Submitted:
29 July 2024
Posted:
30 July 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Data Source
2.2. Variables
2.3. Statistical Analysis: Two-Sample Mendelian Randomization Analysis
Outler Detection
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Declarations
References
- Poikolainen, K. Alcohol and mortality: a review. *J. Clin. Epidemiol.* 1995, 48, 455–465. [CrossRef]
- Volkow, N.D.; Wang, G.J.; Baler, R.D. Reward, dopamine and the control of food intake: Implications for obesity. *Trends Cogn. Sci.* 2011, 15, 37–46. [CrossRef]
- Seitz, H.K.; Stickel, F. Molecular mechanisms of alcohol-mediated carcinogenesis. *Nat. Rev. Cancer* 2007, 7, 599–612. [CrossRef]
- Annunziata, G.; Ciampaglia, R.; Schisano, C. Alcohol-induced oxidative stress and its impact on liver injury. *Nutrients* 2018, 10, 65.
- Manthey, J.; Rehm, J.; Gmel, G. The burden of alcohol use disorders in 195 countries and territories, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. *Lancet Psychiatry* 2017, 4, 987–996.
- Chen, W.Y.; Rosner, B.; Hankinson, S.E.; Colditz, G.A.; Willett, W.C. Moderate alcohol consumption during adult life, drinking patterns, and breast cancer risk. *JAMA* 2011, 306, 1884–1890. [CrossRef]
- Balbo, S.; Meng, L.; Bliss, R.L.; Jensen, J.A.; Hatsukami, D.K.; Hecht, S.S.; Upadhyaya, P. Folate deficiency and alcohol-induced cancer: Implications for DNA methylation. *Cancer Res.* 2009, 69, 9365–9372.
- Lamy, L. Clinical and statistical study of 134 cases of cancer of the oesophagus and of the cardia. *Arch. Mal. App. Dig.* 1910, 4, 451–475.
- Secretan, B.; Straif, K.; Baan, R.; Grosse, Y.; El Ghissassi, F.; Bouvard, V.; et al. A review of human carcinogens--Part E: tobacco, areca nut, alcohol, coal smoke, and salted fish. *Lancet Oncol.* 2009, 10, 1033–1034.
- World Health Organization (WHO). Global status report on alcohol and health 2018; World Health Organization: Geneva, Switzerland, 2018.
- Brien, S.E.; Ronksley, P.E.; Turner, B.J.; Mukamal, K.J.; Ghali, W.A. Effect of alcohol consumption on biological markers associated with risk of coronary heart disease: systematic review and meta-analysis of interventional studies. *BMJ* 2011, 342, d636. [CrossRef]
- Allen, N.E.; Beral, V.; Casabonne, D.; Kan, S.W.; Reeves, G.K.; Brown, A.; et al. Moderate alcohol intake and cancer incidence in women. *J. Natl. Cancer Inst.* 2009, 101, 296–305. [CrossRef]
- Jung, S.; Wang, M.; Anderson, K.; Baglietto, L.; Bergkvist, L.; Bernstein, L.; et al. Alcohol consumption and breast cancer risk by estrogen receptor status: in a pooled analysis of 20 studies. *Int. J. Epidemiol.* 2016, 45, 916–928. [CrossRef]
- Bagnardi, V.; Rota, M.; Botteri, E.; Tramacere, I.; Islami, F.; Fedirko, V.; et al. Alcohol consumption and site-specific cancer risk: a comprehensive dose-response meta-analysis. *Br. J. Cancer* 2015, 112, 580–593. [CrossRef]
- McNabb, S.; Harrison, T.A.; Albanes, D.; Berndt, S.I.; Brenner, H.; Caan, B.J.; et al. Meta-analysis of 16 studies of the association of alcohol with colorectal cancer. *Int. J. Cancer* 2020, 146, 861–873. [CrossRef]
- Walters, R.K.; Polimanti, R.; Johnson, E.C.; McClintick, J.N.; Adams, M.J.; Adkins, A.E.; et al. Transancestral GWAS of alcohol dependence reveals common genetic underpinnings with psychiatric disorders. *Nat. Neurosci.* 2018, 21, 1656–1669. [CrossRef]
- Howe, L.J.; Lawson, D.J.; Davies, N.M.; St Pourcain, B.; Lewis, S.J.; Davey Smith, G.; et al. Genetic evidence for assortative mating on alcohol consumption in the UK Biobank. *Nat. Commun.* 2019, 10, 5039. [CrossRef]
- Larsson, S.C.; Carter, P.; Kar, S.; Vithayathil, M.; Mason, A.M.; Michaëlsson, K.; et al. Smoking, alcohol consumption, and cancer: A mendelian randomisation study in UK Biobank and international genetic consortia participants. *PLoS Med.* 2020, 17, e1003178.
