Submitted:
29 July 2024
Posted:
30 July 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. CBX3/HP1γ Protein in Cancer Proliferation
3. CBX3 as a Multiplayer in Lung Cancer Progression
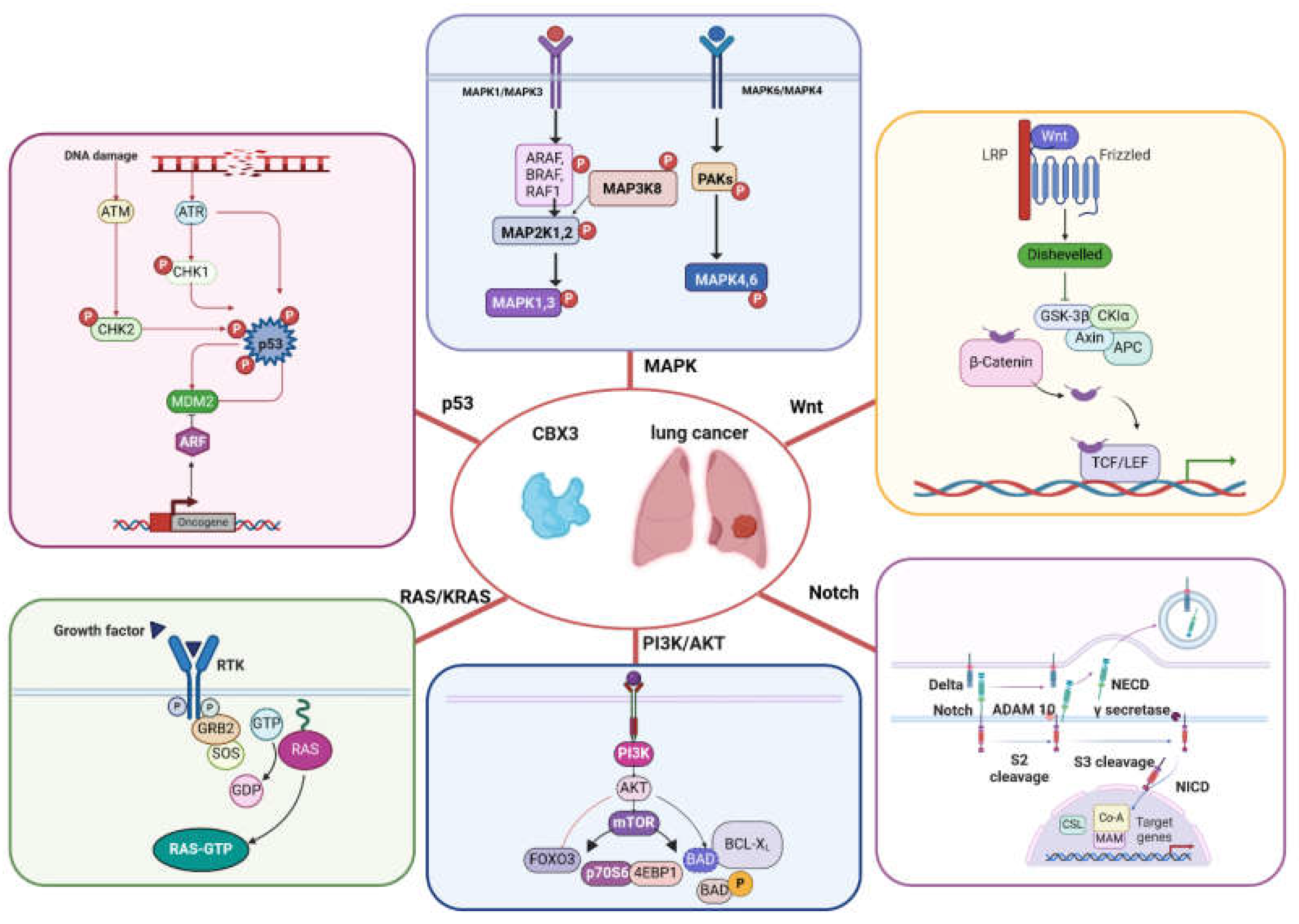
4. Involvement of CBX3 in Pathways Leading to Lung Cancer
4.1. Role of CBX3 in PI3K-AKT and (K)Ras Signaling Pathways
4.2. Role of CBX3 in Notch Signaling Pathway
4.3. Role of CBX3 in Wnt Pathway
4.4. Role of CBX3 in p53 Pathway
4.5. Role of CBX3 in ErbB Pathway
4.6. Role of CBX3 in MAPK Pathway
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ARHGAP24 | rho GTPase Activating Protein 24 |
| BAX | bcl2-associated X protein |
| BRCA1 | breast cancer gene 1 |
| CBX | chromobox |
| CHD | chromodomain |
| CSD | chromoshadow domain |
| DSBs | DNA double-strand breaks |
| DSBs | double-strand breaks |
| EGFR | epidermal growth factor receptor |
| EZH1/2 | enhancer of zeste homologous 1/2 |
| HR | homologous recombination |
| LUAD | lung adenocarcinoma |
| MDM2 | mouse double minute 2 homolog |
| mRNA | messenger RNA |
| SMC | structural maintenance of chromosomes |
| NCAPG | non-SMC Condensin I Complex Subunit G |
| NICD | notch intracellular domain |
| NSCLC | non-small cell lung cancer |
| PI3K | phosphoinositide 3-kinase |
| PMAIP1 | phorbol-12-myristate-13-acetate-induced protein 1 |
| PRC1 | polycomb (Pc) inhibitory complex 1 |
| PRC2 | polycomb Repressive Complex 2 |
| PUMA p53 | upregulated modulator of apoptosis |
| RAC1 | active Ras-related C3 botulinum toxin substrate 1 |
| RBBP4/7 | retinoblastoma binding protein 4/7 |
| STAT3 | signal transducer and activator of transcription 3 |
| TRIM | tripartite motif-containing |
| ZBTB7A | zinc finger and BTB domain containing 7A |
References
- Tang, Q.; Cheng, J.; Cao, X.; Surowy, H.; Burwinkel, B. Blood-Based DNA Methylation as Biomarker for Breast Cancer: A Systematic Review. Clin Epigenetics 2016, 8, 115. [Google Scholar] [CrossRef]
- Chan, S.C.H.; Liang, J.Q. Advances in Tests for Colorectal Cancer Screening and Diagnosis. Expert Rev Mol Diagn 2022, 22, 449–460. [Google Scholar] [CrossRef]
- Tobi, E.W.; Slieker, R.C.; Luijk, R.; Dekkers, K.F.; Stein, A.D.; Xu, K.M.; Biobank-based Integrative Omics Studies Consortium; Slagboom, P. E.; van Zwet, E.W.; Lumey, L.H.; et al. DNA Methylation as a Mediator of the Association between Prenatal Adversity and Risk Factors for Metabolic Disease in Adulthood. Sci Adv 2018, 4, eaao4364. [Google Scholar] [CrossRef]
- Nicetto, D.; Zaret, K.S. Role of H3K9me3 Heterochromatin in Cell Identity Establishment and Maintenance. Curr Opin Genet Dev 2019, 55, 1–10. [Google Scholar] [CrossRef]
- Ho, L.; Crabtree, G.R. Chromatin Remodelling during Development. Nature 2010, 463, 474–484. [Google Scholar] [CrossRef]
- Goldberg, A.D.; Allis, C.D.; Bernstein, E. Epigenetics: A Landscape Takes Shape. Cell 2007, 128, 635–638. [Google Scholar] [CrossRef]
- Allis, C.D.; Jenuwein, T. The Molecular Hallmarks of Epigenetic Control. Nat Rev Genet 2016, 17, 487–500. [Google Scholar] [CrossRef]
- Wu, D.; Liu, T. Studying Reversible Protein Post-Translational Modification through Co-Translational Modification. Chembiochem 2023, 24, e202200716. [Google Scholar] [CrossRef]
- Biswas, S.; Rao, C.M. Epigenetic Tools (The Writers, The Readers and The Erasers) and Their Implications in Cancer Therapy. Eur J Pharmacol 2018, 837, 8–24. [Google Scholar] [CrossRef]
- Vann, K.R.; Klein, B.J.; Kutateladze, T.G. Mechanistic Similarities in Recognition of Histone Tails and DNA by Epigenetic Readers. Curr Opin Struct Biol 2021, 71, 1–6. [Google Scholar] [CrossRef]
