Submitted:
17 August 2024
Posted:
19 August 2024
You are already at the latest version
Abstract
Keywords:
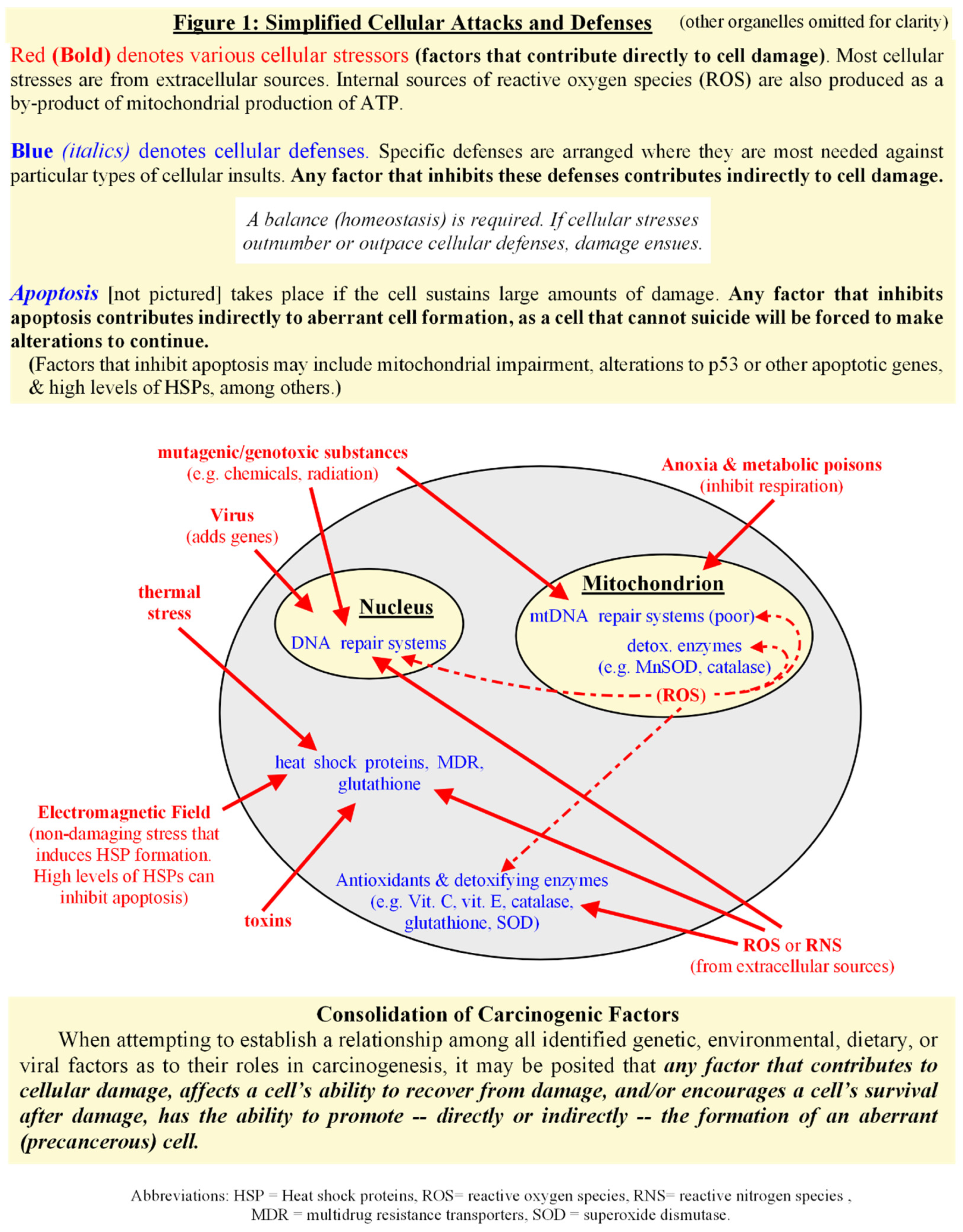
Introduction
Consolidation of Carcinogenic Factors
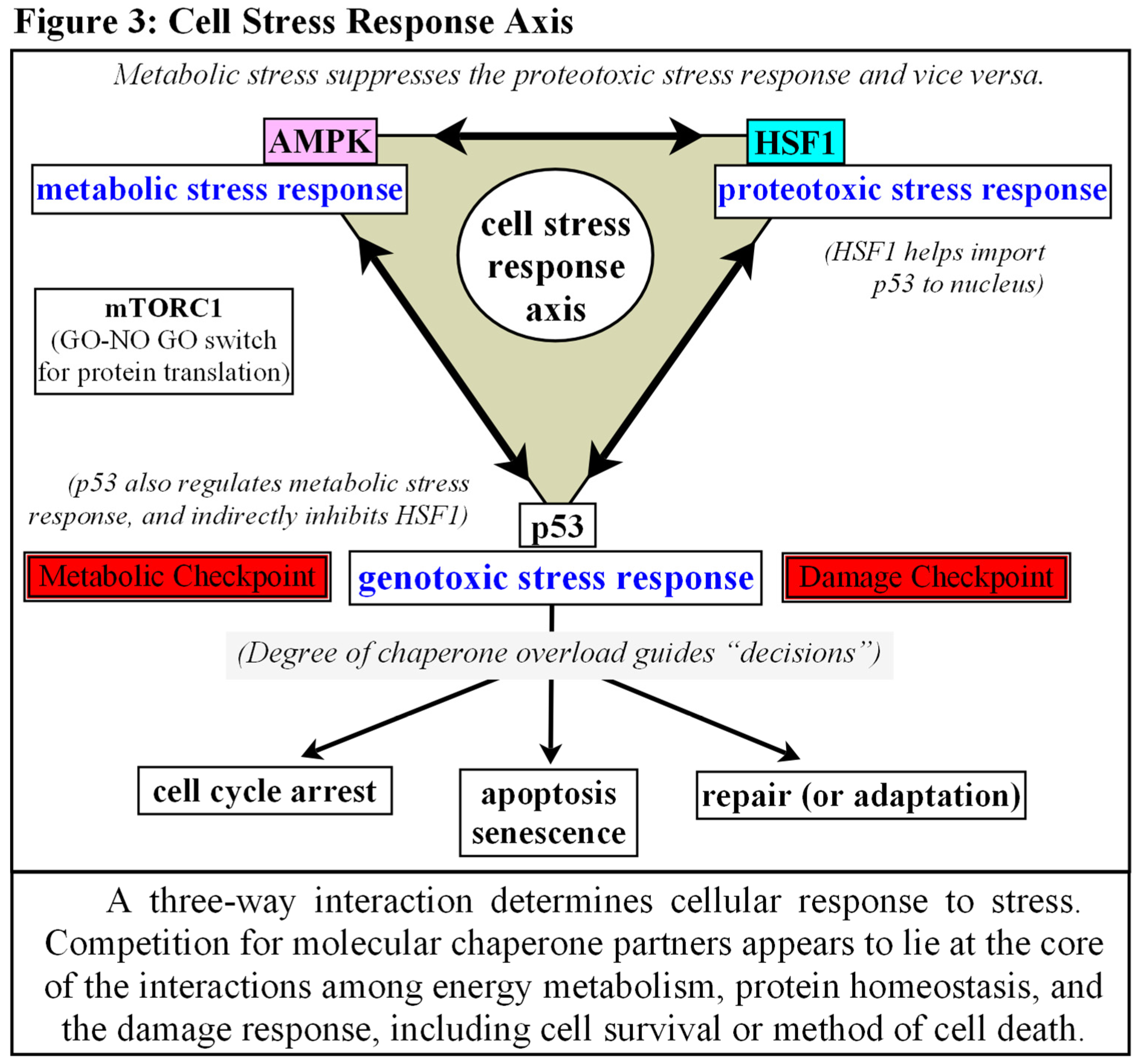
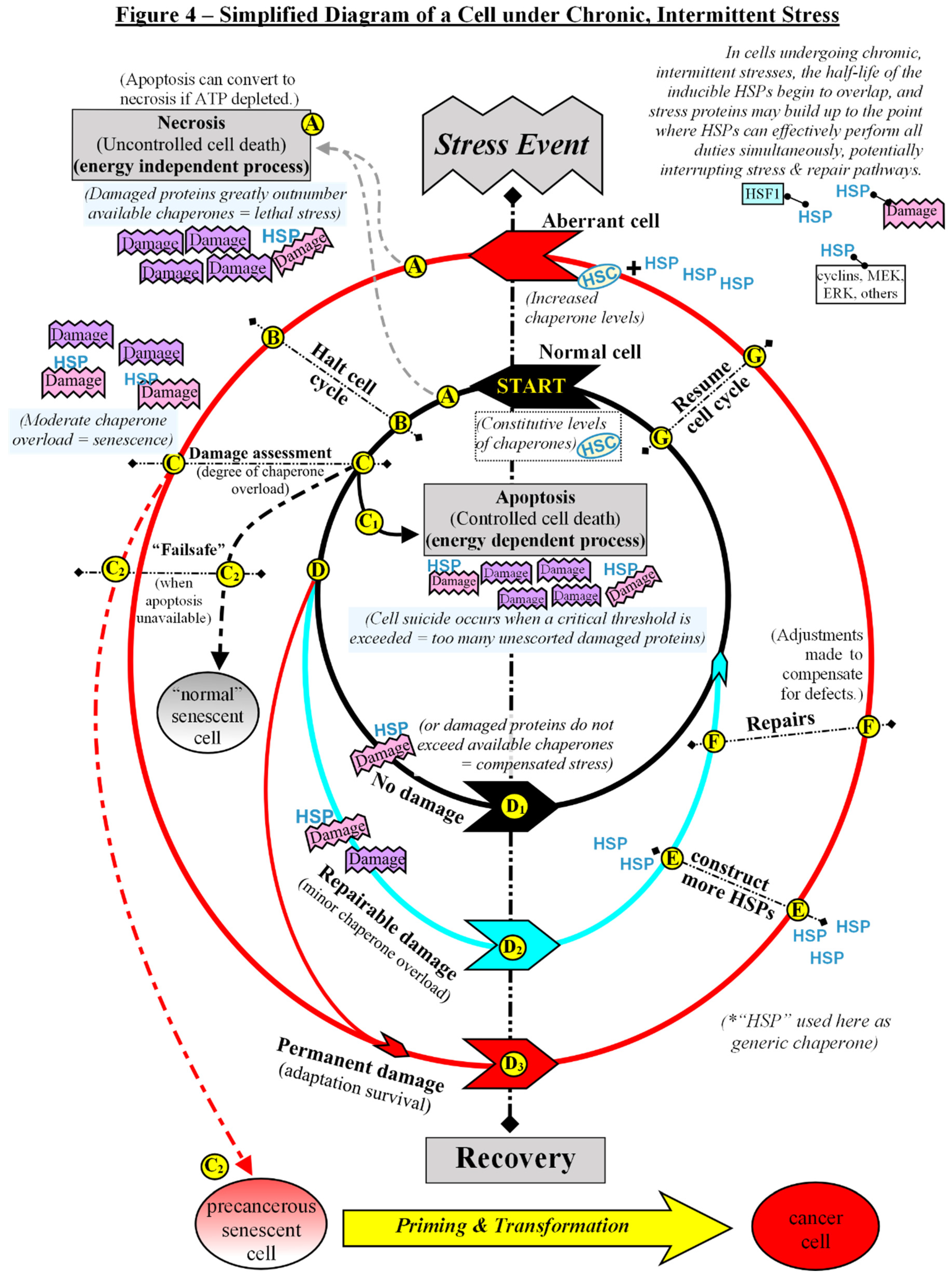
Chaperones as Protectors, Regulators, and Autonomic Decision-Makers.

