derivatives as potential inhibitors of SARS-CoV-2: DFT, QSAR, molecular docking, molecular dynamics simulation, and in-silico pharmacokinetic and toxicity studies by R. K. Mohaptra, K. Dhama, A. A. El-Arabey, A. K. Sarangi, R. Tiwari, T. Bin Emran, et al. Journal of King Saud University-Science, 2021. 33(8), used under Creative Commons Attribution 4.0 International License. No changes were made.
- Elgohary, A.M., A.A. Elfiky, F. Pereira, T.M. Abd El-Aziz, M. Sobeh, R.K. Arafa, and A. El-Demerdash, Investigating the structure-activity relationship of marine polycyclic batzelladine alkaloids as promising inhibitors for SARS-CoV2 main protease (Mpro). Computers in biology and medicine, 2022. 147: p. 105738-105738. [CrossRef]
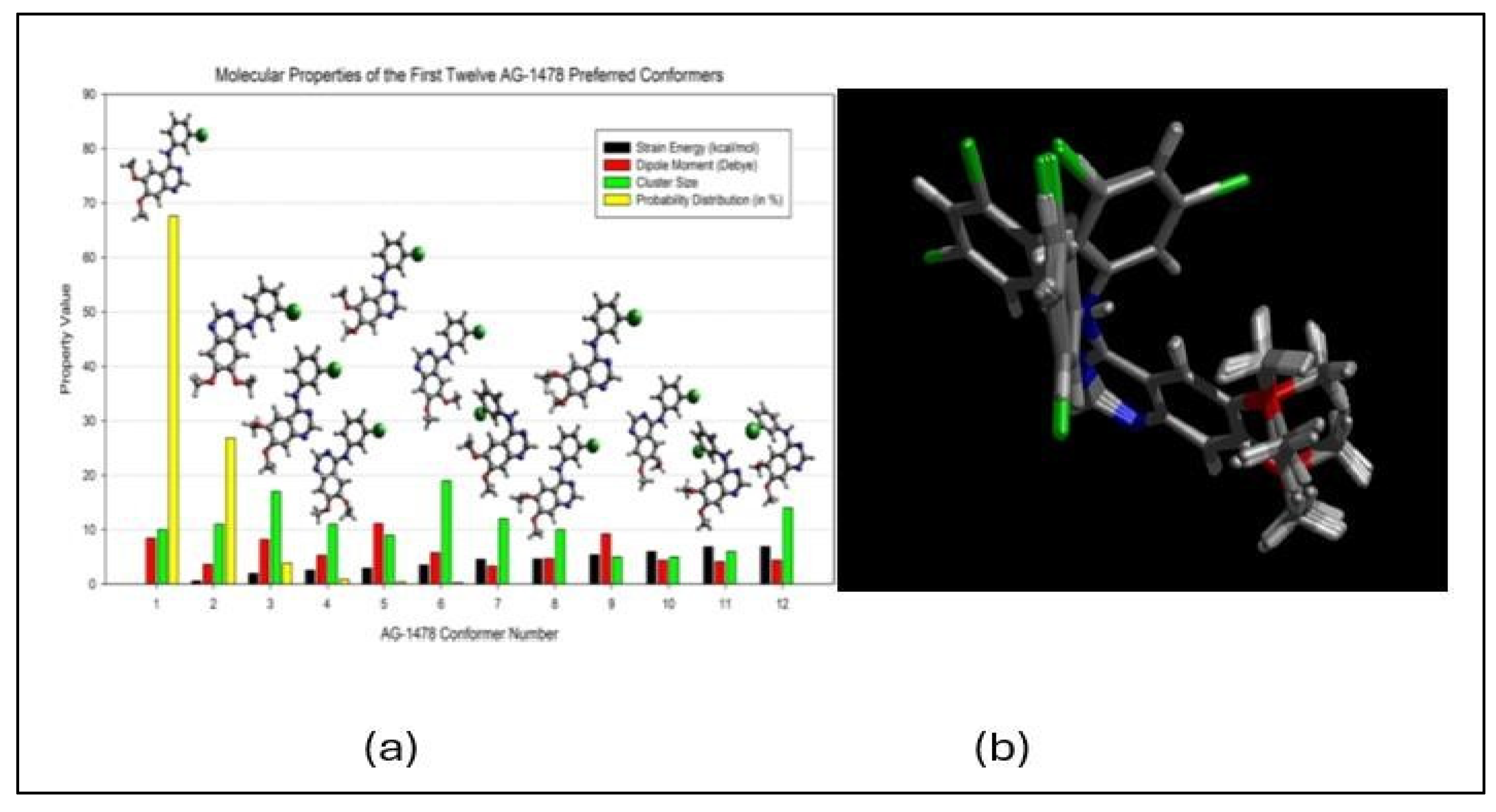
- Khattab, M., C. Chatterjee, A. Clayton, and F. Wang, Tyrosine kinase inhibitor (AG-1478): two conformers of a tyrosine kinase inhibitor (AG-1478) disclosed using simulated UV-vis absorption spectroscopy. New J. Chem, 2016. 40: p. 8296.
