Submitted:
19 July 2024
Posted:
22 July 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Reagents and Solvents
2.2. Cells, Wild Type YFV, and Interferon alfa-2a
2.3. Plant Material
2.4. Ethanolic Extract Preparation
2.5. Bioassay-Guided Chromatographic Fractionation
2.6. Cytotoxicity Assay and wt-YFV Replication Inhibition Assay
2.7. In Silico ADMET Prediction
2.8. Statistical Analyses
3. Results
3.1. Crude Extract Screening against wt-YFV
3.2. Bioassay-Guided Fractionation of H. puniceum Bulb Extract against wt-YFV
3.3. In Silico ADMET Properties
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Thomas, C.; Michaud, C.; Gaillet, M.; Carrión-Nessi, F.S.; Forero-Peña, D.A.; Lacerda, M.V.G.; Duchemin, J.-B.; Rodovalho, S.; Vreden, S.; Ramos, R. Yellow Fever Reemergence Risk in the Guiana Shield: A Comprehensive Review of Cases Between 1990 and 2022. Current Tropical Medicine Reports 2023, 10, 138–145. [Google Scholar] [CrossRef]
- Stanzani, L.M. de A.; Motta, M. de A.; Erbisti, R.S.; Abreu, F.V.S. de; Nascimento-Pereira, A.C.; Ferreira-de-Brito, A.; Neves, M.S.A.S.; Pereira, G.R.; Pereira, G.R.; Santos, C.B. dos; et al. Back to Where It Was First Described: Vectors of Sylvatic Yellow Fever Transmission in the 2017 Outbreak in Espírito Santo, Brazil. Viruses 2022, 14, 2805. [Google Scholar] [CrossRef]
- Taxon Details | ICTV. Available online: https://ictv.global/taxonomy/taxondetails?taxnode_id=202303121&taxon_name=Orthoflavivirus%20flavi (accessed on 25 June 2024).
- Postler, T.S.; Beer, M.; Blitvich, B.J.; Bukh, J.; de Lamballerie, X.; Drexler, J.F.; Imrie, A.; Kapoor, A.; Karganova, G.G.; Lemey, P.; et al. Renaming of the Genus Flavivirus to Orthoflavivirus and Extension of Binomial Species Names within the Family Flaviviridae. Arch Virol 2023, 168, 224. [Google Scholar] [CrossRef] [PubMed]
- Yellow Fever - Number of Reported Cases. Available online: https://www.who.int/data/gho/data/indicators/indicator-details/GHO/yellow-fever---number-of-reported-cases (accessed on 14 December 2023).
- Klitting, R.; Roth, L.; Rey, F.A.; de Lamballerie, X. Molecular Determinants of Yellow Fever Virus Pathogenicity in Syrian Golden Hamsters: One Mutation Away from Virulence. Emerg Microbes Infect 2018, 7, 51. [Google Scholar] [CrossRef] [PubMed]
- Phan, M.V.T.; Mendonca Melo, M.; van Nood, E.; Aron, G.; Kreeft-Voermans, J.J.C.; Koopmans, M.P.G.; Reusken, C.; GeurtsvanKessel, C.H.; Cotten, M. Shedding of Yellow Fever Virus from an Imported Case in the Netherlands After Travel to Brazil. Open Forum Infectious Diseases 2020, 7, ofaa020. [Google Scholar] [CrossRef] [PubMed]
- Cancado, B.; Aranda, C.; Mallozi, M.; Weckx, L.; Sole, D. Yellow Fever Vaccine and Egg Allergy. The Lancet Infectious Diseases 2019, 19, 812. [Google Scholar] [CrossRef] [PubMed]
- Vacina Febre Amarela (Atenuada) 5 e 10 Doses. [Bula] Rio de Janeiro: Instituto de Tecnologia Em Imunobiológicos (BIO-MANGUINHOS) – Fundação Oswaldo Cruz (FIOCRUZ). Available online: https://www.bio.fiocruz.br/en/images/stories/pdfs/bulas/fa/BM_BUL_045_00_V_190702_FA10Nacional.pdf (accessed on 7 August 2023).