- Beyer, J.N.; Raniszewski, N.R.; Burslem, G.M. Advances and Opportunities in Epigenetic Chemical Biology. ChemBioChem 2021, 22, 17–42. [Google Scholar] [CrossRef]
- Cao, G.; Xuan, X.; Hu, J.; Zhang, R.; Jin, H.; Dong, H. How Vascular Smooth Muscle Cell Phenotype Switching Contributes to Vascular Disease. Cell Commun Signal 2022, 20, 180. [Google Scholar] [CrossRef]
- Yang, A.Y.; Kim, H.; Li, W.; Kong, A.-N.T. Natural Compound-Derived Epigenetic Regulators Targeting Epigenetic Readers, Writers and Erasers. Curr Top Med Chem 2016, 16, 697–713. [Google Scholar] [CrossRef]
- Shukla, S.; Ying, W.; Gray, F.; Yao, Y.; Simes, M.L.; Zhao, Q.; Miao, H.; Cho, H.J.; González-Alonso, P.; Winkler, A.; et al. Small-Molecule Inhibitors Targeting Polycomb Repressive Complex 1 RING Domain. Nat Chem Biol 2021, 17, 784–793. [Google Scholar] [CrossRef]
- Vidal, M.; Starowicz, K. Polycomb Complexes PRC1 and Their Function in Hematopoiesis. Exp Hematol 2017, 48, 12–31. [Google Scholar] [CrossRef]
- Ma, R.; Zhang, Y.; Sun, T.; Cheng, B. Epigenetic Regulation by Polycomb Group Complexes: Focus on Roles of CBX Proteins. J Zhejiang Univ Sci B 2014, 15, 412–428. [Google Scholar] [CrossRef]
- Comet, I.; Riising, E.M.; Leblanc, B.; Helin, K. Maintaining Cell Identity: PRC2-Mediated Regulation of Transcription and Cancer. Nat Rev Cancer 2016, 16, 803–810. [Google Scholar] [CrossRef]
- Zoroddu, S.; Marchesi, I.; Bagella, L. PRC2: An Epigenetic Multiprotein Complex with a Key Role in the Development of Rhabdomyosarcoma Carcinogenesis. Clin Epigenetics 2021, 13, 156. [Google Scholar] [CrossRef]
- Ntziachristos, P.; Mullenders, J.; Trimarchi, T.; Aifantis, I. Mechanisms of Epigenetic Regulation of Leukemia Onset and Progression. Adv Immunol 2013, 117, 1–38. [Google Scholar] [CrossRef]
- Parreno, V.; Martinez, A.-M.; Cavalli, G. Mechanisms of Polycomb Group Protein Function in Cancer. Cell Res 2022, 32, 231–253. [Google Scholar] [CrossRef]
- Brockman, Q.R.; Scherer, A.; McGivney, G.R.; Gutierrez, W.R.; Voigt, A.P.; Isaacson, A.L.; Laverty, E.A.; Roughton, G.; Knepper-Adrian, V.; Darbro, B.; et al. PRC2 Loss Drives MPNST Metastasis and Matrix Remodeling. JCI Insight 2022, 7. [Google Scholar] [CrossRef]
- Duan, R.; Du, W.; Guo, W. EZH2: A Novel Target for Cancer Treatment. J Hematol Oncol 2020, 13, 104. [Google Scholar] [CrossRef]
- Kim, J.; Kingston, R.E. The CBX Family of Proteins in Transcriptional Repression and Memory. J Biosci 2020, 45. [Google Scholar] [CrossRef]
- Vincenz, C.; Kerppola, T.K. Different Polycomb Group CBX Family Proteins Associate with Distinct Regions of Chromatin Using Nonhomologous Protein Sequences. Proc Natl Acad Sci U S A 2008, 105, 16572–16577. [Google Scholar] [CrossRef]
- Jaensch, E.S.; Zhu, J.; Cochrane, J.C.; Marr, S.K.; Oei, T.A.; Damle, M.; McCaslin, E.Z.; Kingston, R.E. A Polycomb Domain Found in Committed Cells Impairs Differentiation When Introduced into PRC1 in Pluripotent Cells. Mol Cell 2021, 81, 4677–4691. [Google Scholar] [CrossRef]
- Zhang, X.; Zhou, W.; Zhang, Y.; Liu, Z. CBX3 Is a Prognostic Biomarker Correlated with ATR Activation and Immune Infiltration in Head and Neck Squamous Cell Carcinoma. Int J Gen Med 2022, 15, 1497–1508. [Google Scholar] [CrossRef]
- Xie, X.; Ning, Y.; Long, J.; Wang, H.; Chen, X. Diverse CBX Family Members as Potential Prognostic Biomarkers in Non-Small-Cell Lung Cancer. FEBS Open Bio 2020, 10, 2206–2215. [Google Scholar] [CrossRef]