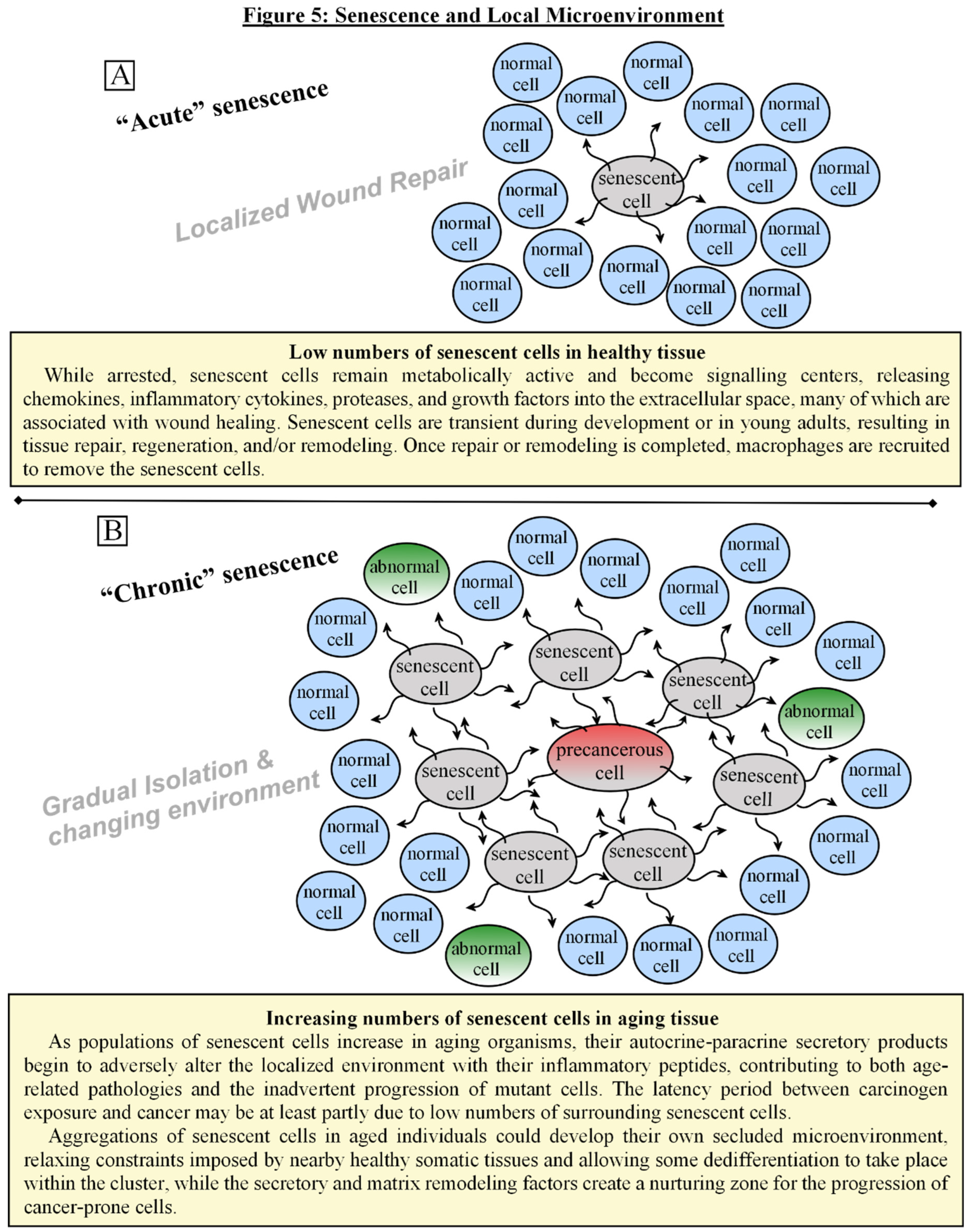
The Role of Senescence
Model for the Development of a Senescent Cell with Malignant Potential
Latency, Microenvironment, and the “Priming” of a Precancerous Senescent Cell
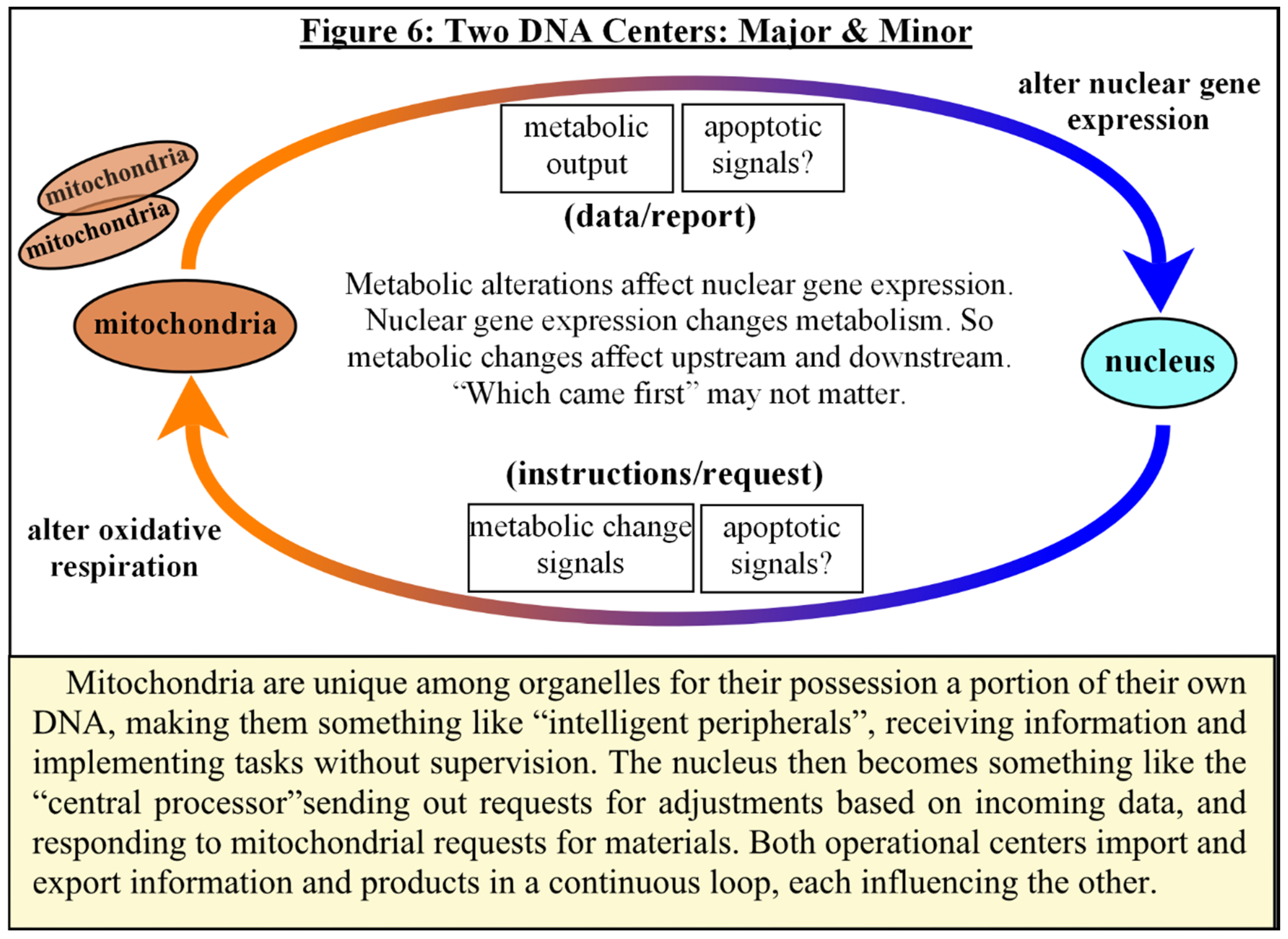
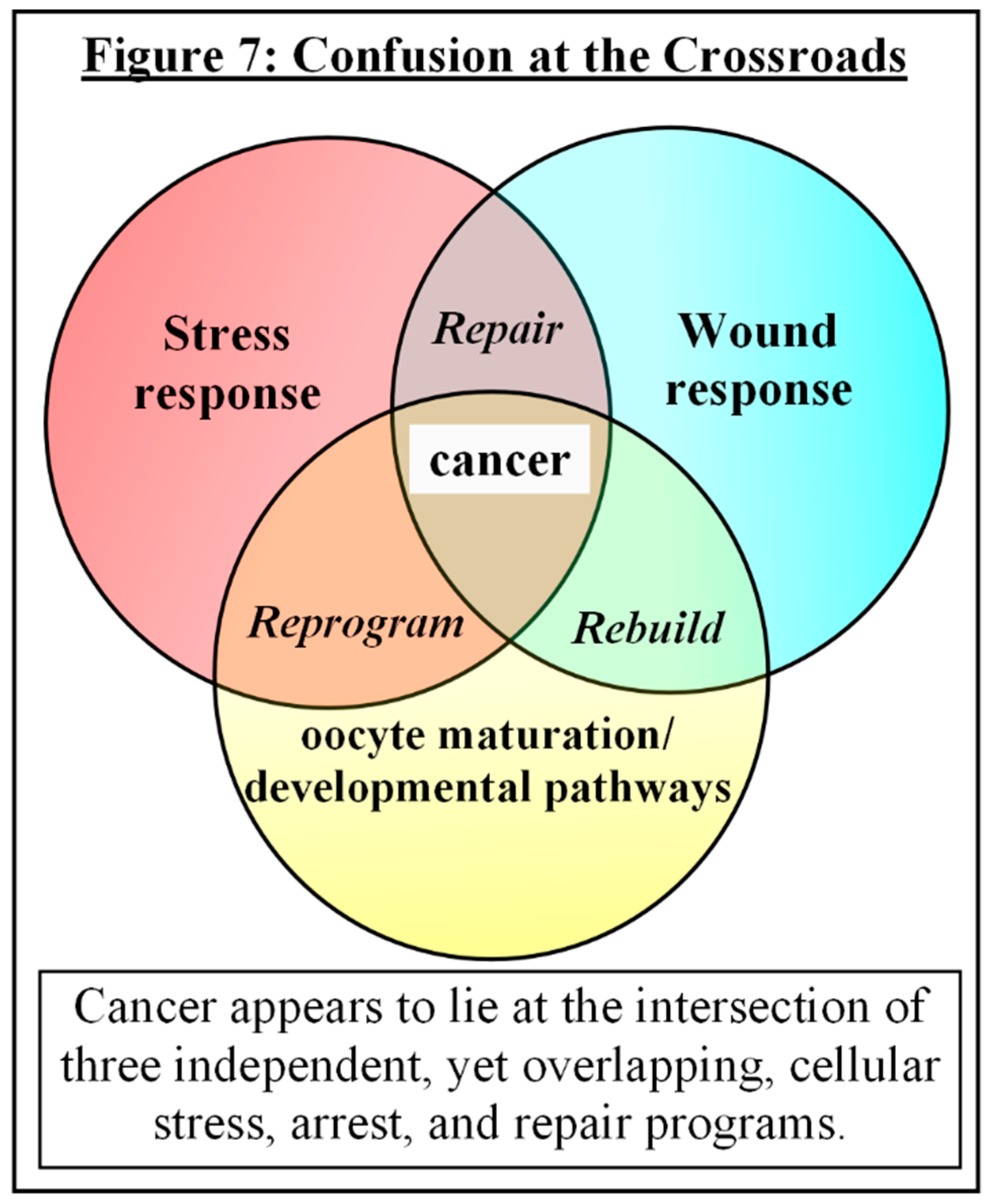
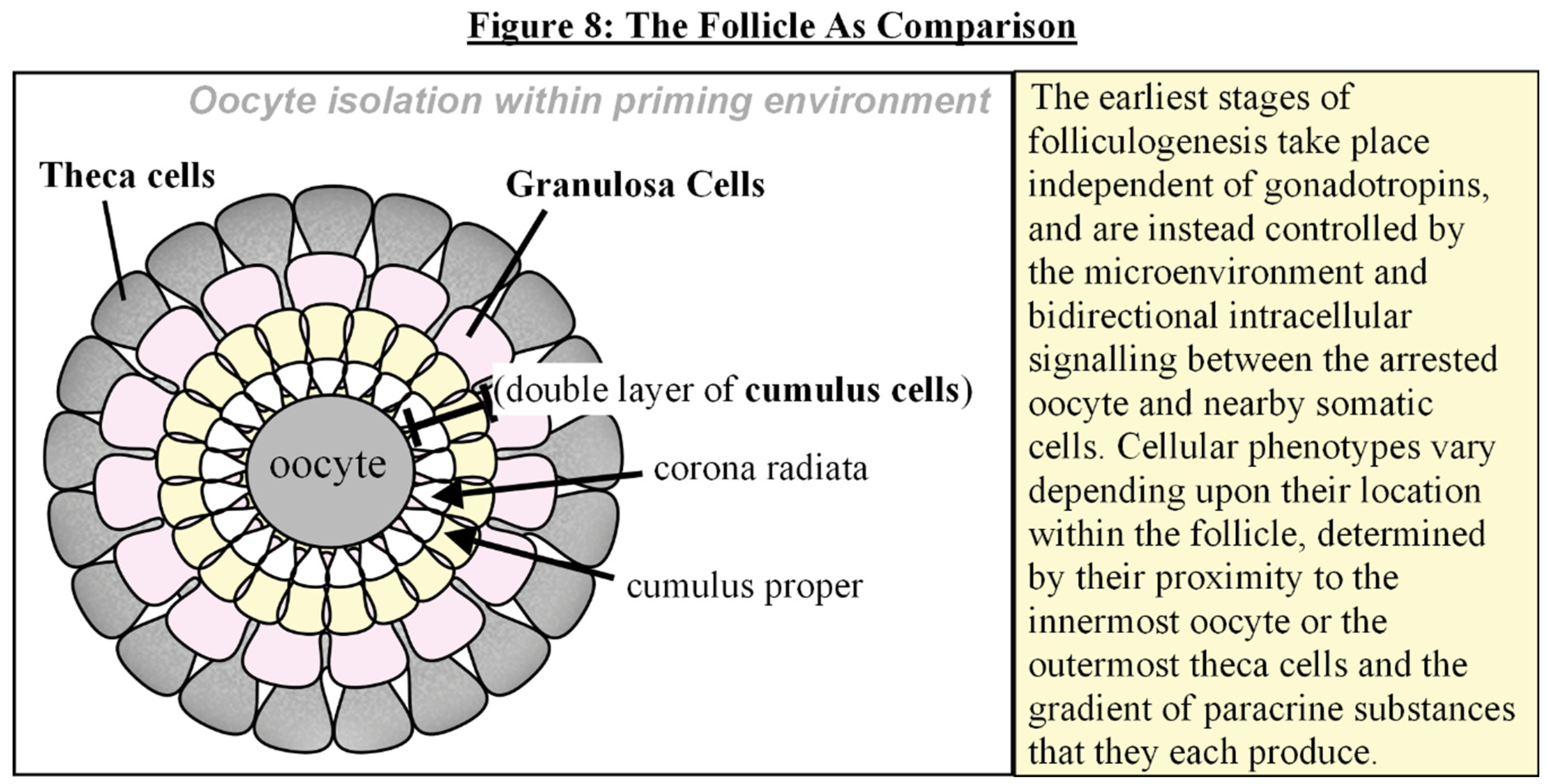
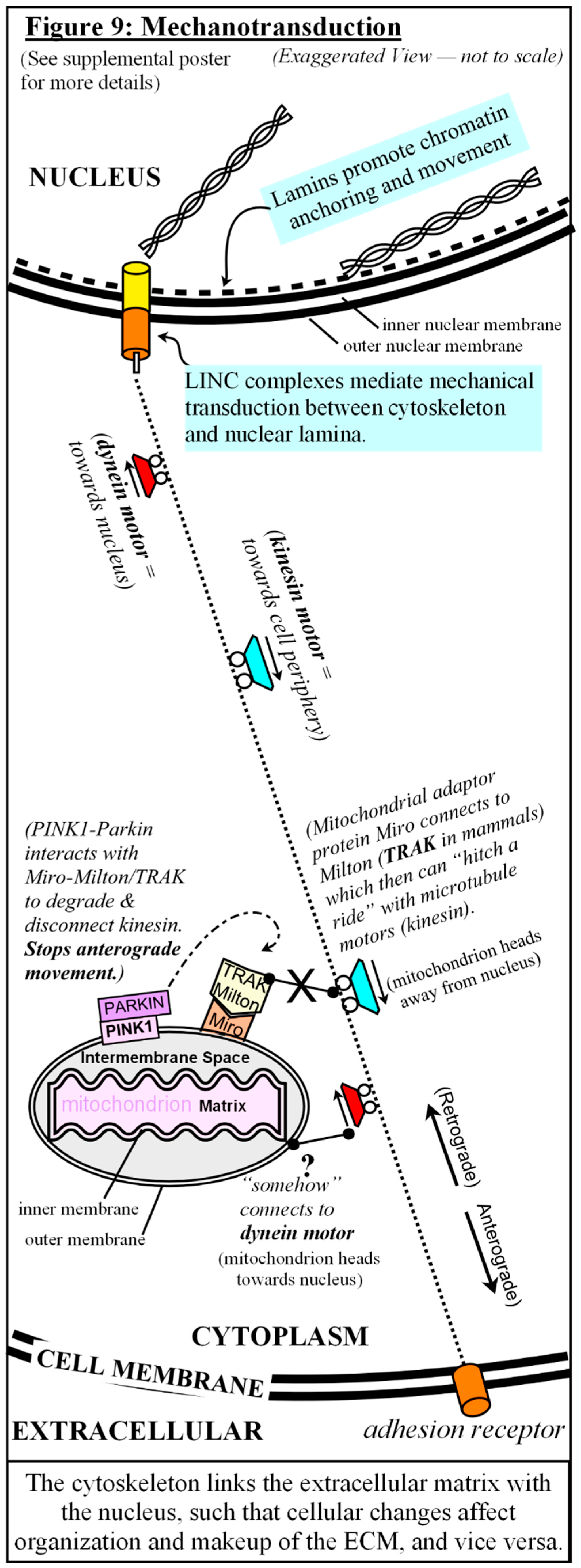
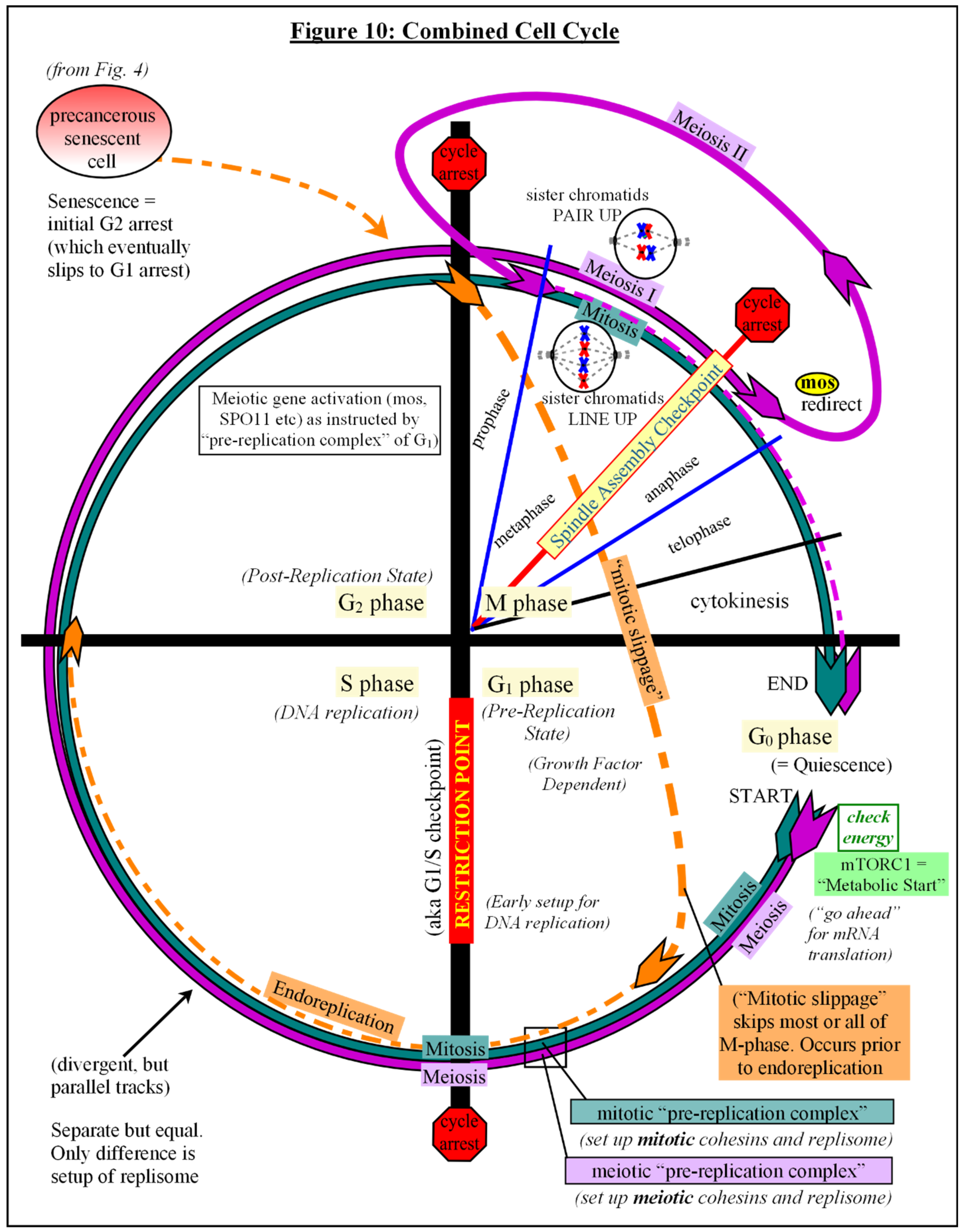
Putting It All Together
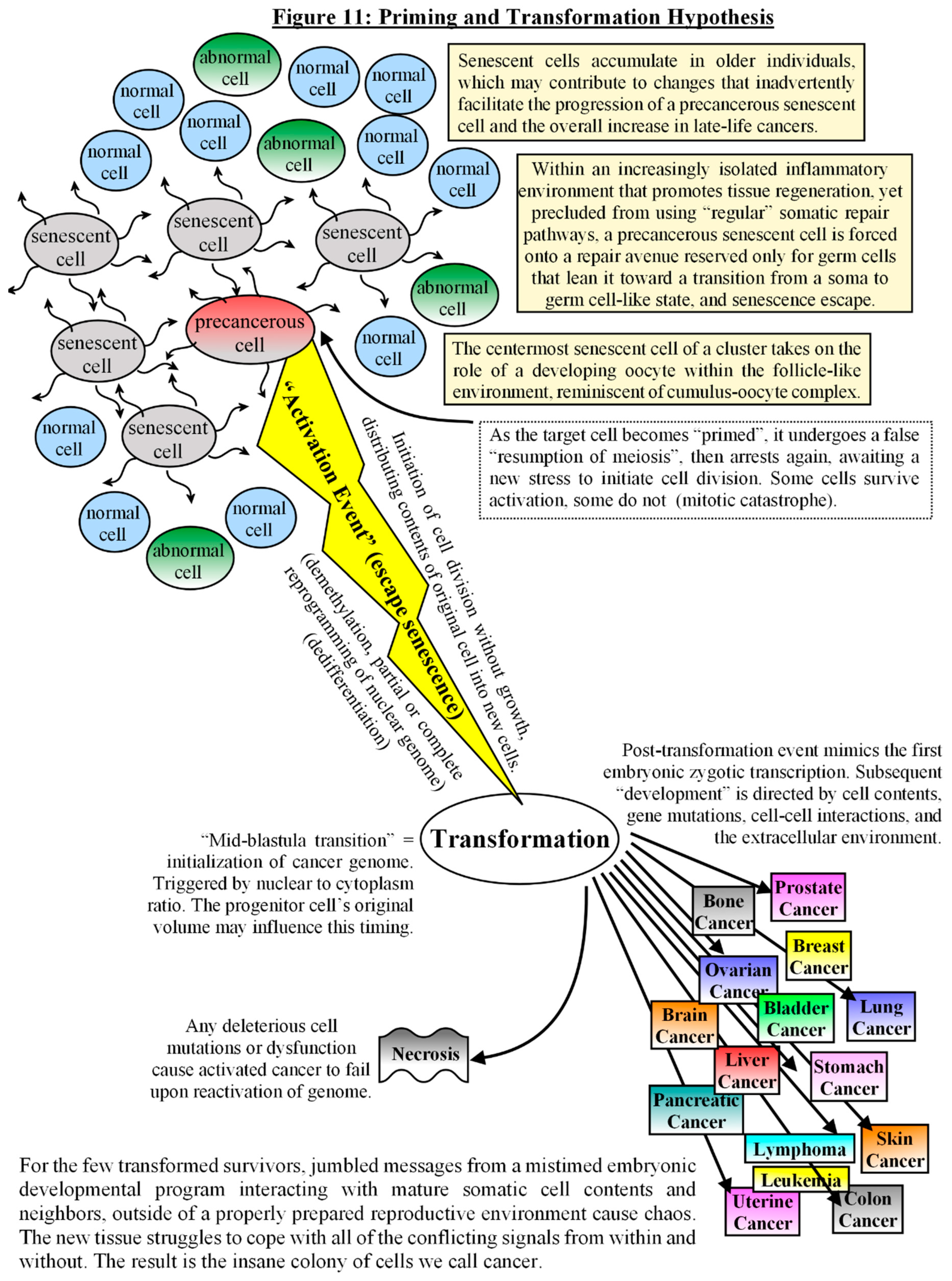
Discussion
Funding Sources
Acknowledgments
Conflict of Interest
References
- 1) Weinberg RA. 2014. Coming Full Circle — From Endless Complexity to Simplicity and Back Again. Cell, 157: 267-271. [CrossRef]
- 2) Pérez-Losada J, Castellanos-Martína A, Mao J-H. 2011. Cancer Evolution and Individual Susceptibility. Integr Biol (Camb),3(4): 316–328. [CrossRef]
- 3) Carbone M, Arron ST, Beutler B, Bononi A, Cavenee W, Cleaver JE, et al. 2020. Tumour Predisposition and Cancer Syndromes as Models to Study Gene X Environment Interactions. Nat Rev Cancer, 20(9):533-549. [CrossRef]
- 4) Adjiri A. 2017. DNA Mutations May Not Be the Cause of Cancer. Oncol Ther, 5:85-101. [CrossRef]
- 5) Krzyszczyk P, Acevedo A, Davidoff EJ, Timmins LM, Marrero-Berrios I, et al. 2018. The Growing Role of Precision and Personalized Medicine for Cancer Treatment. Technology (Singap World Sci), 6(3-4):79-100. [CrossRef]
- 6) Hoeben A, Joosten EA, van den Beuken-van Everdingen MHJ. 2021. Personalized Medicine: Recent Progress in Cancer Therapy. Cancers, 13:242. [CrossRef]
- 7) Fouad YA, Aanei C. 2017. Revisiting the Hallmarks of Cancer. Am J Cancer Res, 7(5):1016-1036.
- 8) Edelman EJ, Guinney J, Chi JT, Febbo PG, Mukherjee S. (2008) Modeling Cancer Progression via Pathway Dependencies. PLoS Comput Biol 4(2): e28. [CrossRef]
- 9) Gabriela Jiménez-Valerio G, Casanovas O. 2013. Anti-Angiogenic Therapy for Cancer and the Mechanisms of Tumor Resistance Contributions to Science, 9:67-73. [CrossRef]
- 10) Bergers G, Hanahan D. 2008. Modes of Resistance to Anti-Angiogenic Therapy. Nat Rev Cancer, 8(8): 592–603. [CrossRef]
- 11) Warburg O. 1956. On The Origin of Cancer Cells. Science, 123(3191):309-314. [CrossRef]