- Pileggi, G.S.; Da Mota, L.M.H.; Kakehasi, A.M.; De Souza, A.W.; Rocha, A.; de Melo, A.K.G.; da Fonte, C.A.M.; Bortoletto, C.; Brenol, C.V.; Marques, C.D.L.; et al. Brazilian Recommendations on the Safety and Effectiveness of the Yellow Fever Vaccination in Patients with Chronic Immune-Mediated Inflammatory Diseases. Advances in Rheumatology 2019, 59, 17. [Google Scholar] [CrossRef] [PubMed]
- Simon, L.V.; Hashmi, M.F.; Torp, K.D. Yellow Fever. In StatPearls; StatPearls Publishing: Treasure Island (FL), 2023. [Google Scholar]
- Leal, C.M.; Leitão, S.G.; de Mello, L.L.O.; Rangel, I. de C.; da Silva, C.V.A.; Miranda, M.D.; Tucci, A.R.; de Assis, C.B.; Sacramento, C. de Q.; Fintelman-Rodrigues, N.; et al. Bioassay-Guided Fractionation of Siparuna Glycycarpa n-Butanol Extract with Inhibitory Activity against Influenza A(H1N1) Pdm09 Virus by Centrifugal Partition Chromatography (CPC). Molecules 2022, 27, 399. [Google Scholar] [CrossRef]
- Hippeastrum Puniceum (Lam.) Voss — Herbário. Available online: https://www.unirio.br/ccbs/ibio/herbariohuni/hippeastrum-puniceum-lam-voss (accessed on 13 December 2023).
- Hippeastrum Puniceum - Useful Tropical Plants. Available online: https://tropical.theferns.info/viewtropical.php?id=Hippeastrum+puniceum (accessed on 13 December 2023).
- Mitchell, S.A.; Ahmad, M.H. A Review of Medicinal Plant Research at the University of the West Indies, Jamaica, 1948-2001. West Indian Med J 2006, 55, 243–269. [Google Scholar] [CrossRef]
- Soprani, L.C.; Andrade, J.P. de; Santos, V.D. dos; Alves-Araújo, A.; Bastida, J.; Silva, C.A.G.; Silveira, D.; Borges, W. de S.; Jamal, C.M. Chemical Evaluation and Anticholinesterase Activity of Hippeastrum Puniceum (Lam.) Kuntz Bulbs (Amaryllidaceae). Braz. J. Pharm. Sci. 2021, 57, e19154. [Google Scholar] [CrossRef]
- Barbosa, E. de C. Avaliação da atividade antiviral de extratos brutos e substâncias, obtidos de plantas e de fungos, contra os vírus Dengue, Zika e Chikungunya. Thesis, 2019.
- de Castro Barbosa, E.; Alves, T.M.A.; Kohlhoff, M.; Jangola, S.T.G.; Pires, D.E.V.; Figueiredo, A.C.C.; Alves, É.A.R.; Calzavara-Silva, C.E.; Sobral, M.; Kroon, E.G.; et al. Searching for Plant-Derived Antivirals against Dengue Virus and Zika Virus. Virology Journal 2022, 19, 31. [Google Scholar] [CrossRef] [PubMed]
- Dulbecco, R.; Vogt, M. Some Problems of Animal Virology as Studied by the Plaque Technique. Cold Spring Harb Symp Quant Biol 1953, 18, 273–279. [Google Scholar] [CrossRef]
- Horai, H.; Arita, M.; Kanaya, S.; Nihei, Y.; Ikeda, T.; Suwa, K.; Ojima, Y.; Tanaka, K.; Tanaka, S.; Aoshima, K.; et al. MassBank: A Public Repository for Sharing Mass Spectral Data for Life Sciences. J Mass Spectrom 2010, 45, 703–714. [Google Scholar] [CrossRef]