- Nielsen, A.L.; Ortiz, J.A.; You, J.; Oulad-Abdelghani, M.; Khechumian, R.; Gansmuller, A.; Chambon, P.; Losson, R. Interaction with Members of the Heterochromatin Protein 1 (HP1) Family and Histone Deacetylation Are Differentially Involved in Transcriptional Silencing by Members of the TIF1 Family. EMBO J 1999, 18, 6385–6395. [Google Scholar] [CrossRef]
- Saunders, W.S.; Chue, C.; Goebl, M.; Craig, C.; Clark, R.F.; Powers, J.A.; Eissenberg, J.C.; Elgin, S.C.; Rothfield, N.F.; Earnshaw, W.C. Molecular Cloning of a Human Homologue of Drosophila Heterochromatin Protein HP1 Using Anti-Centromere Autoantibodies with Anti-Chromo Specificity. J Cell Sci 1993, 104, 573–582. [Google Scholar] [CrossRef]
- Paro, R.; Hogness, D.S. The Polycomb Protein Shares a Homologous Domain with a Heterochromatin-Associated Protein of Drosophila. Proc Natl Acad Sci U S A 1991, 88, 263–267. [Google Scholar] [CrossRef]
- Lachner, M.; O’Carroll, D.; Rea, S.; Mechtler, K.; Jenuwein, T. Methylation of Histone H3 Lysine 9 Creates a Binding Site for HP1 Proteins. Nature 2001, 410, 116–120. [Google Scholar] [CrossRef]
- Nielsen, A.L.; Oulad-Abdelghani, M.; Ortiz, J.A.; Remboutsika, E.; Chambon, P.; Losson, R. Heterochromatin Formation in Mammalian Cells: Interaction between Histones and HP1 Proteins. Mol Cell 2001, 7, 729–739. [Google Scholar] [CrossRef]
- Aasland, R.; Stewart, A.F. The Chromo Shadow Domain, a Second Chromo Domain in Heterochromatin-Binding Protein 1, HP1. Nucleic Acids Res 1995, 23, 3168–3173. [Google Scholar] [CrossRef]
- Cowieson, N.P.; Partridge, J.F.; Allshire, R.C.; McLaughlin, P.J. Dimerisation of a Chromo Shadow Domain and Distinctions from the Chromodomain as Revealed by Structural Analysis. Curr Biol 2000, 10, 517–525. [Google Scholar] [CrossRef]
- Brasher, S. V; Smith, B.O.; Fogh, R.H.; Nietlispach, D.; Thiru, A.; Nielsen, P.R.; Broadhurst, R.W.; Ball, L.J.; Murzina, N. V; Laue, E.D. The Structure of Mouse HP1 Suggests a Unique Mode of Single Peptide Recognition by the Shadow Chromo Domain Dimer. EMBO J 2000, 19, 1587–1597. [Google Scholar] [CrossRef]
- Ruan, J.; Ouyang, H.; Amaya, M.F.; Ravichandran, M.; Loppnau, P.; Min, J.; Zang, J. Structural Basis of the Chromodomain of Cbx3 Bound to Methylated Peptides from Histone H1 and G9a. PLoS One 2012, 7, e35376. [Google Scholar] [CrossRef]
- Lachner, M.; O’Carroll, D.; Rea, S.; Mechtler, K.; Jenuwein, T. Methylation of Histone H3 Lysine 9 Creates a Binding Site for HP1 Proteins. Nature 2001, 410, 116–120. [Google Scholar] [CrossRef]
- Ligresti, G.; Caporarello, N.; Meridew, J.A.; Jones, D.L.; Tan, Q.; Choi, K.M.; Haak, A.J.; Aravamudhan, A.; Roden, A.C.; Prakash, Y.S.; et al. CBX5/G9a/H3K9me-Mediated Gene Repression Is Essential to Fibroblast Activation during Lung Fibrosis. JCI Insight 2019, 5. [Google Scholar] [CrossRef]
- Casale, A.M.; Cappucci, U.; Fanti, L.; Piacentini, L. Heterochromatin Protein 1 (HP1) Is Intrinsically Required for Post-Transcriptional Regulation of Drosophila Germline Stem Cell (GSC) Maintenance. Sci Rep 2019, 9, 4372. [Google Scholar] [CrossRef]
- Azzaz, A.M.; Vitalini, M.W.; Thomas, A.S.; Price, J.P.; Blacketer, M.J.; Cryderman, D.E.; Zirbel, L.N.; Woodcock, C.L.; Elcock, A.H.; Wallrath, L.L.; et al. Human Heterochromatin Protein 1α Promotes Nucleosome Associations That Drive Chromatin Condensation. J Biol Chem 2014, 289, 6850–6861. [Google Scholar] [CrossRef]
- Lomberk, G.; Wallrath, L.; Urrutia, R. The Heterochromatin Protein 1 Family. Genome Biol 2006, 7, 228. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Li, H.; Liang, X.; Xiang, Z. CBX3 Promotes Colon Cancer Cell Proliferation by CDK6 Kinase-Independent Function during Cell Cycle. Oncotarget 2017, 8, 19934–19946. [Google Scholar] [CrossRef] [PubMed]