- Khattab, M., F. Wang, and A.H. Clayton, UV–Vis spectroscopy and solvatochromism of the tyrosine kinase inhibitor AG-1478. Spectrochimica Acta Part A: Molecular and Biomolecular Spectroscopy, 2016. 164: p. 128-132. [CrossRef]
- Khattab, M., F. Wang, and A.H. Clayton, A pH-induced conformational switch in a tyrosine kinase inhibitor identified by electronic spectroscopy and quantum chemical calculations. Scientific Reports, 2017. 7(1): p. 16271. [CrossRef]
- Wang, F. and V. Vasilyev, Accelerating optical reporting for conformation of tyrosine kinase inhibitors in solutions. International Journal of Quantum Chemistry, 2021. 121(20): p. e26765. [CrossRef]
- Alagawani, S., V. Vasilyev, and F. Wang, Optical spectra of EGFR inhibitor AG-1478 for benchmarking DFT functionals. Electronic Structure, 2023. 5(2): p. 024011. [CrossRef]
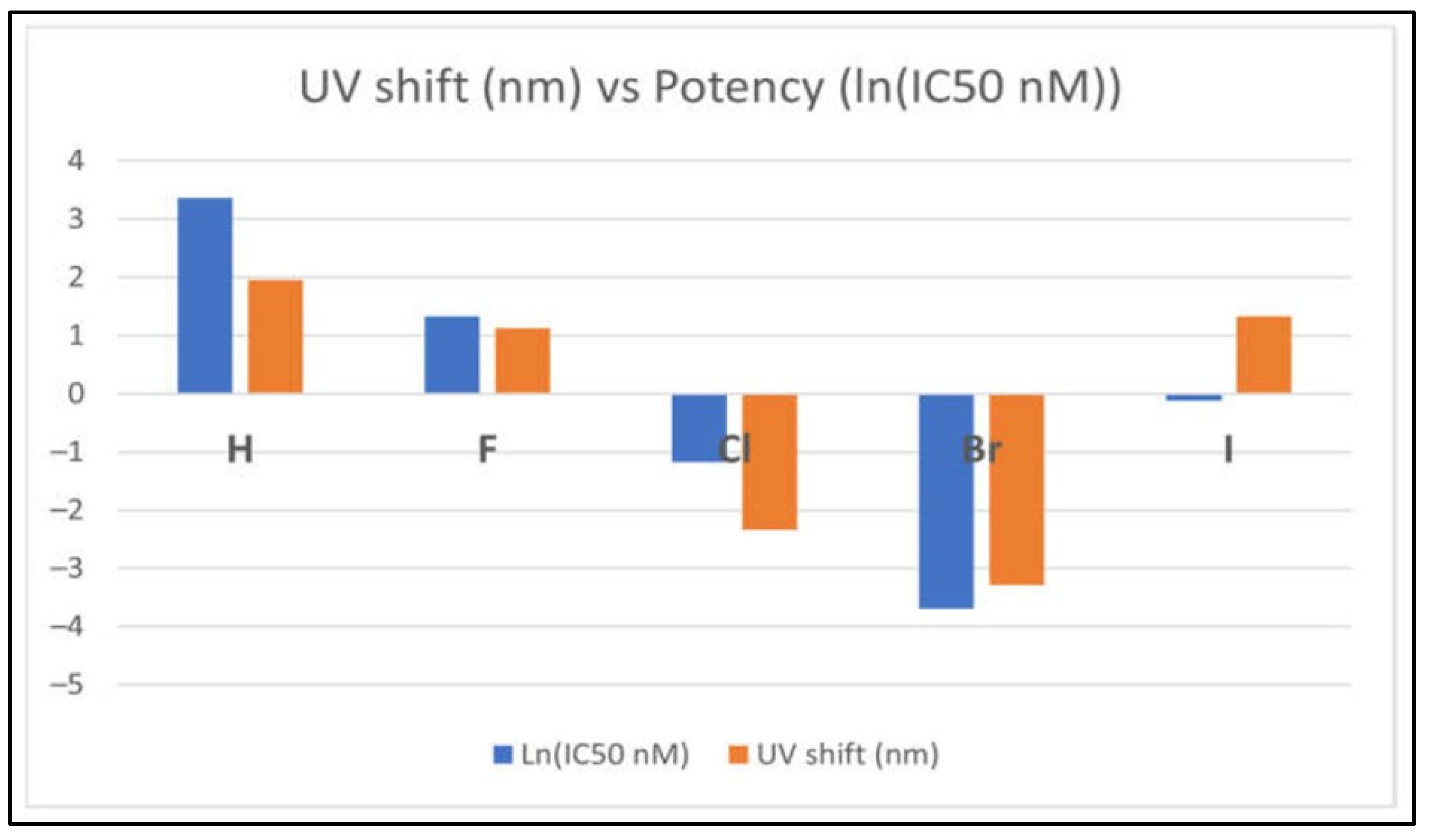
- Alagawani, S., V. Vasilyev, A.H. Clayton, and F. Wang, Insights into Halogen-Induced Changes in 4-Anilinoquinazoline EGFR Inhibitors: A Computational Spectroscopic Study. Molecules, 2024. 29(12): p. 2800. [CrossRef]