- Xiong, J.; Yang, L.; Deng, Y.-Q.; Yan, S.-Y.; Gu, J.-M.; Li, B.-H.; et al. The causal association between smoking, alcohol consumption and risk of bladder cancer: A univariable and multivariable Mendelian randomization study. *Int. J. Cancer* 2022, 151, 2136–2143. [CrossRef]
- Howe, L.J.; Nivard, M.G.; Morris, T.T.; Chen, Z.; Lin, K.; Mills, M.C.; Millwood, I.; Walters, R.; et al. Within-sibship GWAS of 25 phenotypes improves estimates of direct genetic effects. *Nat. Genet.* 2022, 54, 581–592.
- Zhou, X.; Wang, L.; Xiao, J.; Sun, J.; Yu, L.; Zhang, H.; et al. Alcohol consumption, DNA methylation and colorectal cancer risk: Results from pooled cohort studies and Mendelian randomization analysis. *Int. J. Cancer* 2022, 151, 83–94. [CrossRef]
- Wang, J.; Wang, H.; Chen, Y.; Hao, P.; Zhang, Y. Alcohol ingestion and colorectal neoplasia: a meta-analysis based on a Mendelian randomization approach. *Colorectal Dis.* 2011, 13, e71–e78. [CrossRef]
- Brunner, C.; Davies, N.M.; Martin, R.M.; Eeles, R.; Easton, D.; Kote-Jarai, Z.; et al. Alcohol consumption and prostate cancer incidence and progression: A Mendelian randomisation study. *Int. J. Cancer* 2017, 140, 75–85. [CrossRef]
- Boccia, S.; Hashibe, M.; Gallì, P.; De Feo, E.; Asakage, T.; Hashimoto, T.; et al. Aldehyde dehydrogenase 2 and head and neck cancer: a meta-analysis implementing a Mendelian randomization approach. *Cancer Epidemiol. Biomarkers Prev.* 2009, 18, 248–254. [CrossRef]
- Dumitrescu, R.G.; Shields, P.G. The etiology of alcohol-induced breast cancer. *Alcohol* 2005, 35, 213–225. [CrossRef]
- Lee, J.E.; Hunter, D.J.; Spiegelman, D.; Adami, H.-O.; Albanes, D.; Bernstein, L.; et al. Alcohol intake and renal cell cancer in a pooled analysis of 12 prospective studies. *J. Natl. Cancer Inst.* 2007, 99, 801–810. [CrossRef]
- Terry, M.B.; Gammon, M.D.; Zhang, F.F.; Knight, J.A.; Wang, Q.; Britton, J.A.; et al. ADH3 genotype, alcohol intake and breast cancer risk. *Carcinogenesis* 2006, 27, 840–847. [CrossRef]
- Lorenti Garcia, C.; Mechilli, M.; Proietti De Santis, L.; Schinoppi, A.; Kobos, K.; Palitti, F. Relationship between DNA lesions, DNA repair and chromosomal damage induced by acetaldehyde. *Mutat. Res.* 2009, 662, 3–9.