- Smallwood, A.; Hon, G.C.; Jin, F.; Henry, R.E.; Espinosa, J.M.; Ren, B. CBX3 Regulates Efficient RNA Processing Genome-Wide. Genome Res 2012, 22, 1426–1436. [Google Scholar] [CrossRef] [PubMed]
- Niu, H.; Chen, P.; Fan, L.; Sun, B. Comprehensive Pan-Cancer Analysis on CBX3 as a Prognostic and Immunological Biomarker. BMC Med Genomics 2022, 15, 29. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Su, T.; Xue, Y.; Cheng, C.; Lay, F.D.; McKee, R.A.; Li, M.; Vashisht, A.; Wohlschlegel, J.; Novitch, B.G.; et al. Cbx3 Maintains Lineage Specificity during Neural Differentiation. Genes Dev 2017, 31, 241–246. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Shilatifard, A. Epigenetic Modifications of Histones in Cancer. Genome Biol 2019, 20, 245. [Google Scholar] [CrossRef] [PubMed]
- Saldi, T.; Cortazar, M.A.; Sheridan, R.M.; Bentley, D.L. Coupling of RNA Polymerase II Transcription Elongation with Pre-MRNA Splicing. J Mol Biol 2016, 428, 2623–2635. [Google Scholar] [CrossRef] [PubMed]
- Sheldon, L.A. Inhibition of E2F1 Activity and Cell Cycle Progression by Arsenic via Retinoblastoma Protein. Cell Cycle 2017, 16, 2058–2072. [Google Scholar] [CrossRef] [PubMed]
- Pei, Y.; Banerjee, S.; Sun, Z.; Jha, H.C.; Saha, A.; Robertson, E.S. EBV Nuclear Antigen 3C Mediates Regulation of E2F6 to Inhibit E2F1 Transcription and Promote Cell Proliferation. PLoS Pathog 2016, 12, e1005844. [Google Scholar] [CrossRef]
- Marei, H.E.; Althani, A.; Afifi, N.; Hasan, A.; Caceci, T.; Pozzoli, G.; Morrione, A.; Giordano, A.; Cenciarelli, C. P53 Signaling in Cancer Progression and Therapy. Cancer Cell Int 2021, 21, 703. [Google Scholar] [CrossRef]
- Wang, Z.; Jia, R.; Wang, L.; Yang, Q.; Hu, X.; Fu, Q.; Zhang, X.; Li, W.; Ren, Y. The Emerging Roles of Rad51 in Cancer and Its Potential as a Therapeutic Target. Front Oncol 2022, 12. [Google Scholar] [CrossRef]
- Orhan, E.; Velazquez, C.; Tabet, I.; Sardet, C.; Theillet, C. Regulation of RAD51 at the Transcriptional and Functional Levels: What Prospects for Cancer Therapy? Cancers 2021, 13, 2930. [Google Scholar] [CrossRef] [PubMed]
- Li, E.; Xia, M.; Du, Y.; Long, K.; Ji, F.; Pan, F.; He, L.; Hu, Z.; Guo, Z. METTL3 Promotes Homologous Recombination Repair and Modulates Chemotherapeutic Response in Breast Cancer by Regulating the EGF/RAD51 Axis. Elife 2022, 11. [Google Scholar] [CrossRef]
- Wang, H.; Zhao, W.; Wang, J.; Zhang, Z. Clinicopathological Significance of CBX3 in Colorectal Cancer: An Intensive Expression Study Based on Formalin-fixed and Paraffin-embedded Tissues. Pathol Int 2022, 72, 107–116. [Google Scholar] [CrossRef]
- Li, E.; Xia, M.; Du, Y.; Long, K.; Ji, F.; Pan, F.; He, L.; Hu, Z.; Guo, Z. METTL3 Promotes Homologous Recombination Repair and Modulates Chemotherapeutic Response in Breast Cancer by Regulating the EGF/RAD51 Axis. Elife 2022, 11. [Google Scholar] [CrossRef]
- Wang, Z.; Jia, R.; Wang, L.; Yang, Q.; Hu, X.; Fu, Q.; Zhang, X.; Li, W.; Ren, Y. The Emerging Roles of Rad51 in Cancer and Its Potential as a Therapeutic Target. Front Oncol 2022, 12. [Google Scholar] [CrossRef]
- Orhan, E.; Velazquez, C.; Tabet, I.; Sardet, C.; Theillet, C. Regulation of RAD51 at the Transcriptional and Functional Levels: What Prospects for Cancer Therapy? Cancers 2021, 13, 2930. [Google Scholar] [CrossRef]
- CBX3 Promotes Gastric Cancer Progression and Affects Factors Related to Immunotherapeutic Responses.