- 12) Warburg O. 1966. The Prime Cause and Prevention of Cancer — Part 2. Revised lecture at the meeting of the Nobel-Laureates on June 30, 1966 at Lindau, Lake Constance, Germany.
- 13) Enns GM. 2003. The Contribution of Mitochondria to Common Disorders. Mol. Genet. Metab., 80(1-2):11-26. [CrossRef]
- 14) Bartnik E, Lorenc A, Mroczek K. 2001. Human Mitochondria in Health, Disease, Ageing, and Cancer. J. Appl. Genet., 42(1): 65-71. [PubMed]
- 15) Carew JS, Huang P. 2002. Mitochondrial Defects in Cancer. Mol. Cancer, 1(1):9. [CrossRef]
- 16) Wei Y-H., Ma Y-S, Lee H-C, Lee CF, Lu C-Y. 2001. Mitochondrial Theory of Aging Matures — Roles of mtDNA Mutation and Oxidative Stress in Human Aging. Zhonghua Yi Xue Za Zhi, 64(5):259-270. [PubMed]
- 17) Galluzzi L, Kepp O, Trojel-Hansen C, Kroemer G. 2012. Mitochondrial Control of Cellular Life, Stress, and Death. Circ Res. 00:1198-1207. [CrossRef]
- 18) Zamzami N, Susin SA, Marchetti P, Hirsch T, Gomez-Monterray I, Castedo M, Kroemer G. 1996. Mitochondrial Control of Nuclear Apoptosis. Jour. Exp. Med., 183(4):1533-1544. [CrossRef]
- 19) Singh KK, Russell J, Sigala B, Zhang Y, Williams J, Keshav KF. 1999. Mitochondrial DNA Determines the Cellular Response to Cancer Therapeutic Agents. Oncogene, 18(48): 6641-6646. [CrossRef]
- 20) Vaupel P, Multhoff G. 2021. Revisiting the Warburg effect: Historical Dogma Versus Current Understanding. J Physiol 599.6: 1745–1757. [CrossRef]
- 21) Monk M, Holding C. 2001. Human Embryonic Genes Re-Expressed in Cancer Cells. Oncogene, 20(56):8085-8091. [CrossRef]
- 22) Aiello NM., Stanger BZ. 2016. Echoes of the Embryo: Using the Developmental Biology Toolkit to Study Cancer. Dis. Model Mech, 9(2): 105-114. [CrossRef]
- 23) Dreesen O, Brivanlou AH. 2007. Signaling Pathways in Cancer and Embryonic Stem Cells. Stem Cell Rev. [CrossRef]
- 24) Costanzo V, Bardelli A, Siena S, Abrignani S. 2018. Exploring the Links Between Cancer and Placenta Development. Open Biol., 8:180081. [CrossRef]