- Kudi, A.C.; Myint, S.H. Antiviral Activity of Some Nigerian Medicinal Plant Extracts. J Ethnopharmacol 1999, 68, 289–294. [Google Scholar] [CrossRef] [PubMed]
- Mosmann, T. Rapid Colorimetric Assay for Cellular Growth and Survival: Application to Proliferation and Cytotoxicity Assays. J Immunol Methods 1983, 65, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Pires, D.E.V.; Blundell, T.L.; Ascher, D.B. PkCSM: Predicting Small-Molecule Pharmacokinetic and Toxicity Properties Using Graph-Based Signatures. J Med Chem 2015, 58, 4066–4072. [Google Scholar] [CrossRef]
- Pires, D.E.V.; Kaminskas, L.M.; Ascher, D.B. Prediction and Optimization of Pharmacokinetic and Toxicity Properties of the Ligand. Methods Mol Biol 2018, 1762, 271–284. [Google Scholar] [CrossRef]
- Wagner, G.J. Content and Vacuole/Extravacuole Distribution of Neutral Sugars, Free Amino Acids, and Anthocyanin in Protoplasts 1. Plant Physiol 1979, 64, 88–93. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.-J.; Li, X.-F.; Hu, J.-Q.; Ni, X.-J.; Lu, H.-Y.; Wang, J.-J.; Huang, X.-N.; Lin, C.-X.; Shang, D.-W.; Wen, Y.-G. A Simple HPLC–MS/MS Method for Determination of Tryptophan, Kynurenine and Kynurenic Acid in Human Serum and Its Potential for Monitoring Antidepressant Therapy. Journal of Analytical Toxicology 2017, 41, 37–44. [Google Scholar] [CrossRef]
- Guo, N.; Yang, D.; Yang, X.; Yan, H.; Fan, B.; Dai, J.; Lei, Y.; Yan, D. A Rapid, Sensitive, and Widely Applicable Method for Quantitative Analysis of Underivatized Amino Acids in Different Biological Matrices by UHPLC-MS/MS. Journal of Separation Science 2019, 42, 3173–3181. [Google Scholar] [CrossRef]
- Tömösi, F.; Kecskeméti, G.; Cseh, E.K.; Szabó, E.; Rajda, C.; Kormány, R.; Szabó, Z.; Vécsei, L.; Janáky, T. A Validated UHPLC-MS Method for Tryptophan Metabolites: Application in the Diagnosis of Multiple Sclerosis. Journal of Pharmaceutical and Biomedical Analysis 2020, 185, 113246. [Google Scholar] [CrossRef]
- More, G.K.; Vervoort, J.; Steenkamp, P.A.; Prinsloo, G. Metabolomic Profile of Medicinal Plants with Anti-RVFV Activity. Heliyon 2022, 8, e08936. [Google Scholar] [CrossRef] [PubMed]
- Giordani, R.B.; de Andrade, J.P.; Verli, H.; Dutilh, J.H.; Henriques, A.T.; Berkov, S.; Bastida, J.; Zuanazzi, J.A.S. Alkaloids from Hippeastrum Morelianum Lem.(Amaryllidaceae). Magnetic Resonance in Chemistry 2011, 49, 668–672. [Google Scholar] [CrossRef]
- Catherine, S. Lane Rapid LC-MS-MS Analysis of Free Amino Acids in Extracellular Matrix.
- Lopez, M.H.M. Potencial de Inibição de Enzimas de Interesse Farmacêutico Por Espécies de Amaryllidaceae. 2018.