- CBX3 Predicts an Unfavorable Prognosis and Promotes Tumorigenesis in Osteosarcoma.
- Alam, H.; Li, N.; Dhar, S.S.; Wu, S.J.; Lv, J.; Chen, K.; Flores, E.R.; Baseler, L.; Lee, M.G. HP1γ Promotes Lung Adenocarcinoma by Downregulating the Transcription-Repressive Regulators NCOR2 and ZBTB7A. Cancer Res 2018, 78, 3834–3848. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Zhao, X.; Xia, L.; Lian, J.; You, J. Clinicopathological and Prognostic Significance of CBX3 Expression in Human Cancer: A Systematic Review and Meta-Analysis. Dis Markers 2020, 2020, 1–11. [Google Scholar] [CrossRef]
- CBX3/Heterochromatin Protein 1 Gamma Is Significantly Upregulated in Patients with Non-Small Cell Lung Cancer.
- Jin, X.; Zhang, B.; Zhang, H.; Yu, H. Smoking-Associated Upregulation of CBX3 Suppresses ARHGAP24 Expression to Activate Rac1 Signaling and Promote Tumor Progression in Lung Adenocarcinoma. Oncogene 2022, 41, 538–549. [Google Scholar] [CrossRef]
- Wang, J.; Yang, B.; Zhang, X.; Liu, S.; Pan, X.; Ma, C.; Ma, S.; Yu, D.; Wu, W. Chromobox Proteins in Cancer: Multifaceted Functions and Strategies for Modulation (Review). Int J Oncol 2023, 62. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Chang, L.; Yao, Y.; Chao, C.; Ge, Z.; Fan, C.; Yu, H.; Wang, B.; Yang, J. Role of the CBX Molecular Family in Lung Adenocarcinoma Tumorigenesis and Immune Infiltration. Front Genet 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- Hu, K.; Yao, L.; Xu, Z.; Yan, Y.; Li, J. Prognostic Value and Therapeutic Potential of CBX Family Members in Ovarian Cancer. Front Cell Dev Biol 2022, 10, 832354. [Google Scholar] [CrossRef] [PubMed]
- Czerwinska, P.; Mackiewicz, A.A. Mining Transcriptomic Data to Uncover the Association between CBX Family Members and Cancer Stemness. Int J Mol Sci 2022, 23. [Google Scholar] [CrossRef] [PubMed]
- Sundar, I.K.; Nevid, M.Z.; Friedman, A.E.; Rahman, I. Cigarette Smoke Induces Distinct Histone Modifications in Lung Cells: Implications for the Pathogenesis of COPD and Lung Cancer. J Proteome Res 2014, 13, 982–996. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yu, H.; Ren, D.; Sun, Y.; Guo, F.; Cai, H.; Zhou, C.; Zhou, Y.; Jin, X.; Wu, H. CBX3 Regulated By YBX1 Promotes Smoking-Induced Pancreatic Cancer Progression via Inhibiting SMURF2 Expression. Int J Biol Sci 2022, 18, 3484–3497. [Google Scholar] [CrossRef] [PubMed]
- Haley, J.A.; Haughney, E.; Ullman, E.; Bean, J.; Haley, J.D.; Fink, M.Y. Altered Transcriptional Control Networks with Trans-Differentiation of Isogenic Mutant-KRas NSCLC Models. Front Oncol 2014, 4, 344. [Google Scholar] [CrossRef] [PubMed]
- Bosso, G.; Cipressa, F.; Tullo, L.; Cenci, G. Co-Amplification of CBX3 with EGFR or RAC1 in Human Cancers Corroborated by a Conserved Genetic Interaction among the Genes. Cell Death Discov 2023, 9, 317. [Google Scholar] [CrossRef]
- Xu, H.; Jiang, C.; Chen, D.; Wu, Y.; Lu, J.; Zhong, L.; Yao, F. Analysis of Pan-Cancer Revealed the Immunological and Prognostic Potential of CBX3 in Human Tumors. Front Med (Lausanne) 2022, 9, 869994. [Google Scholar] [CrossRef]
- Chen, J.; Lin, Y.; Zheng, S.; Chen, Q.; Tang, S.; Zhong, X. CBX3 Promotes Clear Cell Renal Carcinoma through PI3K/AKT Activation and Aberrant Immunity. J Transl Med 2023, 21, 600. [Google Scholar] [CrossRef]