- 25) Feichtinger J, Aldeailij I, Anderson R, Almutairi M, Almatrafi A, Alsiwiehri N, et al. 2012. Meta-Analysis of Clinical Data Using Human Meiotic Genes Identifies a Novel Cohort of Highly-Restricted Cancer-Specific Marker Genes. Oncotarget, 3(8): 843-853. [CrossRef]
- 26) Wepsic HT. 1983. Overview of Oncofetal Antigens in Cancer. Ann.Clin.Lab.Sci., 13(4): 261-266. [PubMed]
- 27) Hall C, Clarke L, Pal A, Buchwald P, Eglinton T, Wakeman C, et al. 2019. A Review of the Role of Carcinoembryonic Antigen in Clinical Practice. Ann Coloproctol, 35(6): 294-305. [CrossRef]
- 28) Beard J. 1902. Embryological Aspects and Etiology of Carcinoma. Lancet, 1:1758-1761. [CrossRef]
- 29) Leach SD. 2005. Epithelial Differentiation in Pancreatic Development and Neoplasia. J Clin Gastroenterol, 39(2): S78-S82. [CrossRef]
- 30) Murray MJ, Lessey BA. 1999. Embryo Implantation and Tumor Metastasis: Common Pathways of Invasion and Angiogenesis. Sem Reprod. Endocrin., 17(3): 275-290. [CrossRef]
- 31) Manzo G. 2019. Similarities Between Embryo Development and Cancer Process Suggest New Strategies for Research and Therapy of Tumors: A New Point of View. Front. Cell Dev. Biol., 7:20. [CrossRef]
- 32) Barrett JC. 1993. Mechanisms of Multistep Carcinogenesis and Carcinogen Risk Assessment. Environmental Health Perspectives, 100:9-20. [CrossRef]
- 33) Braakhuis BJM, Brakenhoff RH, Leemans CR. 2005. Second Field Tumors: A New Opportunity for Cancer Prevention? The Oncologist,10:493–500. [CrossRef]
- 34) Ahmad AS, Ormiston-Smith N, Sasieni PD. 2015. Trends in the Lifetime Risk of Developing Cancer in Great Britain: Comparison of Risk for Those Born From 1930 to 1960. BJCancer, 112: 943-947. [CrossRef]
- 35) Huang R-X, Zhou P-K. 2020. DNA Damage Response Signaling Pathways and Targets for Radiotherapy Sensitization in Cancer. Signal Transduction and Targeted Therapy 5:60. [CrossRef]
- 36) Stewart BW. 2019. Mechanisms of Carcinogenesis: from Initiation and Promotion to the Hallmarks. In: Tumour Site Concordance and Mechanisms of Carcinogenesis. Lyon (FR): International Agency for Research on Cancer; 2019. (IARC Scientific Publications, No. 165.) Chapter 11. Baan RA, Stewart BW, Straif K, editors. Available from: https://www.ncbi.nlm.nih.gov/books/NBK570326/.