- Klupczynska, A.; Misiura, M.; Miltyk, W.; Oscilowska, I.; Palka, J.; Kokot, Z.J.; Matysiak, J. Development of an LC-MS Targeted Metabolomics Methodology to Study Proline Metabolism in Mammalian Cell Cultures. Molecules 2020, 25, 4639. [Google Scholar] [CrossRef] [PubMed]
- Bowerbank, S.L.; Gallidabino, M.D.; Dean, J.R. Plant Poisons in the Garden: A Human Risk Assessment. Separations 2022, 9, 308. [Google Scholar] [CrossRef]
- Santana, O.; Reinab, M.; Anaya, A.L.; Hernández, F.; Izquierdo, M.E.; González-Coloma, A. 3-O-Acetyl-Narcissidine, a Bioactive Alkaloid from Hippeastrum Puniceum Lam. (Amaryllidaceae). Z Naturforsch C J Biosci 2008, 63, 639–643. [Google Scholar] [CrossRef] [PubMed]
- Dührkop, K.; Fleischauer, M.; Ludwig, M.; Aksenov, A.A.; Melnik, A.V.; Meusel, M.; Dorrestein, P.C.; Rousu, J.; Böcker, S. SIRIUS 4: A Rapid Tool for Turning Tandem Mass Spectra into Metabolite Structure Information. Nat Methods 2019, 16, 299–302. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, C.A. Lead- and Drug-like Compounds: The Rule-of-Five Revolution. Drug Discov Today Technol 2004, 1, 337–341. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and Computational Approaches to Estimate Solubility and Permeability in Drug Discovery and Development Settings. Advanced Drug Delivery Reviews 1997, 23, 3–25. [Google Scholar] [CrossRef]
- Santos, V.L. dos A.; Gonsalves, A. de A.; Araújo, C.R.M. ABORDAGEM DIDÁTICA PARA O DESENVOLVIMENTO DE MOLÉCULAS BIOATIVAS: REGRA DOS CINCO DE LIPINSKI E PREPARAÇÃO DE HETEROCICLO 1,3,4-OXADIAZOL EM FORNO DE MICRO-ONDAS DOMÉSTICO. Quím. Nova 2018, 41, 110–115. [Google Scholar] [CrossRef]
- Barros, A.G. Avaliação ADMET de substâncias. BIOINFO 2023, 3, 25. [Google Scholar] [CrossRef]
- Mandal, S.K.; Rehman, M.M.-U.; Katyal, A.; Rajvanshi, K.; Kannan, M.; Garg, M.; Murugesan, S.; Deepa, P.R. In Silico Anti-Viral Assessment of Phytoconstituents in a Traditional (Siddha Medicine) Polyherbal Formulation - Targeting Mpro and Pan-Coronavirus Post-Fusion Spike Protein. J Tradit Complement Med 2024, 14, 55–69. [Google Scholar] [CrossRef]
- Roskoski, R. Rule of Five Violations among the FDA-Approved Small Molecule Protein Kinase Inhibitors. Pharmacol Res 2023, 191, 106774. [Google Scholar] [CrossRef]