- Seitz, H.K.; Stickel, F. Molecular mechanisms of alcohol-mediated carcinogenesis. *Nat. Rev. Cancer* 2007, 7, 599–612. [CrossRef]
- Garaycoechea, J.I.; Crossan, G.P.; Langevin, F.; Mulderrig, L.; Louzada, S.; Yang, F.; et al. Alcohol and endogenous aldehydes damage chromosomes and mutate stem cells. *Nature* 2018, 553, 171–177. [CrossRef]

| Trait Name | Sample Size | Number of Variants | First Author | Year | Consortium | Sex | ||
|---|---|---|---|---|---|---|---|---|
| Exposure | ||||||||
| ukb-a-25 | Alcohol intake frequency | 336,965 | 10,894,599 | Neale BM | 2017 | Neale lab | M, F | |
| ieu-b-4834 | Alcohol consumption | 83,626 | 7,914,362 | Howe LJ | 2022 | WFGC | M, F | |
| Outcome | Cancer site | |||||||
| finn-b-c3-stomach | Stomach | NA | 16,380,305 | NA | 2021 | NA | M, F | |
| finn-b-C3-liver | Liver | NA | 16,380,303 | NA | 2021 | NA | M, F | |
| ieu-b-4955 | Lung | 374,687 | 11,078,115 | Burrows | 2021 | UK Biobank | M, F | |
| finn-b-c3-colon | Colon | NA | 16,380,317 | NA | 2021 | NA | M, F | |
| finn-b-c3-thyroid | Thyroid | NA | 16,380,316 | NA | 2021 | NA | M, F | |
| finn-b-c3-cervix | Cervix | NA | 16,378,927 | NA | 2021 | NA | M, F | |
| ieu-a-1168 | Breast | 33,832 | 13,011,123 | Michailidou K | 2015 | BCAC | F | |
| finn-b-c3-prostate | Prostate | NA | 16,377,987 | NA | 2021 | NA | M | |
| finn-b-c3-colorectal | Colorectal | NA | 16,380,321 | NA | 2021 | NA | M, F | |
| finn-b-c3-bladder | Bladder | NA | 16,380,305 | NA | 2021 | NA | M, F | |
| finn-b-c3-larynx | Larynx | NA | 16,380,304 | NA | 2021 | NA | M, F | |
| finn-b-c3-lip-oral | Lip-Oral-Pharynx | NA | 16,380,304 | NA | 2021 | NA | M, F | |