- 37) Hajri QA, Dash S, Feng W, Garner HR, Anandakrishnan R. 2020. Identifying Multi-Hit Carcinogenic Gene Combinations: Scaling Up a Weighted Set Cover Algorithm Using Compressed Binary Matrix Representation on a GPU. Scientific Reports,10:2022. [CrossRef]
- 38) Li R, Sonik A, Stindl R, Rasnick D, Duesberg P. 2000. Aneuploid -vs- Gene Mutation Hypothesis of Cancer: Recent Study Claims Mutation But is Found to Support Aneuploidy. PNAS, 97(7): 3236-3241. [CrossRef]
- 39) Zhu S, Wang J, Zellmer L, Xu N, Liu M, Hu Y, et al. 2022. Mutation or Not, What Directly Establishes a Neoplastic State, Namely Cellular Immortalilty and Autonomy, Still Remains Unknown and Should Be Prioritized in our Research. J. Cancer, 13(9):2810-2843. [CrossRef]
- 40) Nohmi T. 2018. Thresholds of Genotoxic and Non-Genotoxic Carcinogens. Toxicol Res, 34(4):281-290. [CrossRef]
- 41) Goldblatt H, Cameron G. 1953. Induced Malignancy in Cells From Rat Myocardium Subjected to Intermittent Anaerobiosis during Long Propagation in vitro. J.Exp.Med., 97(4):525-552. [CrossRef]
- 42) Poljsak B, Milisav I. 2012. Clinical Implications of Cellular Stress Responses. Basic Med Sci, 12(2): 122-126. [CrossRef]
- 43) Li D, Duncan RF. 1995. Transient Acquired Thermotolerance in Drosophila, Correlated with Rapid Degradation of Hsp70 During Recovery. Eur J Biochem., 231:454-465. [CrossRef]
- 44) George I, Geddis MS, Lill Z, Lin H, Gomez T, Blank M, et al. 2008. Myocardial Function Improved by Electromagnetic Field Induction of Stress Protein hsp70. J Cell Physiol, 216(3): 816-823. [CrossRef]
- 45) Soti C, Sreedhart AS, Csermely P. 2003. Apoptosis, Necrosis, and Cellular Senescence: Chaperone Occupancy as a Potential Switch. Aging Cell, 2:39-45. [CrossRef]
- 46) Ishikawa T, Zhang SS-M, Qin X, Takahashi Y, Oda H, Nakatsure Y, et al. 2003. DNA Repair and Cancer: Lessons from Mutant Mouse Models. Cancer Sci, 95(2): 112-117. [CrossRef]
- 47) Knudson AG. 1971. Mutation and Cancer: Statistical Study of Retinoblastoma. Proc Nat Acad Sci, 68(4): 820-823. [CrossRef]
- 48) Lamech LT, Haynes CM. 2015. The Unpredictability of Prolonged Activation of Stress Response Pathways. J Cell Biol, 209(6): 781-787. [CrossRef]
- 49) Ameya G, Birri DJ. 2023. The Molecular Mechanisms of Virus-Induced Human Cancers. J Mic. Path, 183:106292. [CrossRef]
- 50) Hatano Y, Ideta T, Hirata A, Hatano K, Tomita H, Okada H, et al. 2021. Virus-Driven Carcinogenesis. Cancers, 13:2625. [CrossRef]
- 51) Sree BR, Joy JM. 2011. A Review on Molecular Chaperones and Chaperone Overload. IJPDT, 1(2): 58-65.
- 52) Jeng W, Lee S, Sung N, Lee J, Tsai FTF. 2015. Molecular Chaperones: Guardians of the Proteome in Normal and Disease States. F1000 Research2015, 4(F1000 Faculty Rev):1448. [CrossRef]
- 53) Moreno DF, Parisi E, Yahya G, Vaggi F, Csikasz-Nagy A, Aldea M. 2019. Competition in the Chaperone-Client Network Subordinates Cell-Cycle Entry to Growth and Stress. Life Science Alliance, 2(2):201800277. [CrossRef]
- 54) Truman AW, Kristjansdottir K, Wolfgeher D, Hasin N, Polier S, Zhang H, et al. 2012. CDK-Dependent Hsp70 Phosphorylation Controls G1 Cyclin Abundance and Cell-Cycle Progression. Cell, 151: 1308-1318. [CrossRef]
- 55) Ali A, Bharadwaj S, O’Carroll R, Ovsenek N. 1998. HSP90 Interacts with and Regulates the Activity of Heat Shock Factor 1 in Xenopus Oocytes. Mol. Cell. Biol., 18(9): 4949-4960. [CrossRef]
- 56) Masser AE, Ciccarelli M, Andreasson C. 2020. Hsf1 on a Leash – Controlling the Heat Shock Response by Chaperone Titration. Exp. Cell Res., 396: 112246. [CrossRef]
- 57) Taylor RC, Cullen SP, Martin SJ. 2008. Apoptosis: Controlled Demolition at the Cellular Level. Nat. Rev. Mol. Cell Biol., 9: 231-241. [CrossRef]
- 58) Stracker TH. 2017. Chaperoning the DNA Damage Response. The FEBS Jour, 284:2375-2377. [CrossRef]
- 59) Knighton LE, Truman AW. 2019. Role of the Molecular Chaperones Hsp70 and Hsp90 in the DNA Damage Response. In: Heat Shock Proteins in Signaling Pathways. Heat Shock Proteins, vol. 17: 345-358. Springer, Cham. [CrossRef]
- 60) Pennisi R, Ascenzi P, di Masi A. 2015. Hsp90: A New Player in DNA Repair? Biomolecules, 5:2589-2618. [CrossRef]
- 61) Dote H, Burgan WE, Camphausen K, Tofilon PJ. 2006. Inhibition of Hsp90 Compromises the DNA Damage Response to Radiation. Cancer Res, 66(18): 9211-9220. [CrossRef]
- 62) Huiting W, Dekker SL, van der Lienden JCJ, Mergener R, Musskopf MK, Furtado GV, et al. 2022. Targeting DNA Topoisomerases or Checkpoint Kinases Results in an Overload of Chaperone Systems, Triggering Aggregation of a Metastable Subproteome. eLife, 11:e70726. [CrossRef]
- 63) Su K-H, Dai S, Tang Z, Xu M, Dai C. 2019. Heat Shock Factor 1 is a Direct Antagonist of AMP-Activated Protein Kinase. Molecular Cell, 76: 546-561. [CrossRef]
- 64) Dai S, Tang Z, Cao J, Zhou W, Li H, Sampson S, et al. 2015. Suppression of the HSF1-Mediated Proteotoxic Stress Response by the Metabolic Stress Sensor AMPK. The EMBO Jour, 34(3):275-293. [CrossRef]
- 65) Li J, Labbadia, J, Morimoto RL. 2017. Rethinking HSF1 in Stress, Development and Organismal Health. Trends Cell Biol, 27(12):895-905. [CrossRef]
- 66) Qiao A, Jin X, Pang J, Moskophidis D, Mivechi NF. 2017. The Transcriptional Regulator of the Chaperone Response HSF1 Controls Hepatic Bioenergetics and Protein Homeostasis. J. Cell. Biol., 216(3): 723-741. [CrossRef]
- 67) Canto C. 2017. The Heat Shock Factor HSF1 Juggles Protein Quality Control and Metabolic Regulation. J. Cell. Biol., 216(3): 551-553. [CrossRef]
- 68) Yang Q, Wang Y, Wang H, Li H, Zhu J, Cong L, et al. 2021. NAD+ Repletion Attenuates Obesity-Induced Oocyte Mitochondrial Dysfunction and Offspring Metabolic Abnormalities via a SIRT3-Dependent Pathway. Clin. Transl. Med., 11:e628. [CrossRef]
- 69) Nassour J, Martien S, Martin N, Deruy E, Tomellini E, Malaquin N, et al. 2016. Defective DNA Single-Strand Break Repair is Responsible for Senescence and Neoplastic Escape of Epithelial Cells. Nat. Comm., 7: 10399. [CrossRef]
- 70) Li J, Bonkowski MS,Moniot S, Zhang D, Hubbard BP, Ling AJY, et al. 2017. A Conserved NAD+ Binding Pocket That Regulates Protein-Protein Interactions During Aging. Science, 355(6331): 1312-1317. [CrossRef]
- 71) Li Q, Feldman RA, Radhakrishnan VM, Carey S, Martinez JD. 2008. Hsf1 is Required for the Nuclear Translocation of p53 Tumor Suppressor. Neoplasia, 10(10):1138-1145. [CrossRef]
- 72) Li Q, Martinez, JD. 2011. P53 is Transported into the Nucleus via an Hsf1-Dependent Nuclear Localization Mechanism. Mol Carcinog., 50(2): 143-152. [CrossRef]
- 73) Toma-Jonik A, Vydra N, Janus P, Widlak W. 2019. Interplay Between HSF1 and p53 Signaling Pathways in Cancer Initiation and Progression: Non-Oncogene and Oncogene Addiction. Cell. Onc., 42:579-589. [CrossRef]
- 74) Hasty P, Sharp ZD, Curiel TJ, Campisi J. 2013. MTORC1 and p53. Clash of the Gods? Cell Cycle, 12(1):20-25. [CrossRef]
- 75) Humpton TJ, Vousden KH. 2016. Regulation of Cellular Metabolism and Hypoxia by p53. Cold Spring Harb Perspect Med, 6(7):a026146. [CrossRef]
- 76) Su K-H, Cao J, Tang Z, Dai S, He Y, Sampson SB, et al. 2016. HSF1 Critically Attunes Proteotoxic Stress Sensing by mTORC1 to Combat Stress and Promote Growth. Nat. Cell. Biol., 18(5):527-539. [CrossRef]
- 77) Hayflick L, Moorhead PS. 1961. The Serial Cultivation of Human Diploid Cell Strains. Exp. Cell Res., 25:585-621. [CrossRef]
- 78) Ohtani N, Hara E. 2013. Roles and Mechanisms of Cellular Senescence in Regulation of Tissue Homeostasis. Cancer Sci., 104(5): 525-530. [CrossRef]
- 79) Ben-Porath I, Weinberg RA. 2004. When Cells Get Stressed: An Integrative View of Cellular Senescence. J. Clin. Invest. 113:8–13. [CrossRef]
- 80) Kuilman T, Michaloglou C, Mooi WJ, Peeper DS. 2010. The Essence of Senescence. Genes & Dev., 24:2463-2479. [CrossRef]
- 81) Gire V, Dulic V. 2015. Senescence From G2 Arrest, Revisited. Cell Cycle, 14(3):297-304. [CrossRef]