- Volpato, D.C.; Oliveira, E.A.; Okawa, R.T.; Teixeira, J.J.V. Idade e polifarmácia como fatores de risco para potenciais interações de drogas psicotrópicos via CYP450. Revista Contexto & Saúde 2022, 22, e9543–e9543. [Google Scholar] [CrossRef]
- Audi, E.A.; Pussi, F.D. Isoenzimas Do CYP450 e Biotransformação de Drogas. 2000, 22, 599–604.
- Wagner, G.J. Content and Vacuole/Extravacuole Distribution of Neutral Sugars, Free Amino Acids, and Anthocyanin in Protoplasts 1. Plant Physiol 1979, 64, 88–93. [Google Scholar] [CrossRef]
- Wu, G. Amino Acids: Metabolism, Functions, and Nutrition. Amino Acids 2009, 37, 1–17. [Google Scholar] [CrossRef]
- Shah, R.; Chen, S. Metabolic Signaling Cascades Prompted by Glutaminolysis in Cancer. Cancers 2020, 12, 2624. [Google Scholar] [CrossRef]
- Yoo, H.C.; Yu, Y.C.; Sung, Y.; Han, J.M. Glutamine Reliance in Cell Metabolism. Exp Mol Med 2020, 52, 1496–1516. [Google Scholar] [CrossRef]
- Lee, W.; Lee, S.-H.; Ahn, D.-G.; Cho, H.; Sung, M.-H.; Han, S.H.; Oh, J.-W. The Antiviral Activity of Poly-γ-Glutamic Acid, a Polypeptide Secreted by Bacillus Sp., through Induction of CD14-Dependent Type I Interferon Responses. Biomaterials 2013, 34, 9700–9708. [Google Scholar] [CrossRef]
- Wang, H.; Zheng, X.; Liu, B.; Xia, Y.; Xin, Z.; Deng, B.; He, L.; Deng, J.; Ren, W. Aspartate Metabolism Facilitates IL-1β Production in Inflammatory Macrophages. Front Immunol 2021, 12, 753092. [Google Scholar] [CrossRef]
- Vasconcelos, P.F. da C. Febre amarela. Rev. Soc. Bras. Med. Trop. 2003, 36, 275–293. [Google Scholar] [CrossRef]
- Katoch, D.; Sharma, U. Simultaneous Quantification and Identification of Amaryllidaceae Alkaloids in Narcissus Tazetta by Ultra Performance Liquid Chromatography-Diode Array Detector-Electrospray Ionisation Tandem Mass Spectrometry. Journal of Pharmaceutical and Biomedical Analysis 2019, 175, 112750. [Google Scholar] [CrossRef]
- Faisal, S.; Badshah, S.L.; Kubra, B.; Emwas, A.-H.; Jaremko, M. Alkaloids as Potential Antivirals. A Comprehensive Review. Nat Prod Bioprospect 2023, 13, 4. [Google Scholar] [CrossRef]
- Boshra, Y.R.; Fahim, J.R.; Hamed, A.N.E.; Desoukey, S.Y. Phytochemical and Biological Attributes of Narcissus Pseudonarcissus L. (Amaryllidaceae): A Review. South African Journal of Botany 2022, 146, 437–458. [Google Scholar] [CrossRef]
- Simbala, H.E.I.; Nurkolis, F.; Mayulu, N.; Rotty, L.W.A. Metabolites of Pinang Yaki (Areca Vestiaria) Fruit Extract: A Metabolite Profiling Study. F1000Res 2022, 10, 1021. [Google Scholar] [CrossRef]