| ieu-b-94 | Oral cavity | 4,151 | 7,510,833 | Lesseur | 2016 | OOCOC | M, F | |
| finn-b-c3-brain | Brain | NA | 16,380,308 | NA | 2021 | NA | M, F | |
| finn-b-c3 | Biliary tract | NA | 16,380,304 | NA | 2021 | NA | M, F | |
| finn-b-c3-oesphagus | Oesphagus | NA | 16,380,466 | NA | 2021 | NA | M, F | |
| Cancer site | Sample size | Cases | Before removing outliers | After removing outliers | ||
| OR (95% CI) | P value | OR (95% CI) | P value | |||
| Stomach | 174,006 | 633 | 1.06 (0.56-1.98) | 0.86300 | 1.06 (0.73-1.54) | 0.7739 |
| Liver | 218,488 | 304 | 1.14 (0.46-2.83) | 0.7704 | 0.73 (0.27-1.94) | 0.4401 |
| Lung | 217,165 | 2671 | 0.99 (0.99-1.00) | 0.4408 | 0.99 (0.99-1.00) | 0.4584 |
| Colon | 174,006 | 1,803 | 1.13 (0.77-1.66) | 0.5098 | 0.848 (0.620-1.157) | 0.2978 |
| Thyroid | 174,006 | 989 | 1.18 (0.70-1.97) | 0.5616 | 2.39 (2.07-2.74) | ≤0.0001 |
| Cervix | 99,321 | 1648 | 0.98 (0.62-1.55) | 0.9260 | 0.98 (0.73- 1.32) | 0.9096 |
| Breast | 115,178 | 15748 | 0.93 (0.81-1.63) | 0.2660 | 0.90 (0.82-0.99) | 0.0159 |
| Prostate | 74865 | 6311 | 1.09 (0.80-1.48) | 0.5630 | 1.05 (0.86-1.26) | 0.6568 |
| Colorectal | 215,770 | 3022 | 0.94 (0.70-1.27) | 0.6844 | 0.87 (0.72-1.06) | 0.2467 |
| Bladder | 174,006 | 1115 | 1.29 (0.80-2.09) | 0.2906 | 1.17 (0.99-1.38) | 0.1062 |
| Larynx | 218,612 | 180 | 2.12 (0.44-10.13) | 0.3357 | NA | NA |
| Lip-Oral-Pharynx | 218,666 | 126 | 1.16 (0.28-4.74) | 0.8317 | 1.53 (0.64- 3.62) | 0.3661 |
| Oral cavity | 4151 | 1223 | 2.33 (0.91-5.92) | 0.07685 | NA | NA |
| Brain | 218,328 | 464 | 0.98 (0.45-2.13) | 0.9604 | 1.04 (0.65-1.67) | 0.8965 |
| Biliary tract | 174,006 | 109 | 0.15 (0.01-3.97) | 0.30220 | 0.60 (0.28-1.27) | 0.3423 |