- Ashraf-Uz-Zaman, M., T.K. Chua, X. Li, Y. Yao, B.K. Moku, C.B. Mishra, V. Avadhanula, P.A. Piedra, and Y. Song, Design, Synthesis, X-ray Crystallography, and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease. ACS Infectious Diseases, 2024. 10(2): p. 715-731. [CrossRef]
- Wang, K., J. Gao, S. Shen, J.A. Tuszynski, J. Ruan, and G. Hu, An Accurate Method for Prediction of Protein-Ligand Binding Site on Protein Surface Using SVM and Statistical Depth Function. BioMed Research International, 2013. 2013(1): p. 409658. [CrossRef]
- Habgood, M., T. James, and A. Heifetz, Conformational searching with quantum mechanics. Quantum Mechanics in Drug Discovery, 2020: p. 207-229. [CrossRef]
- Reprinted (adapted) with permission from Wang, Vasilyev., Accelerating optical reporting for conformation of tyrosine kinase inhibitors in solutions, International Journal of Quantum Chemistry, copyright © 2021 Wiley Periodicals LLC. All rights are preserved.

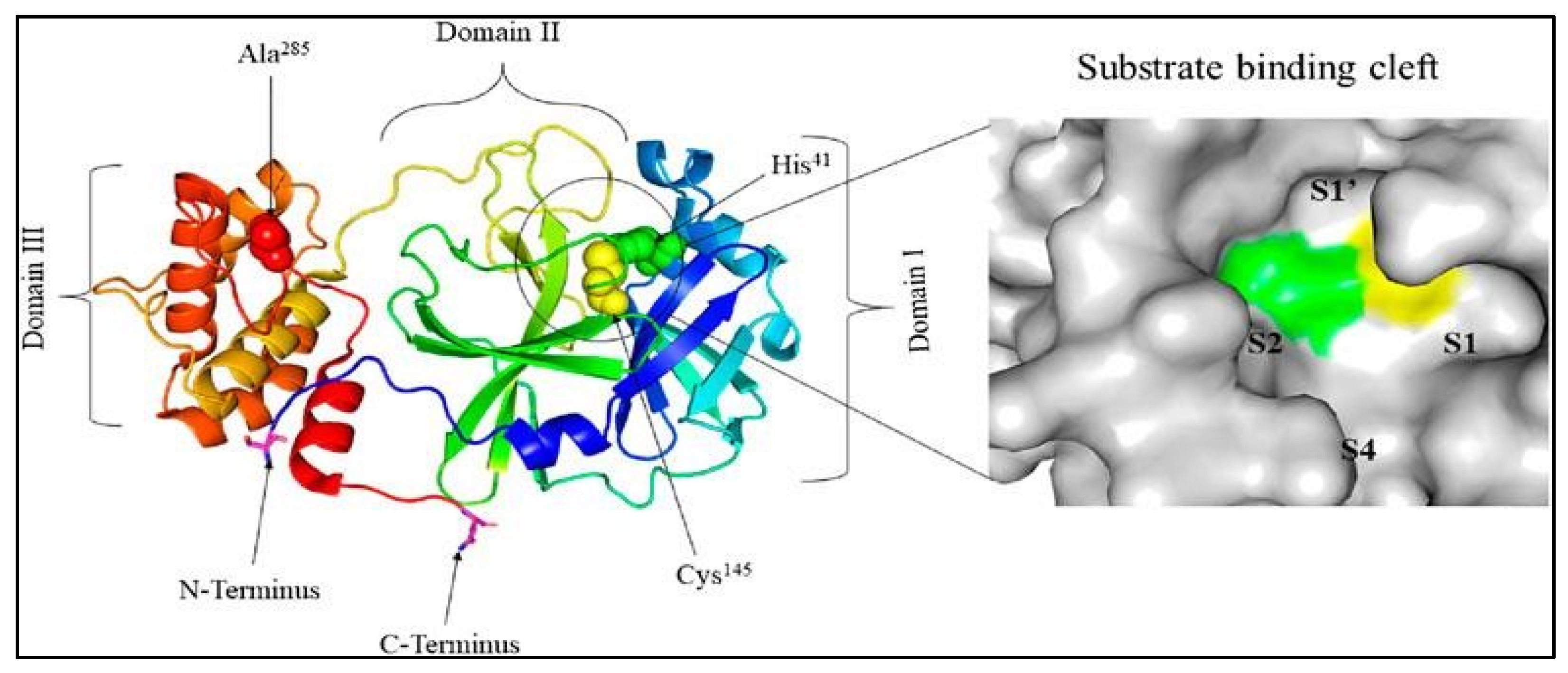