- Chen, H.; Han, C.; Liu, D.; Wang, F.; Ha, C. CBX3 Promotes Ovarian Cancer Progression by Regulating P53/P21-Mediated Glucose Metabolism via Inhibiting NCOR2. Archives of Medical Science 2022. [Google Scholar] [CrossRef]
- Taguchi, Y.-H.; Wang, H. Genetic Association between Amyotrophic Lateral Sclerosis and Cancer. Genes (Basel) 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Loboda, A.; Nebozhyn, M.; Klinghoffer, R.; Frazier, J.; Chastain, M.; Arthur, W.; Roberts, B.; Zhang, T.; Chenard, M.; Haines, B.; et al. A Gene Expression Signature of RAS Pathway Dependence Predicts Response to PI3K and RAS Pathway Inhibitors and Expands the Population of RAS Pathway Activated Tumors. BMC Med Genomics 2010, 3, 26. [Google Scholar] [CrossRef] [PubMed]
- Pavan, A.; Bragadin, A.B.; Calvetti, L.; Ferro, A.; Zulato, E.; Attili, I.; Nardo, G.; Dal Maso, A.; Frega, S.; Menin, A.G.; et al. Role of next Generation Sequencing-Based Liquid Biopsy in Advanced Non-Small Cell Lung Cancer Patients Treated with Immune Checkpoint Inhibitors: Impact of STK11, KRAS and TP53 Mutations and Co-Mutations on Outcome. Transl Lung Cancer Res 2021, 10, 202–220. [Google Scholar] [CrossRef] [PubMed]
- de Jesus, V.H.F.; Mathias-Machado, M.C.; de Farias, J.P.F.; Aruquipa, M.P.S.; Jácome, A.A.; Peixoto, R.D. Targeting KRAS in Pancreatic Ductal Adenocarcinoma: The Long Road to Cure. Cancers (Basel) 2023, 15. [Google Scholar] [CrossRef] [PubMed]
- Kalungi, F.; Nsubuga, A.; Anywar, G. Network Analysis and Molecular Docking Studies of Quercetin as a Potential Treatment for Prostate Cancer. In Silico Pharmacol 2023, 11, 24. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.S.; Ibaseta, A.; Fischer, M.M.; Cancilla, B.; O’Young, G.; Cristea, S.; Luca, V.C.; Yang, D.; Jahchan, N.S.; Hamard, C.; et al. Intratumoural Heterogeneity Generated by Notch Signalling Promotes Small-Cell Lung Cancer. Nature 2017, 545, 360–364. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Chen, D.; Maguire, E.M.; He, S.; Chen, J.; An, W.; Yang, M.; Afzal, T.A.; Luong, L.A.; Zhang, L.; et al. Cbx3 Inhibits Vascular Smooth Muscle Cell Proliferation, Migration, and Neointima Formation. Cardiovasc Res 2018, 114, 443–455. [Google Scholar] [CrossRef] [PubMed]
- Kopan, R. Notch Signaling. Cold Spring Harb Perspect Biol 2012, 4. [Google Scholar] [CrossRef]
- Zhou, B.; Lin, W.; Long, Y.; Yang, Y.; Zhang, H.; Wu, K.; Chu, Q. Notch Signaling Pathway: Architecture, Disease, and Therapeutics. Signal Transduct Target Ther 2022, 7, 95. [Google Scholar] [CrossRef]
- Ragot, H.; Monfort, A.; Baudet, M.; Azibani, F.; Fazal, L.; Merval, R.; Polidano, E.; Cohen-Solal, A.; Delcayre, C.; Vodovar, N.; et al. Loss of Notch3 Signaling in Vascular Smooth Muscle Cells Promotes Severe Heart Failure Upon Hypertension. Hypertension 2016, 68, 392–400. [Google Scholar] [CrossRef]
- Zou, B.; Zhou, X.-L.; Lai, S.-Q.; Liu, J.-C. Notch Signaling and Non-Small Cell Lung Cancer. Oncol Lett 2018, 15, 3415–3421. [Google Scholar] [CrossRef] [PubMed]
- Janghorban, M.; Xin, L.; Rosen, J.M.; Zhang, X.H.-F. Notch Signaling as a Regulator of the Tumor Immune Response: To Target or Not To Target? Front Immunol 2018, 9, 1649. [Google Scholar] [CrossRef]
- Koni, M.; Pinnarò, V.; Brizzi, M.F. The Wnt Signalling Pathway: A Tailored Target in Cancer. Int J Mol Sci 2020, 21, 7697. [Google Scholar] [CrossRef]