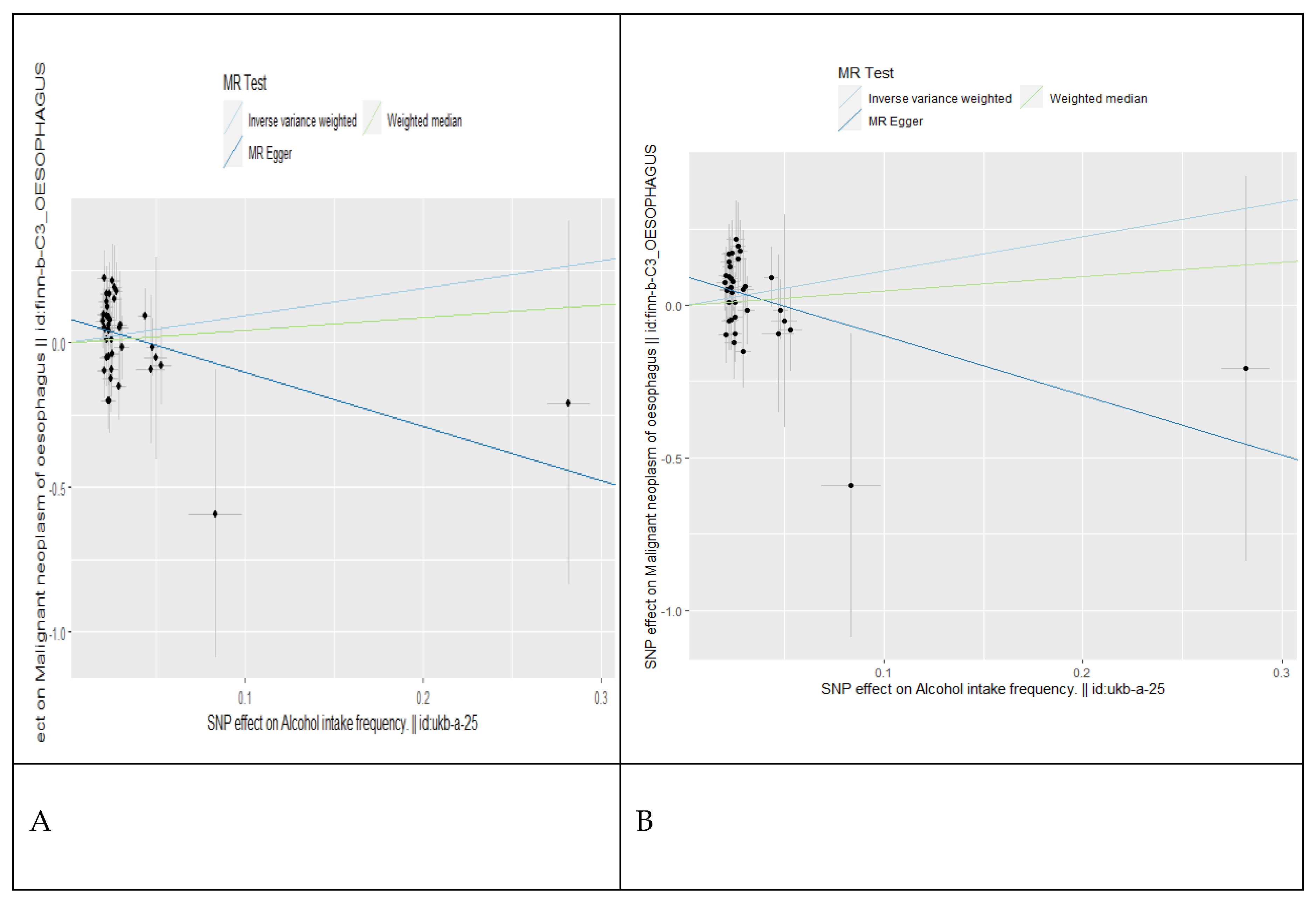
| Oesophagus | 218,560 | 232 | 1.61 (0.58-4.48) | 0.3590 | 1.83 (0.90-3.75) | 0.0449 |
| Cancer site | Sample size | Cases | Before removing outliers | After removing outliers | ||
| OR (95% CI) | P value | OR (95% CI) | P value | |||
| Stomach | 174,006 | 633 | 0.48 (0.23-1.00) | 0.05 | 1.77 (0.97-3.23) | 0.0036 |
| Liver | 218,488 | 304 | 0.96 (0.34-2.71) | 0.9386 | 0.82 (0.33-2.07) | 0.6813 |
| Lung | 217,165 | 1,627 | 0.92 (0.58-1.45) | 0.7169 | 1.08 (0.73-1.58) | 0.7088 |
| Colon | 174,006 | 1,803 | 1.11 (0.768-1.616) | 5.69E-01 | 1.83 (0.99-3.35) | 0.0022 |
| Thyroid | 174,006 | 989 | 1.12 (0.57-2.18) | 0.7411 | 1.15 (0.65-2.04) | 0.6219 |
| Cervix | 99,321 | 1,648 | 0.99 (0.54-1.80) | 0.9665 | 0.85 (0.55-1.30) | 0.4488 |
| Breast | 115,178 | 8,401 | 0.84 (0.61-1.15) | 0.2786 | 0.93 (0.84-1.02) | 0.1189 |
| Prostate | 88,902 | 6,311 | 0.99 (0.71-1.38) | 0.9552 | 0.93 (0.72-1.21) | 0.6027 |
| Colorectal | 215,770 | 3,022 | 0.79 (0.57-1.11) | 0.1822 | 0.85 (0.67-1.09) | 0.1666 |
| Bladder | 174,006 | 1,115 | 1.19 (0.69-2.05) | 0.5414 | 1.08 (0.88-1.30) | 0.5002431 |
| Larynx | 218,612 | 180 | 0.59 (0.15-1.27) | 0.4409 | 0.69 (0.19-2.45) | 0.5743 |
| Lip-Oral-Pharynx | 218,666 | 126 | 1.14 (0.20-6.64) | 0.8808 | 1.02 (0.28-3.78) | 0.7621 |
| Oral cavity | 4151 | 1223 | 2.32 (0.91-5.92) | 0.0768 | NA | NA |
| Brain | 218,328 | 464 | 1.35 (0.58-3.14) | 0.4905 | 1.16 (0.57-2.36) | 0.6839 |
| Biliary tract | 174,006 | 109 | 1.44 (0.25-8.43) | 0.6776 | 3.86 (0.89-16.85) | 0.0722 |
| Oesophagus | 218,560 | 232 | 2.58 (0.74-9.00) | 0.1380 | 3.44 (1.19-9.89) | 0.0217 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