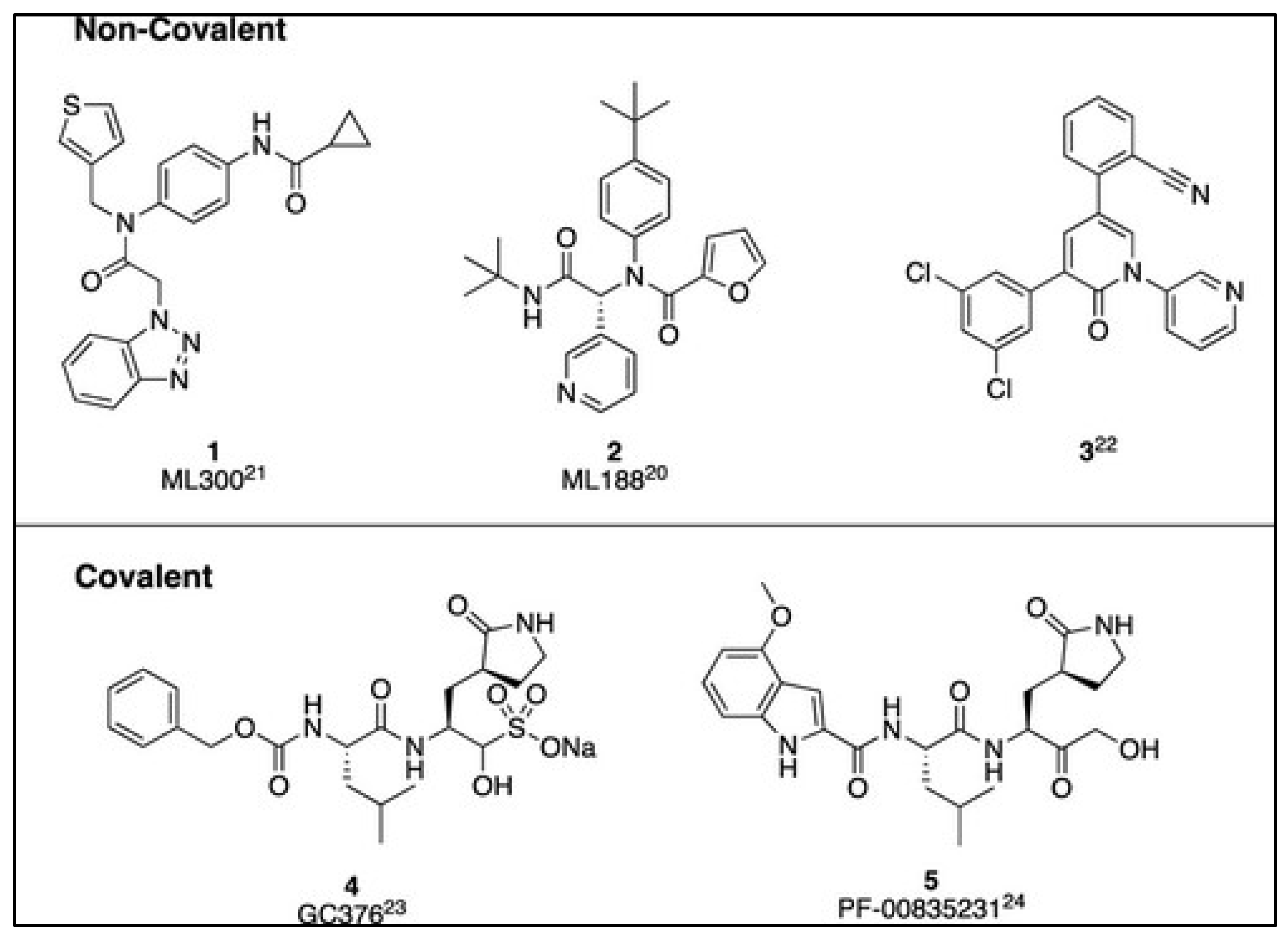
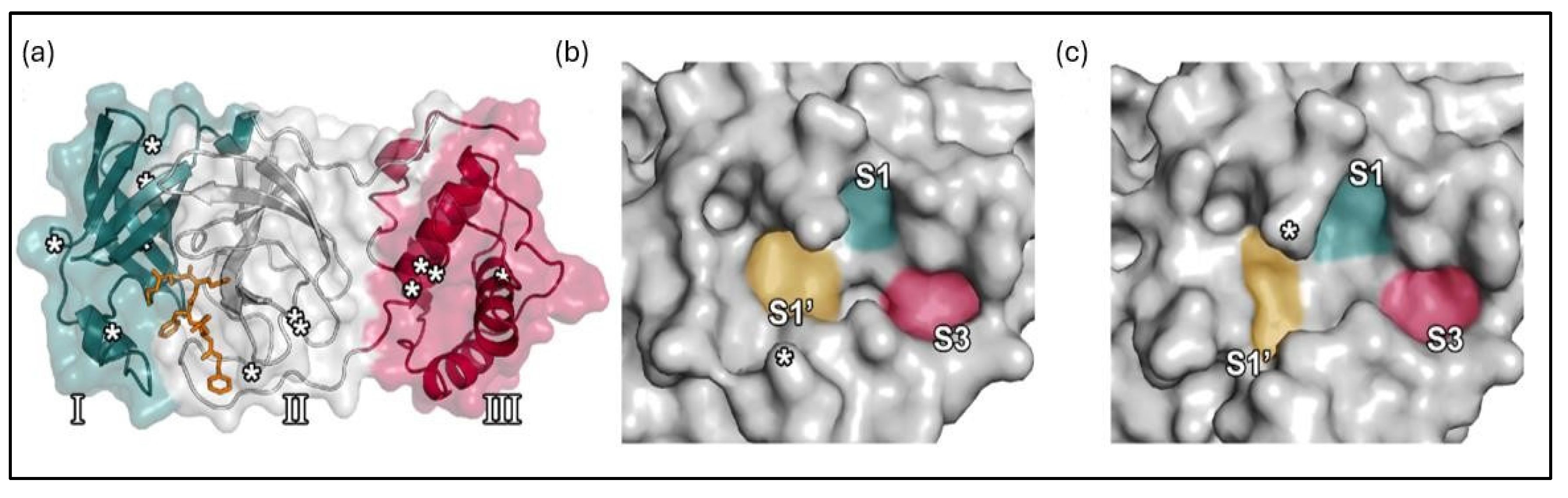
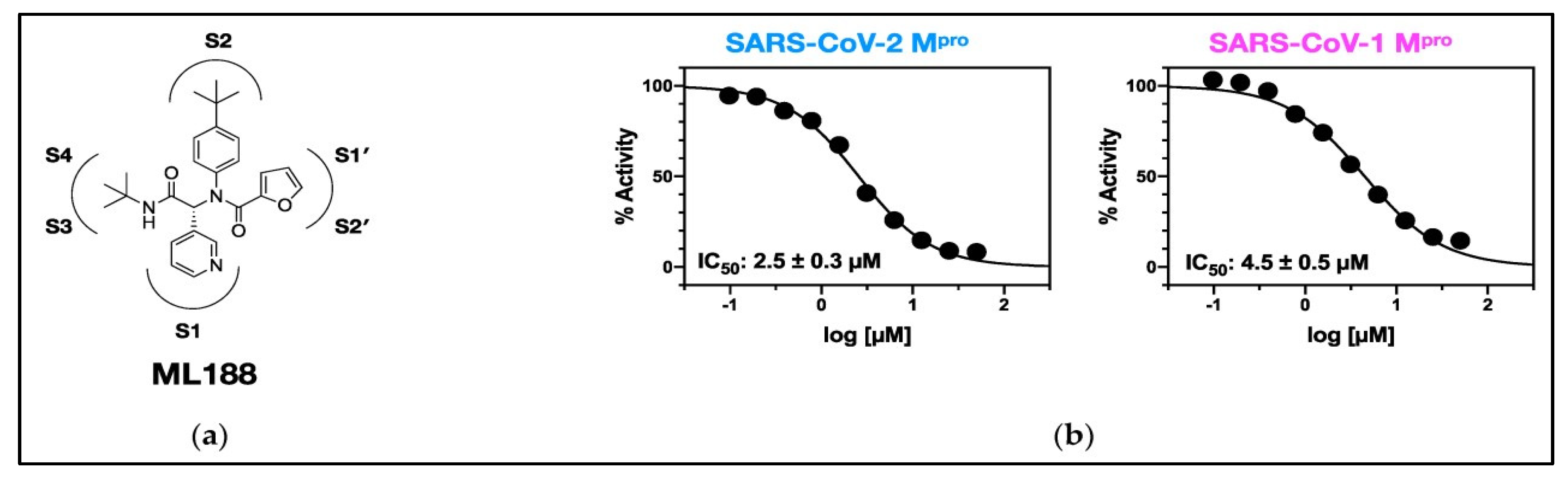
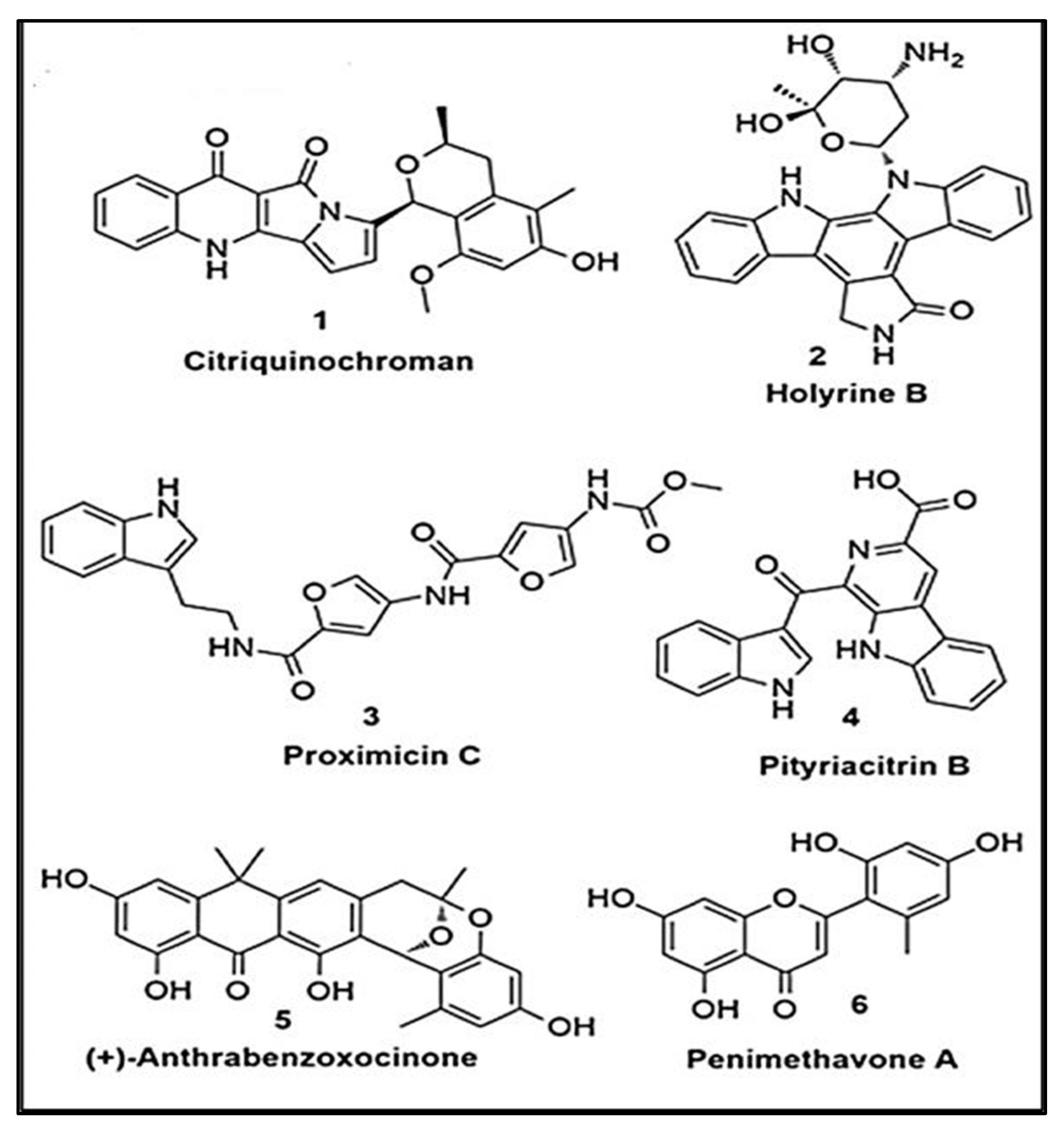
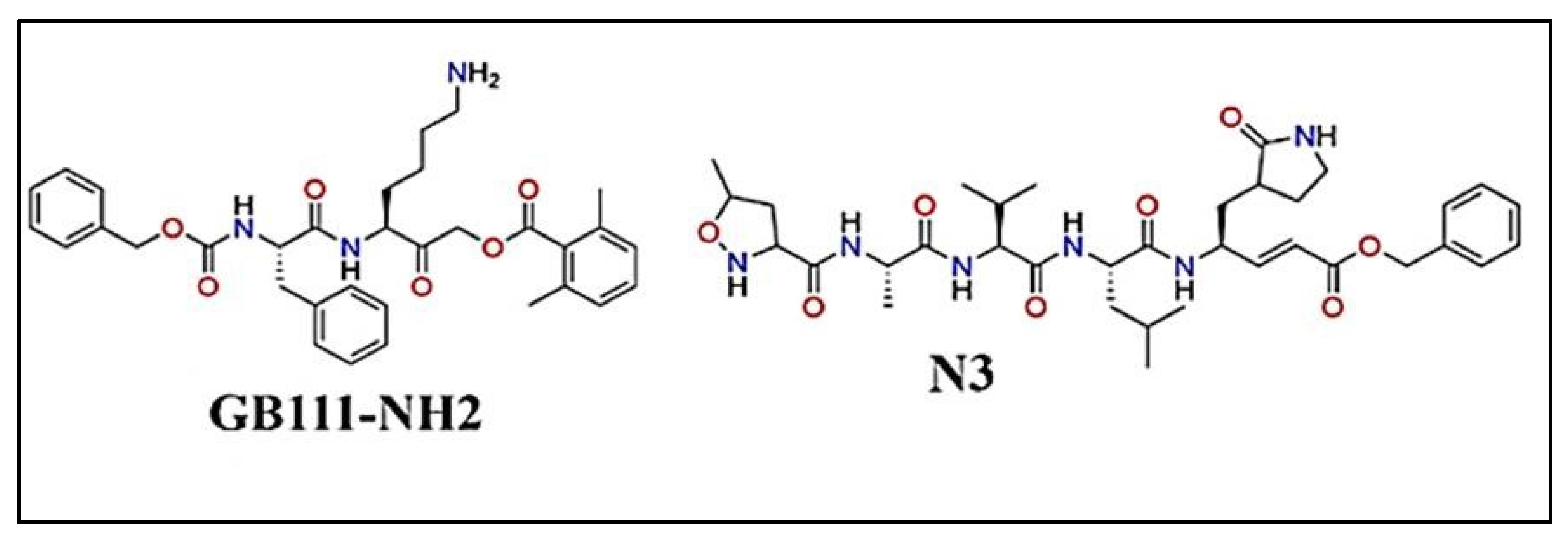
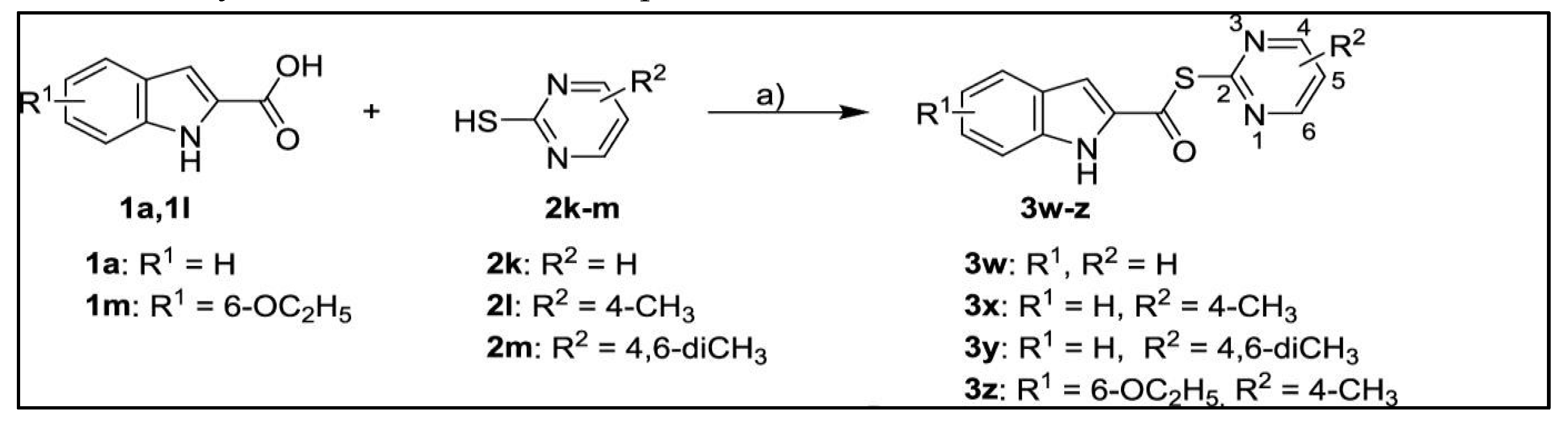
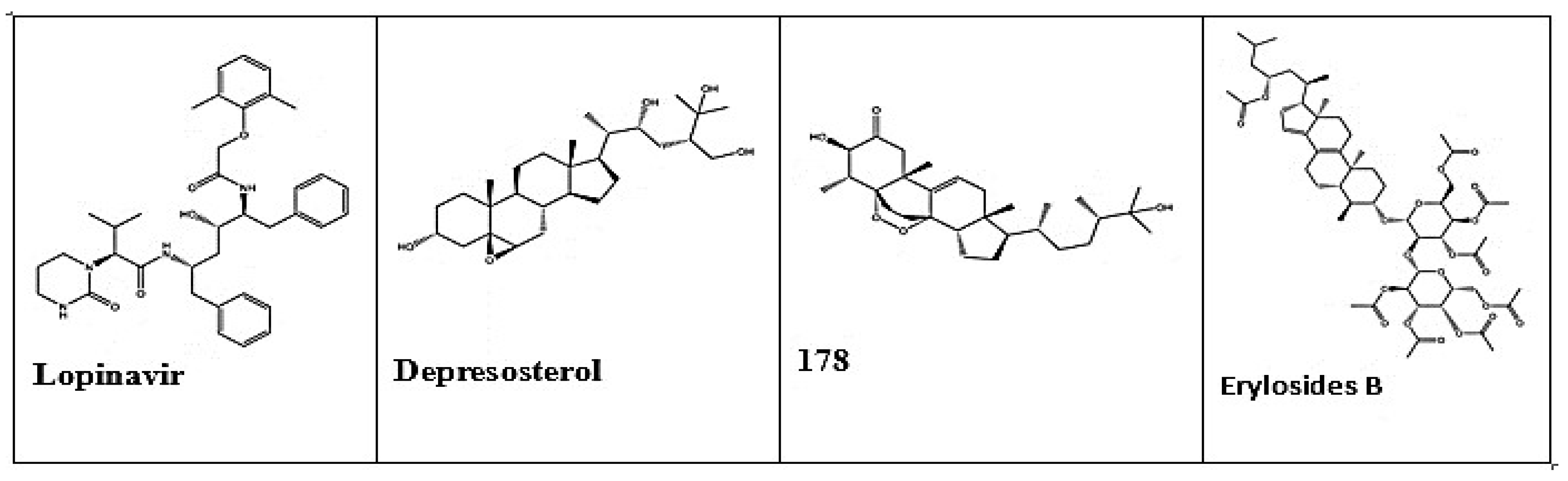



 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