- 82) Soto-Gamez A, Quax WJ, Demaria M. 2019. Regulation of Survival Networks in Senescent Cells: From Mechanisms to Interventions. J. Mol. Biol., 431:2629-2643. [CrossRef]
- 83) Krtolica A, Parrinello S, Locket S, Desprez P-Y, Campisi J. 2001. Senescent Fibroblasts Promote Epithelial Cell Growth and Tumorigenesis: A Link Between Cancer and Aging. PNAS, 98(21): 12072-12077. [CrossRef]
- 84) van Deursen JM. 2014. The Role of Senescent Cells in Ageing. Nature, 509(7501): 439-446. [CrossRef]
- 85) Campisi J. 2013. Aging, Cellular Senescence, and Cancer. Annu Rev Physiol.,75:685–705. [CrossRef]
- 86) Demaria M, Ohtani N, Youssef SA, Rodier F, Toussaint W, Mitchell JR, et al. 2014. An Essential Role for Senescent Cells in Optimal Wound Healing through Secretion of PDGF-AA. Develop. Cell, 31:722-733. [CrossRef]
- 87) Jun J-I, Lau LF. 2010. Cellular Senescence Controls Fibrosis in Wound Healing. Aging, 2(9): 627-631. [CrossRef]
- 88) Stewart SA, Weinberg RA. 2000. Telomerase and Human Tumorigenesis. Sem in Canc. Biol., 10(6):399-406. [CrossRef]
- 89) Schwarze SR, DePrimo SE, Grabert LM, Fu VX, Brooks JD, Jarrard DF. 2002. Novel Pathways Associated with Bypassing Cellular Senescence in Human Prostate Epithelial Cells. J. Biol. Chem., 277(17):14877-14883. [CrossRef]
- 90) Shay JW, Wright WE. 2005. Senescence and Immortalization: Role of Telomeres and Telomerase. Carcinogenesis, 26(5): 867-874. [CrossRef]
- 91) Morel Y, Barouki R. 1999. Repression of Gene Expression by Oxidative Stress. Biochem. J., 342:481-496. [CrossRef]
- 92) Childs BG, Baker DJ, Kirkland JL, Campisi J, van Deursen JM. 2014. Senescence and Apoptosis: Dueling or Complementary Cell Fates? EMBO Reports,15(11):1139-1153. [CrossRef]
- 93) von Zglinicki T, Wan T, Miwa S. 2021. Senescence in Post-Mitotic Cells: A Driver of Aging? Antioxidants & Redox Signaling, 34(4):308-323. [CrossRef]
- 94) DiCarlo AL, Farrell JM, Litovitz TA. 1999. Myocardial Protection Conferred by Electromagnetic Fields. Circulation, 99:813-816. [CrossRef]
- 95) Wiese M, Bannister AJ. 2020. Two Genomes, One cell: Mitochondrial-Nuclear Coordination via Epigenetic Pathways. Molec. Metab., 38:100942. [CrossRef]
- 96) Walker BR, Moraes CT. 2022. Nuclear-Mitochondrial Interactions. Biomolecules, 12:427. [CrossRef]
- 97) Herranz N, Gil J. 2018. Mechanisms and Functions of Cellular Senescence. J. Clin. Invest., 128(4): 1238-1246. [CrossRef]
- 98) Maciel-Baron LA, Morales-Rosales SL, Aquino-Cruz AA, Triana-Martínez F, Galván-Arzate S, Luna-López A, et al. 2016. Senescence Associated Secretory Phenotype Profile from Primary Lung Mice Fibroblasts Depends on the Senescence Induction Stimuli. Age,38:26. [CrossRef]
- 99) Young ARJ, Narita M. 2009. SASP Reflects Senescence. EMBO Reports, 10(3):228-230. [CrossRef]
- 100) Ritschka B, Storer M, Mas A, Heinzmann F, Ortells MC, Morton JP, et al. 2017. The Senescence-Associated Secretory Phenotype Induces Cellular Plasticity and Tissue Regeneration. Genes & Develop., 31:172-183. [CrossRef]
- 101) Rhinn M, Ritschka B, Keyes WM. 2019. Cellular Senescence in Development, Regeneration and Disease. Development, 146:dev151837. [CrossRef]
- 102) Nelson G, Wordsworth J, Wang C, Jurk D, Lawless C, Martin-Ruiz C, et al. 2012. A Senescent Cell Bystander Effect: Senescence-Induced Senescence. Aging Cell,11:345-349. [CrossRef]
- 103) Olivieri F, Albertini MC, Orciani M, Ceka A, Cricca M, Procopio AD, et al. 2015. DNA Damage Response (DDR) and Senescence: Shuttled Inflamm-miRNAs on the Stage of Inflamm-aging. Oncotarget, 6(34): 35509-33521. [CrossRef]
- 104) Parrinello S, Coppe J-P, Krtolica A, Campisi J.2004. Stromal-Epithelial Interactions in Aging and Cancer: Senescent Fibroblasts Alter Epithelial Cell Differentiation. Journal of Cell Science, 118: 485-496. [CrossRef]
- 105) Ovadya Y, Landsberger T, Leins H, Vadai E, Gal H, Biran A, et al. 2018. Impaired Immune Surveillance Accelerates Accumulation of Senescent Cells and Aging. Nature Communications, 9:5435. [CrossRef]
- 106) Sprenger CC, Plymate SR, Reed MJ. 2010. Aging-Related Alterations in the Extracellular Matrix Modulate the Microenvironment and Influence Tumor Progression. Int. J. Cancer, 127(12): 2739-2748. [CrossRef]
- 107) Mosteiro L, Pantoja C, Alcazar N, Marión RM, Chondronasiou D, Rovira M, et al. 2016. Tissue Damage and Senescence Provide Critical Signals for Cellular Reprogramming in vivo. Science, 354(6315). [CrossRef]
- 108) Von Aderkas P, Bonga JM. 2000. Influencing Micropropagation and Somatic Embryogenesis in Mature Trees by Manipulation of Phase Change, Stress and Culture Environment. Tree Physiol., 20:921-928. [CrossRef]
- 109) Feher A, Pasternak T, Miskolczi P, Ayaydin F, Dudits D. 2000. Induction of the Embryogenic Pathway in Somatic Plant Cells. Acta Hort., 560:293-298. [CrossRef]
- 110) Dudits D, Bogre L, Gyorgyey J. 1991. Molecular and Cellular Approaches to the Analysis of Plant Embryo Development from Somatic Cells in vitro. J. Cell. Sci., 99:475-484. [CrossRef]
- 111) Ikeda-Iwai M, Umehara M, Satoh S, Kamada H. 2003. Stress-Induced Somatic Embryogenesis in Vegetative Tissues of Arabidopsis thaliana. The Plant Jour., 34:107-114. [CrossRef]
- 112) Mordhorst BR, Benne JA, Cecil RF, Whitworth KM, Samuel MS, Spate LD, et al. 2019. Improvement of in vitro and Early in utero Porcine Clone Development after Somatic Donor Cells are Cultured Under Hypoxia. Mol Reprod Dev., 86:558-565. [CrossRef]
- 113) Liu J-L, Wang M-K, Sun Q-Y, Zhang X-R, Jiang L-K, Chen D-Y. 2001. Refrigeration of Donor Cells in Preparation for Bovine Somatic Nuclear Transfer. Reproduction, 122:801-808. [CrossRef]
- 114) Wilmut I, Beaujean N, de Sousa PA, Dinnyes AA, King TJ, Paterson LA, et al. 2002. Somatic Cell Nuclear Transfer. Nature, 419(6907):583-587. [CrossRef]
- 115) Takahashi K, Yamanaka S. 2006. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell, 126:663-676. [CrossRef]
- 116) Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. 2007. Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell, 131:861-872. [CrossRef]
- 117) Folmes CDL, Nelson TJ, Martinez-Fernandez A, Arrell DK, Lindor JZ, Dzeja PP, et al. 2011. Somatic Oxidative Bioenergetics Transitions into Pluripotency-Dependent Glycolysisto Facilitate Nuclear Reprogramming. Cell Metab., 14(2): 264–271. [CrossRef]
- 118) Folmes CDL, Nelson TJ, Terzic A. 2011. Energy Metabolism in Nuclear Reprogramming. Biomark. Med., 5(6):715-729. [CrossRef]
- 119) Ishida T, Nakao S, Ueyama T, Harada Y, Kawamura T. 2020. Metabolic Remodeling During Somatic Cell Reprogramming to Induced Pluripotent Stem Cells: Involvement of Hypoxia-Inducible Factor 1. Inflammation and Regeneration, 40:8. [CrossRef]
- 120) Sinclair JW, Hoying DR, Bresciani E, Nogare DD, Needle CD, Berger A, et al. 2021. The Warburg Effect is Necessary to Promote Glycosylation in the Blastema During Zebrafish Tail Regeneration. Npj Regenerative Medicine, 6:55. [CrossRef]
- 121) Forristal CE, Christensen DR, Chinnery FE, Petruzzelli R, Parry KL, Sanchez-Elsner T, et al. 2013. Environmental Oxygen Tension Regulates the Energy Metabolism and Self-Renewal of Human Embryonic Stem Cells. PLoS ONE 8(5): e62507. [CrossRef]
- 122) McCusker C, Bryant SV, Gardiner DM. 2015. The Axolotl Limb Blastema: Cellular and Molecular Mechanisms Driving Blastema Formation and Limb Regeneration in Tetrapods. Regeneration:54-71. [CrossRef]
- 123) Simkin J, Sammarco MC, Dawson LA, Schanes PP, Yu L, Muneoka K. 2015. The Mammalian Blastema: Regeneration at our Fingertips. Regeneration:93-105. [CrossRef]
- 124) Elder SS, Emmerson E. 2020. Senescent Cells and Macrophages: Key Players for Regeneration? Open Biol., 10:200309. [CrossRef]
- 125) Mosser DM, Edwards JP. 2008. Exploring the Full Spectrum of Macrophage Activation. Nat Rev Immunol., 8(12): 958–969. [CrossRef]
- 126) Wu R, Van der Hoek KH, Ryan NK, Norman RJ, Robker RL. 2004. Macrophage Contributions to Ovarian Function. Human Reproduction Update, 10(2): 119-133. [CrossRef]