- Yang, H.; Pu, L.; Li, R.; Zhu, R. NCAPG Is Transcriptionally Regulated by CBX3 and Activates the Wnt/β-Catenin Signaling Pathway to Promote Proliferation and the Cell Cycle and Inhibit Apoptosis in Colorectal Cancer. J Gastrointest Oncol 2023, 14, 900–912. [Google Scholar] [CrossRef]
- Mauser, R.; Kungulovski, G.; Keup, C.; Reinhardt, R.; Jeltsch, A. Application of Dual Reading Domains as Novel Reagents in Chromatin Biology Reveals a New H3K9me3 and H3K36me2/3 Bivalent Chromatin State. Epigenetics Chromatin 2017, 10, 45. [Google Scholar] [CrossRef]
- Fan, Y.; Li, H.; Liang, X.; Xiang, Z. CBX3 Promotes Colon Cancer Cell Proliferation by CDK6 Kinase-Independent Function during Cell Cycle. Oncotarget 2017, 8, 19934–19946. [Google Scholar] [CrossRef]
- Fan, Y.; Li, H.; Liang, X.; Xiang, Z. CBX3 Promotes Colon Cancer Cell Proliferation by CDK6 Kinase-Independent Function during Cell Cycle. Oncotarget 2017, 8, 19934–19946. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-Y.; Cheng, C.-S.; Qu, C.; Wang, P.; Chen, H.; Meng, Z.-Q.; Chen, Z. Overexpression of CBX3 in Pancreatic Adenocarcinoma Promotes Cell Cycle Transition-Associated Tumor Progression. Int J Mol Sci 2018, 19. [Google Scholar] [CrossRef] [PubMed]
- Saxena, K.; Konopleva, M. An Expert Overview of Emerging Therapies for Acute Myeloid Leukemia: Novel Small Molecules Targeting Apoptosis, P53, Transcriptional Regulation and Metabolism. Expert Opin Investig Drugs 2020, 29, 973–988. [Google Scholar] [CrossRef]
- Sandy, Z.; da Costa, I.C.; Schmidt, C.K. More than Meets the ISG15: Emerging Roles in the DNA Damage Response and Beyond. Biomolecules 2020, 10. [Google Scholar] [CrossRef] [PubMed]
- Carr, S.M.; Munro, S.; La Thangue, N.B. Lysine Methylation and the Regulation of P53. Essays Biochem 2012, 52, 79–92. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Yang, N.; Chen, Y.; Zhu, M.; Lian, Y.; Xiong, Z.; Wang, B.; Feng, L.; Jia, X. An Integrated Strategy for Effective-Component Discovery of Astragali Radix in the Treatment of Lung Cancer. Front Pharmacol 2020, 11, 580978. [Google Scholar] [CrossRef] [PubMed]
- Hirata, H.; Hinoda, Y.; Kikuno, N.; Kawamoto, K.; Suehiro, Y.; Tanaka, Y.; Dahiya, R. MDM2 SNP309 Polymorphism as Risk Factor for Susceptibility and Poor Prognosis in Renal Cell Carcinoma. Clin Cancer Res 2007, 13, 4123–4129. [Google Scholar] [CrossRef] [PubMed]
- Appert-Collin, A.; Hubert, P.; Crémel, G.; Bennasroune, A. Role of ErbB Receptors in Cancer Cell Migration and Invasion. Front Pharmacol 2015, 6, 283. [Google Scholar] [CrossRef] [PubMed]
- Fry, W.H.D.; Kotelawala, L.; Sweeney, C.; Carraway, K.L. Mechanisms of ErbB Receptor Negative Regulation and Relevance in Cancer. Exp Cell Res 2009, 315, 697–706. [Google Scholar] [CrossRef] [PubMed]
- Black, L.E.; Longo, J.F.; Carroll, S.L. Mechanisms of Receptor Tyrosine-Protein Kinase ErbB-3 (ERBB3) Action in Human Neoplasia. Am J Pathol 2019, 189, 1898–1912. [Google Scholar] [CrossRef] [PubMed]
- Cragg, M.S.; Kuroda, J.; Puthalakath, H.; Huang, D.C.S.; Strasser, A. Gefitinib-Induced Killing of NSCLC Cell Lines Expressing Mutant EGFR Requires BIM and Can Be Enhanced by BH3 Mimetics. PLoS Med 2007, 4, 1681–1689. [Google Scholar] [CrossRef] [PubMed]
- Uimari, O.; Rahmioglu, N.; Nyholt, D.R.; Vincent, K.; Missmer, S.A.; Becker, C.; Morris, A.P.; Montgomery, G.W.; Zondervan, K.T. Genome-Wide Genetic Analyses Highlight Mitogen-Activated Protein Kinase (MAPK) Signaling in the Pathogenesis of Endometriosis. Hum Reprod 2017, 32, 780–793. [Google Scholar] [CrossRef]