- 127) Espey LL. 1980. Ovulation as an Inflammatory Reaction — A Hypothesis. Biol. Reprod., 22:73-106. [CrossRef]
- 128) Duffy DM, Ko C, Jo M, Brannstrom M, Curry, Jr. TE. 2019.Ovulation: Parallels with Inflammatory Process. Endocrine Reviews, 40:369-416. [CrossRef]
- 129) Masuda M, Wakasaki T, Toh S. 2016. Stress-Triggered Atavistic Reprogramming (STAR) Addiction: Driving Force Behind Head and Neck Cancer? Am. J. Cancer Res., 6(6):1149-1166. [PubMed]
- 130) Feichtinger J, McFarlane RJ. 2019. Meiotic Gene Activation in Somatic and Germ Cell Tumours. Andrology, 7:415–427. [CrossRef]
- 131) Gantchev J, Villarreal AM, Gunn S, Zetka M, Ødum N, Litvinov IV. 2020. The Ectopic Expression of meiCT Genes Promotes Meiomitosis and May Facilitate Carcinogenesis. Cell Cycle, 19(8):837–854. [CrossRef]
- 132) Hosoya N, Miyagawa K. 2020. Synaptonemal Complex Proteins Modulate the Level of Genome Integrity in Cancers. Cancer Science, 112:989–996. [CrossRef]
- 133) Uyar A, Torrealday S, Seli E. 2013. Cumulus and Granulosa Cell Markers of Oocyte and Embryo Quality. Fert. Steril., 99(4):979-997. [CrossRef]
- 134) Jones ASK, Shikanov A. 2019. Follicle Development as an Orchestrated Signaling Network in a 3D Organoid. Jour. Biol. Eng., 13(2). [CrossRef]
- 135) Kreeger PK, Deck JW, Woodruff TK, Shea LD. 2006. The in vitro Regulation of Ovarian Follicle Development Using Alginate-Extracellular Matrix Gels. Biomaterials, 27(5):714-723. [CrossRef]
- 136) Horandl E, Hadacek F. 2013. The Oxidative Damage Initiation Hypothesis for Meiosis. Plant Reprod, 26: 351–367. [CrossRef]
- 137) Mirzaghaderi G, Horandl E. 2016. The Evolution of Meiotic Sex and its Alternatives. Proc. R. Soc. B, 283: 20161221. [CrossRef]
- 138) Bernstein H, Bernstein C. 2010. Evolutionary Origin of Recombination During Meiosis. BioScience, 60(7):498-505. [CrossRef]
- 139) Lenormand T, Engelstadter J, Johnston SE, Wijnker E, Haag CR. 2016. Evolutionary Mysteries in Meiosis. Trans. R. Soc. B, 371:20160001. [CrossRef]
- 140) Garg SG, Martin WF. 2016. Mitochondria, the Cell Cycle, and the Origin of Sex via a Syncytial Eukaryote Common Ancestor. Genome Biol. Evol., 8(6):1950–1970. [CrossRef]
- 141) Blagosklonny MV. 2011. Cell Cycle Arrest is Not Senescence. Aging, 3(2):94-101.
- 142) Duckworth BC, Weaver JS, Ruderman JV. 2002. G2 Arrest in Xenopus Oocytes Depends on Phosphorylation of Cdc25 by Protein Kinase A. PNAS, 99(26):16794–16799. [CrossRef]
- 143) Tsang M, Gantchev J, Netchiporouk E, Moreau L, Ghazawi FM, Glassman S, et al. 2018. A Study of Meiomitosis and Novel Pathways of Genomic Instability in Cutaneous T-Cell Lymphomoas (CTCL). Oncotarget, 9(102):37647-37661. [CrossRef]
- 144) Claybon A, Karia B, Bruce C, Bishop AJR. 2010. PARP1 Suppresses Homologous Recombination Events in Mice in vivo. Nucleic Acids Research, 38(21):7538–7545. [CrossRef]
- 145) Lao JP, Hunter N. 2010. Trying to Avoid Your Sister. PLoS Biol., 8(10):e1000519. [CrossRef]
- 146) Okuda A, Suzuki A. 2016. Unexpected Link Between MAX and Meiotic Onset. Cell Cycle, 15(17):2235–2236. [CrossRef]
- 147) Yamashita T, Higashi M, Momose S, Adachi A, Watanabe T, Tanaka Y, et al.2020. Decreased MYC-Associated Factor X (MAX) Expression is a New Potential Biomarker for Adverse Prognosis in Anaplastic Large Cell Lymphoma. Sci Rep., 10:10391. [CrossRef]
- 148) Janisiw E, Raices M, Balmir F, Paulin LF, Baudrimont A, von Haeseler A, et al. 2020. Poly(ADP-ribose) Glycohydrolase Coordinates Meiotic DNA Double-Strand Break Induction and Repair Independent of its Catalytic Activity. Nature Communications, 11:4869. [CrossRef]
- 149) Suzuki, A, Hirasaki M, Okuda A. 2017. Does MAX Open Up a New Avenue for Meiotic Research? Develop. Growth Differ., 59:61–69. [CrossRef]
- 150) Kimble J. 2011. Molecular Regulation of the Mitosis/Meiosis Decision in Multicellular Organisms. Cold Spring Harb Perspect Biol, 3:a002683. [CrossRef]
- 151) Honigberg SM, Purnapatre K. 2003. Signal Pathway Integration in the Switch from the Mitotic Cell Cycle to Meiosis in Yeast. Jour. Cell Sci, 116:2137-2147. [CrossRef]
- 152) Kodiha M, Rassi JG, Brown CM, Stochaj U. 2007. Localization of AMP-Kinase is Regulated by Stress, Cell Density, and Signaling Through the MEK–> ERK1/2 Pathway. Am J Physiol Cell Physiol 293: C1427–C1436. [CrossRef]
- 153) Clements D, Mayer RJ, Johnson SR. 2007. Subcellular Distribution of the TSC2 Gene Product Tuberin in Human Airway Smooth Muscle Cells is Driven by Multiple Localization Sequences and is Cell-Cycle Dependent. Am J Physiol Lung Cell Mol Physiol 292: L258 –L266, 2007. [CrossRef]
- 154) Karni-Schmidt O, Friedler A, Zupnick A, McKinney K, Mattia M, Beckerman R, et al. 2007. Energy-Dependent Nucleolar Localization of p53 in vitro Requires Two Discrete Regions within the p53 Carboxyl Terminus. Oncogene (2007) 26, 3878–3891. [CrossRef]
- 155) Xu-Monette ZY, Medeiros LJ, Li Y, Orlowski RZ, Andreeff M, Bueso-Ramos CE, et al. 2012. Dysfunction of the TP53 Tumor Suppressor Gene in Lymphoid Malignancies. Blood, 119(16):3668-3683. [CrossRef]
- 156) Nilsson K, Landberg G. 2006. Subcellular Localization, Modification and Protein Complex Formation of the Cdk-Inhibitor p16 in Rb-Functional and Rb-Inactivated Tumor Cells. Int. J. Cancer, 118:1120–1125. [CrossRef]
- 157) Apostolova MD, Ivanova IA, Dagnino C, D’Souza SJA, Dagnino L. 2002. Active Nuclear Import and Export Pathways Regulate E2F-5 Subcellular Localization. J Biol Chem, 277(37):34471–34479. [CrossRef]
- 158) Trinh DLN, Elwi AN, Kim S-W. 2010. Direct Interaction Between p53 and Tid1 Proteins Affects p53 Mitochondrial Localization and Apoptosis. Oncotarget, 1(6):396 - 404. [CrossRef]
- 159) Agarwal S, Ganesh S. 2020. Perinuclear Mitochondrial Clustering, Increased ROS Levels, and HIF1are Required for the Activation of HSF1 by Heat Stress. Jour. Cell Sci., 133: jcs245589. [CrossRef]
- 160) Takahashi Y, Hashimoto S, Yamochi T, Goto H, Yamanaka M, Amo A, et al. 2016. Dynamic Changes in Mitochondrial Distribution in Human Oocytes During Meiotic Maturation. J Assist Reprod Genet, 33:929–938. [CrossRef]
- 161) Lees JG, Gardner DK, Harvey AJ. 2017. Pluripotent Stem Cell Metabolism and Mitochondria: Beyond ATP. Stem Cells International, 2017(2874283):17 pages. [CrossRef]
- 162) Moiseeva O, Bourdeau V, Roux A, Deschenes-Simard X, Ferbeyre G. 2009. Mitochondrial Dysfunction Contributes to Oncogene-Induced Senescence. Mol. Cell. Biol, 29(16):4495–4507. [CrossRef]
- 163) Chakrabarty RP, Chandel NS. 2021. Mitochondria as Signaling Organelles Control Mammalian Stem Cell Fate. Cell Stem Cell, 28:394-408. [CrossRef]
- 164) Saxton WM, Hollenbeck PJ. 2012. The Axonal Transport of Mitochondria. J Cell Sci, 125:2095–2104. [CrossRef]
- 165) Voglhuber J, Holzer M, Radulovic S, Thai PN, Djalinac N, Matzer I, et al. 2022. Functional Remodelling of Perinuclear Mitochondria Alters Nucleoplasmic Ca2+ Signalling in Heart Failure. Phil. Trans. R. Soc. B, 377: 20210320. [CrossRef]
- 166) Fenton AR, Jongens TA, Holzbaur ELF. 2021. Mitochondrial Adapter TRAK2 Activates and Functionally Links Opposing Kinesin and Dynein Motors. Nature Comm, 12:4578. [CrossRef]
- 167) Thomas LW, Staples O, Turmaine M, Ashcroft M. 2017. CHCHD4 Regulates Intracellular Oxygenation and Perinuclear Distribution of Mitochondria. Front. Oncol., 7:71. [CrossRef]
- 168) Giardino Torchia ML, Ashwell JD. 2018. Getting MAD at MYC. PNAS,115(40):9821–9823. [CrossRef]
- 169) Kemmerer K, Weigand JE. 2014. Hypoxia Reduces MAX Expression in Endothelial Cells by Unproductive Splicing. FEBS Letters, 588:4784–4790. [CrossRef]
- 170) Suzuki A, Hirasaki M, Hishida T, Wu J, Okamura D, Ueda A, et al. 2016. Loss of MAX Results in Meiotic Entry in Mouse Embryonic and Germline Stem Cells. Nature Comm, 7:11056. [CrossRef]
- 171) Gordon JD, Bertovrt JA, Hu C-J, Diehl JA, Simon MC. 2007. HIF-2∀ Promotes Hypoxic Cell Proliferation by Enhancing c-Myc Transcriptional Activity. Cancer Cell, 11(4):335-347. [CrossRef]
- 172) Yan Y, Liu F, Han L, Zhao L, Chen J, Olopade OI, et al. 2018. HIF-2∀ Promotes Conversion to a Stem Cell Phenotype and Induces Chemoresistance in Breast Cancer Cells by Activating Wnt and Notch Pathways. J Exp. Clin. Canc. Res., 37:256. [CrossRef]
- 173) Tharp KM, Higuchi-Sanabria R, Timblin GA, Ford B, Garzon-Coral C, Schneider C, et al. 2021. Adhesion-Mediated Mechanosignaling Forces Mitohormesis. Cell Metab., 33:1322-1341. [CrossRef]
- 174) Koppers M, Özkan N, Farías GG. 2020. Complex Interactions Between Membrane-Bound Organelles, Biomolecular Condensates and the Cytoskeleton. Front. Cell Dev. Biol., 8:618733. [CrossRef]
- 175) Heffler J, Shah PP, Robison P, Phyo S, Veliz K, Uchida K, et al. 2020. A Balance Between Intermediate Filaments and Microtubules Maintains Nuclear Architecture in the Cardiomyocyte. Circ Res., 126(3): e10–e26. [CrossRef]
- 176) Spichal M, Fabre E. 2017. The Emerging Role of the Cytoskeleton in Chromosome Dynamics. Front. Genet., 8:60. [CrossRef]
- 177) Bissell MJ, Barcellos-Hoff MH. 1987. The Influence of Extracellular Matrix on Gene Expression: Is Structure the Message? J Cell Sci. Suppl., 8: 327-343. [CrossRef]
- 178) Thorne JT, Segal TR, Chang S, Jorge S, Segars JH, Leppert PC. 2015. Dynamic Reciprocity Between Cells and Their Microenvironment in Reproduction. Biol. Reprod., 92(1):25, 1–10. [CrossRef]
- 179) Spencer VA, Xu R, Bissell MJ. 2007. Extracellular Matrix, Nuclear and Chromatin Structure, and Gene Expression in Normal Tissues and Malignant Tumors: A Work in Progress. Adv Cancer Res., 97:275–294. [CrossRef]
- 180) Schultz GS, Davidson JM, Kirsner RS, Bornstein P, Herman IM. 2011. Dynamic Reciprocity in the Wound Microenvironment. Wound Repair Regen., 19(2):134–148. [CrossRef]
- 181) Lenain C, de Graaf CA, Pagie L, Visser NL, de Haas M, de Vries SS, et al. 2017. Massive Reshaping of Genome-Nuclear Lamina Interactions During Oncogene-Induced Senescence. Genome Res, 27:1634–1644. [CrossRef]
- 182) Burla R, La Torre M, Saggio I. 2016. Mammalian Telomeres and their Partnership with Lamins. Nucleus, 7(2):187-202. [CrossRef]
- 183) Meqbel BRM, Gomes M, Omer, A, Gallouzi IE, Horn HF. 2022. LINCing Senescence and Nuclear Envelope Changes. Cells, 11:1787. [CrossRef]
- 184) Link J, Jahn D, Schmitt J, Gob E, Baar J, Ortega S, et al. 2013. The Meiotic Nuclear Lamina Regulates Chromosome Dynamics and Promotes Efficient Homologous Recombination in the Mouse. PloS Genet., 9(1):e1003261. [CrossRef]
- 185) Morimoto, A, Shibuya H, Zhu X, Kim J, Ishiguro K, Han M, et al. 2012. A Conserved KASH Domain Protein Associates with Telomeres, SUN1, and Dynactin During Mammalian Meiosis. J. Cell Biol.,198(2): 165–172. [CrossRef]
- 186) Davis L, Smith GR. 2006. The Meiotic Bouquet Promotes Homolog Interactions and Restricts Ectopic Recombination in Schizosaccharomyces pombe. Genetics, 174:167–177. [CrossRef]
- 187) Zhuang J, Wang P, Huang X, Chen X, Kang J-G, Hwang PM. 2013. Mitochondrial Disulfide Relay Mediates Translocation of p53 and Partitions its Subcellular Activity. PNAS, 110(43):17356–17361. [CrossRef]
- 188) Saleem A, Hood DA. 2013. Acute Exercise Induces Tumor Suppressor p53 Translocation to the Mitochondriaand Promotes a p53-Tfam-Mitochondrial DNA Complex in Skeletal Muscle. J Physiol, 591.14:3625–3636. [CrossRef]
- 189) Brunyanszki A, Szczesny B, Virág L, Szabo C. 2016. Mitochondrial Poly(ADP-Ribose) Polymerase: The Wizard of Oz at Work. Free Radic Biol Med., 100: 257–270. [CrossRef]
- 190) Lee S-J, Lim C-J, Min J-K, Lee J-K, Kim Y-M, Lee J-Y, et al. 2007. Protein Phosphatase 1 Nuclear Targeting Subunit is a Hypoxia Inducible Gene: Its Role in Post-Translational Modification of p53 and MDM2. Cell Death and Differentiation, 14:1106–1116. [CrossRef]
- 191) Landsverk HB, Mora-Bermudez F, Landsverk OJB, Hasvold G, Naderi S, Bakke O, et al. 2010. The Protein Phosphatase 1 Regulator PNUTS is a New Component of the DNA Damage Response. EMBO reports, 11(11): 868–875. [CrossRef]
- 192) Leman AR, Noguchi E. 2013. The Replication Fork: Understanding the Eukaryotic Replication Machinery and the Challenges to Genome Duplication. Genes, 4:1-32. [CrossRef]
- 193) Carson DR, Christman MF. 2001. Evidence That Replication Fork Components Catalyze Establishment of Cohesion Between Sister Chromatids. PNAS, 98(15):8270 – 8275. [CrossRef]
- 194) Forsburg SL. 2002. Only Connect: Linking Meiotic DNA Replication with Chromosome Dynamics. Molecular Cell, 9:703–711. [CrossRef]
- 195) Skibbens RV. 2000. Holding Your Own: Establishing Sister Chromatid Cohesion. Genome Research, 10:1664-1671. [CrossRef]
- 196) Toth A, Rabitsch KP, Galova M, Schleiffer A, Buonomo SBC, Nasmyth K. 2000. Functional Genomics Identifies Monopolin: A Kinetochore Protein Required for Segregation of Homologs During Meiosis I. Cell, 103:1155–1168. [CrossRef]
- 197) Solc P, Schultz RM, Motlik J. 2010. Prophase I Arrest and Progression to Metaphase I in Mouse Oocytes: Comparison of Resumption of Meiosis and Recovery from G2-Arrest in Somatic Cells. Molecular Human Reproduction, 16(9): 654– 664. [CrossRef]
- 198) Sasaki K, Chiba K. 2001. Fertilization Blocks Apoptosis of Starfish Eggs by Inactivation of the MAP Kinase Pathway. Develop Biol, 237:18-28. [CrossRef]
- 199) Harrison RH, Kuo HC, Scriven PN, Handyside AH, Ogilvie CM. 2000. Lack of Cell cycle Checkpoints in Human Cleavage Stage Embryos Revealed by a Clonal Pattern of Chromosomal Mosaicism Analysed by Sequential Multicolour FISH. Zygote, 8(3):217-224. [CrossRef]
- 200) Lee HO, Davidson JM, Duronio RJ. 2009. Endoreplication: Polyploidy with Purpose. Genes & Development, 23:2461–2477. [CrossRef]
- 201) Tan Z, Chu DZV, Chan YJA, Lu YE, Rancati G. 2018. Mammalian Cells Undergo Endoreduplication in Response to Lactic Acidosis. Scientific Reports,8:2890. [CrossRef]
- 202) Walen KH. 2015. Wound Healing is a First Response in a Cancerous Pathway: Hyperplasia Developments to 4n Cell Cycling in Dysplasia Linked to RB-Inactivation. Journal of Cancer Therapy, 2015, 6, 906-916. [CrossRef]
- 203) Salmina K, Huna A, Kalejs M, Pjanova D, Scherthan H, Cragg MS, et al. 2019. The Cancer Aneuploidy Paradox: In the Light of Evolution. Genes, 10: 83. [CrossRef]
- 204) Salmina K, Vainshelbaum NM, Kreishmane M, Inashkina I, Cragg MS, Pjanova D, et al. 2023. The Role of Mitotic Slippage in Creating a “Female Pregnancy-like System” in a Single Polyploid Giant Cancer Cell. Int. J. Mol. Sci., 24:3237. [CrossRef]
- 205) Erenpreisa J, Cragg MS. 2013. Three Steps to the Immortality of Cancer Cells: Senescence, Polyploidy and Self-Renewal. Cancer Cell International, 13:92. [CrossRef]
- 206) Lok TM, Wang Y, Xu WK, Xie S, Ma HT, Poon RYC. 2020. Mitotic Slippage is Determined by p31comet and the Weakening of the Spindle-Assembly Checkpoint. Oncogene, 39:2819–2834. [CrossRef]
- 207) Ianzini F, Kosmacek EA, Nelson ES, Napoli E, Erenpreisa J, Kalejs M, et al. 2009. Activation of Meiosis-Specific Genes is Associated with Depolyploidization of Human Tumor Cells Following Radiation-Induced Mitotic Catastrophe. Cancer Res.,69(6):2296–2304. [CrossRef]
- 208) Mosieniak G, Sliwinska MA, Alster O, Strzeszewska A, Sunderland P, Piechota M, et al. 2015. Polyploidy Formation in Doxorubicin-Treated Cancer Cells Can Favor Escape from Senescence. Neoplasia, 17: 882–893. [CrossRef]
- 209) Amodeo AA, Jukam D, Straight AF, Skotheim JM. 2015. Histone Titration Against the Genome Sets the DNA-to-Cytoplasm Threshold for the Xenopus Midblastula Transition. PNAS:E1086-E1095. [CrossRef]
- 210) Niu N, Mercado-Uribe I, Liu J. 2017. Dedifferentiation into Blastomere-like Cancer Stem Cells via Formation of Polyploid Giant Cancer Cells. Oncogene, 36:4887-4900. [CrossRef]
- 211) Tu, S-M, Zhang, M, Wood, CG, Pisters, LL. 2021. Stem Cell Theory of Cancer: Origin of Tumor Heterogeneity. Cancers, 13:4006. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