- Thandra, K.C.; Barsouk, A.; Saginala, K.; Aluru, J.S.; Barsouk, A. Epidemiology of Lung Cancer. Contemp Oncol (Pozn) 2021, 25, 45–52. [Google Scholar] [CrossRef]
- Chang, S.-C.; Lai, Y.-C.; Chen, Y.-C.; Wang, N.-K.; Wang, W.-S.; Lai, J.-I. CBX3/Heterochromatin Protein 1 Gamma Is Significantly Upregulated in Patients with Non-Small Cell Lung Cancer. Asia Pac J Clin Oncol 2018, 14, e283–e288. [Google Scholar] [CrossRef]
- Czerwinska, P.; Mackiewicz, A.A. Mining Transcriptomic Data to Uncover the Association between CBX Family Members and Cancer Stemness. Int J Mol Sci 2022, 23. [Google Scholar] [CrossRef]
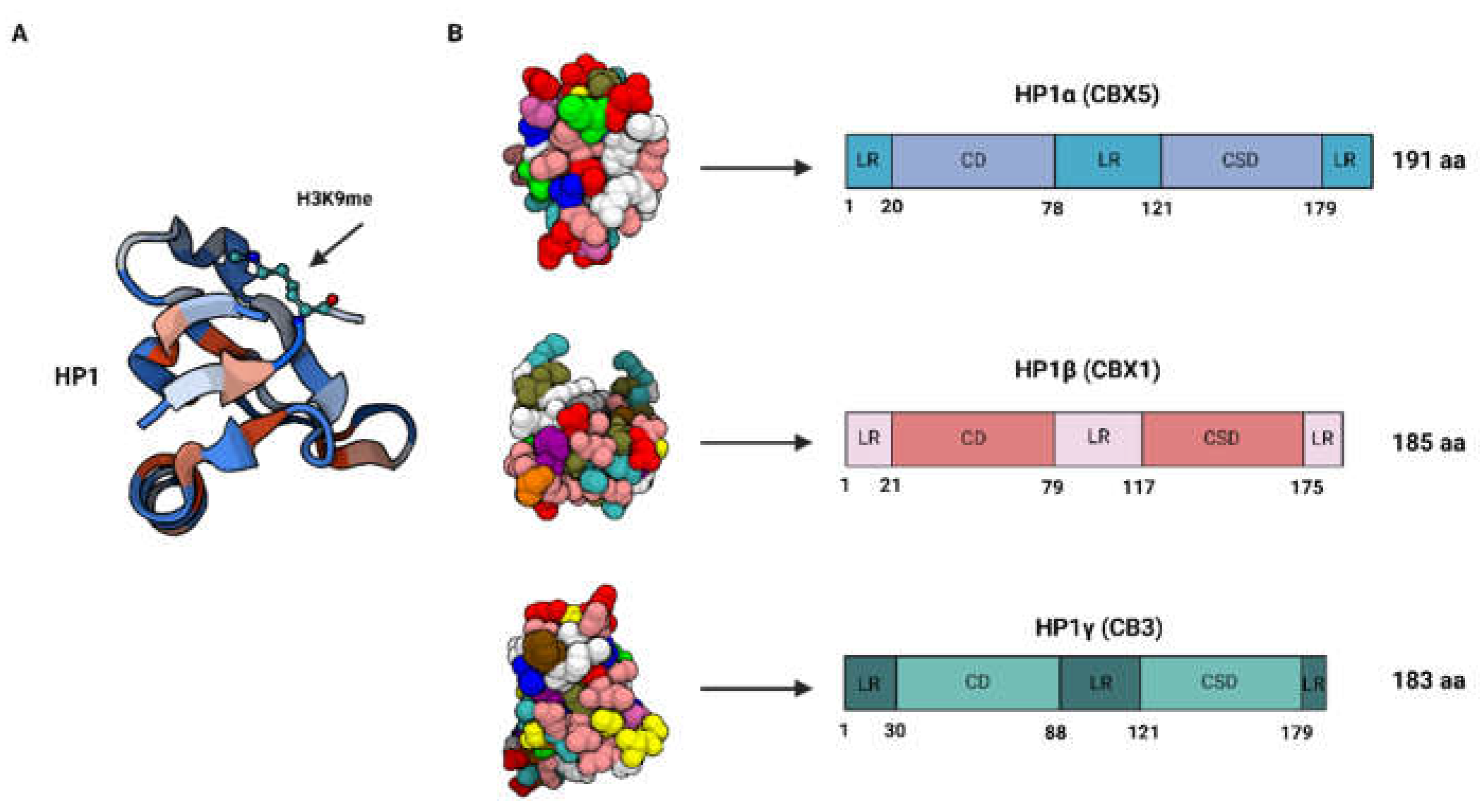
| Interactor | Type of Interaction | Effect on Lung Cancer | Pathway Involvement | Mechanism of Action |
|---|---|---|---|---|
| EGFR/RAC1 | Genetic Interaction | Co-amplification with CBX3 is associated with lung adenocarcinoma proliferation and poor prognosis | Not Specified | Increase in CBX3 mRNA leads to increased EGFR/RAC1 protein levels, promoting cancer cell proliferation [71] |
| CDK6/P21 | Transcriptional Regulation | CBX3 inhibits transcription of negative cell cycle regulators, promoting colorectal cancer cell proliferation; similar mechanisms may be involved in lung cancer | Cell Cycle Regulation |
CBX3 is able to inhibit transcription of CDK6 and p21, promoting cell proliferation [72] |
| CBX Molecular Family (CBX1/2/3/5/7) | Gene Expression | CBX3/5 expression is associated with poor prognosis in lung adenocarcinoma, while CBX7 shows the opposite effect | Tumorigenesis and Immune Infiltration |
Differential expression of CBX family members affects tumor progression and immune response [65] |
| PI3K/AKT Pathway | Activation | Although the study is on renal carcinoma, similar activation by CBX3 may occur in lung cancer, promoting metastasis and invasion | PI3K/AKT Pathway | CBX3 promotes cancer progression through PI3K/AKT activation, which regulates cell metastasis and invasion [73] |
| ARHGAP24 | Suppression | Smoking-associated upregulation of CBX3 suppresses ARHGAP24, activating RAC1 signaling and promoting tumor progression in lung adenocarcinoma | RAC1 Signaling | CBX3 overexpression leads to suppression of ARHGAP24, activating RAC1 and promoting tumor progression [63] |
| NCOR2 | Regulation | In ovarian cancer, CBX3 inhibits NCOR2, affecting p53/p21-mediated glucose metabolism; similar effects may occur in lung cancer | Glucose Metabolism | CBX3 inhibits NCOR2, affecting p53/p21-mediated pathways and potentially promoting cancer metabolism [74] |
| Immune System | Immunological Biomarker | CBX3 expression is related to immune cell infiltration and may serve as an immunological and prognostic biomarker in various cancers, including lung cancer | Immune Response |
CBX3 expression influences immune cell infiltration and tumor immunity, which varies based on tumor type [72] |
| Transcriptome/Metabolome | Biomarker Association | Possible association between CBX3 expression and transcriptome/metabolome changes in cancers, including lung cancer | Various Pathways | CBX3 expression may be linked to changes in the transcriptome and metabolome, affecting multiple cancer-related pathways [75] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