- Özçelik, B.; Kartal, M.; Orhan, I. Cytotoxicity, Antiviral and Antimicrobial Activities of Alkaloids, Flavonoids, and Phenolic Acids. Pharmaceutical Biology 2011, 49, 396–402. [Google Scholar] [CrossRef]
- Bedada, W.; de Andrés, F.; Engidawork, E.; Hussein, J.; LLerena, A.; Aklillu, E. Effects of Khat (Catha Edulis) Use on Catalytic Activities of Major Drug-Metabolizing Cytochrome P450 Enzymes and Implication of Pharmacogenetic Variations. Sci Rep 2018, 8, 12726. [Google Scholar] [CrossRef]
- Hung, T.-C.; Lee, W.-Y.; Chen, K.-B.; Chan, Y.-C.; Chen, C.Y.-C. Lead Screening for HIV-1 Integrase (IN) Inhibited by Traditional Chinese Medicine. Biomed Res Int 2014, 2014, 479367. [Google Scholar] [CrossRef]
- Masi, M.; Cala, A.; Tabanca, N.; Cimmino, A.; Green, I.R.; Bloomquist, J.R.; Van Otterlo, W.A.L.; Macias, F.A.; Evidente, A. Alkaloids with Activity against the Zika Virus Vector Aedes Aegypti (L.)—Crinsarnine and Sarniensinol, Two New Crinine and Mesembrine Type Alkaloids Isolated from the South African Plant Nerine Sarniensis. Molecules 2016, 21, 1432. [Google Scholar] [CrossRef]
- Herraiz, T.; Galisteo, J. Tetrahydro-β-Carboline Alkaloids Occur in Fruits and Fruit Juices. Activity as Antioxidants and Radical Scavengers. J. Agric. Food Chem. 2003, 51, 7156–7161. [Google Scholar] [CrossRef]
- Chen, H.; Lao, Z.; Xu, J.; Li, Z.; Long, H.; Li, D.; Lin, L.; Liu, X.; Yu, L.; Liu, W.; et al. Antiviral Activity of Lycorine against Zika Virus in Vivo and in Vitro. Virology 2020, 546, 88–97. [Google Scholar] [CrossRef] [PubMed]
- Hwang, Y.-C.; Chu, J.J.-H.; Yang, P.L.; Chen, W.; Yates, M.V. Rapid Identification of Inhibitors That Interfere with Poliovirus Replication Using a Cell-Based Assay. Antiviral Res 2008, 77, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Renard-Nozaki, J.; Kim, T.; Imakura, Y.; Kihara, M.; Kobayashi, S. Effect of Alkaloids Isolated from Amaryllidaceae on Herpes Simplex Virus. Research in Virology 1989, 140, 115–128. [Google Scholar] [CrossRef] [PubMed]
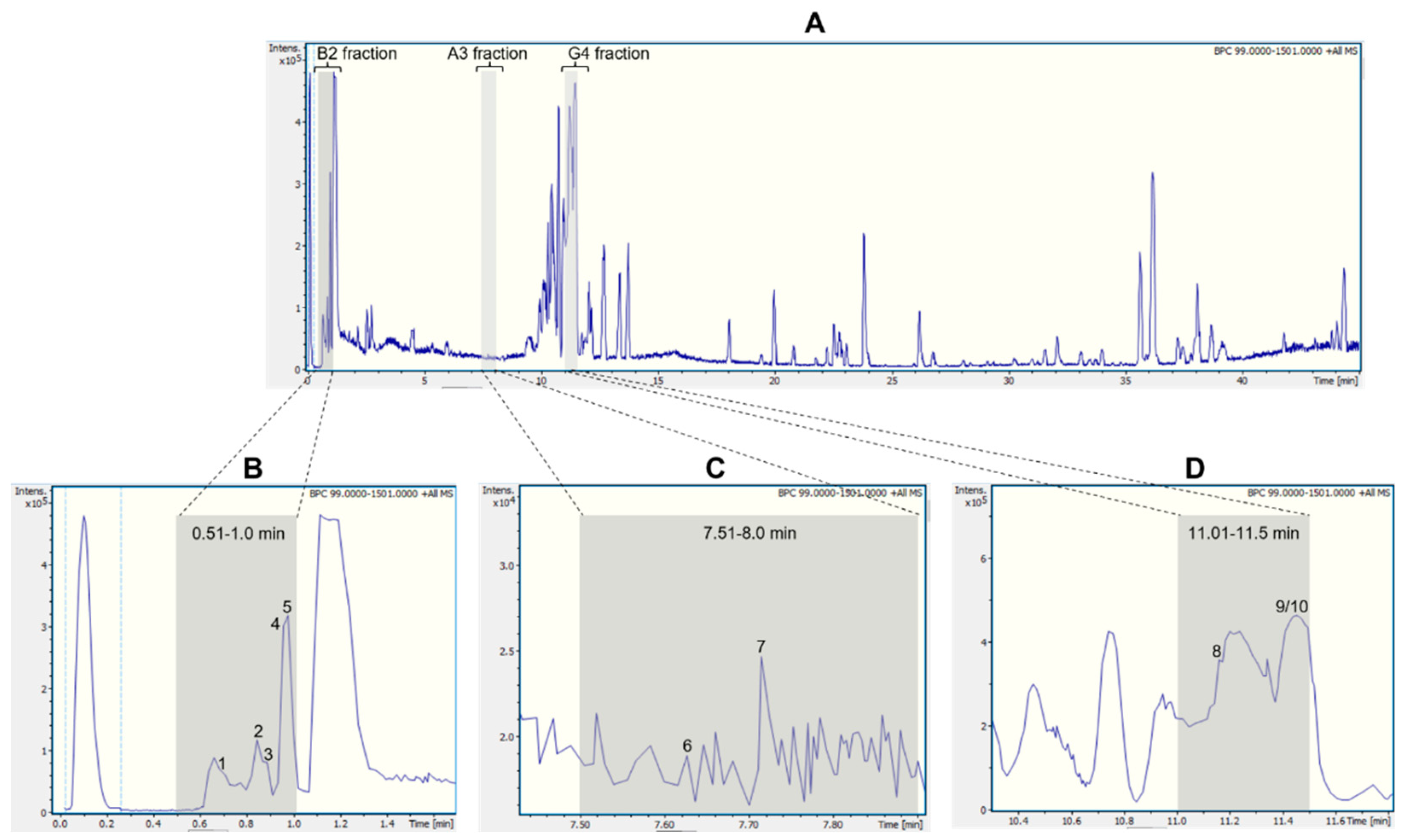
| Fraction | Peak ID | RT (min) |
Parent Ion [M+H]+ (m/z) | MS/MS Fragments (m/z, relative abundance - %) |
Molecular formula |
Annotation | |
|---|---|---|---|---|---|---|---|
| B2 (0.51-1.00 min) |
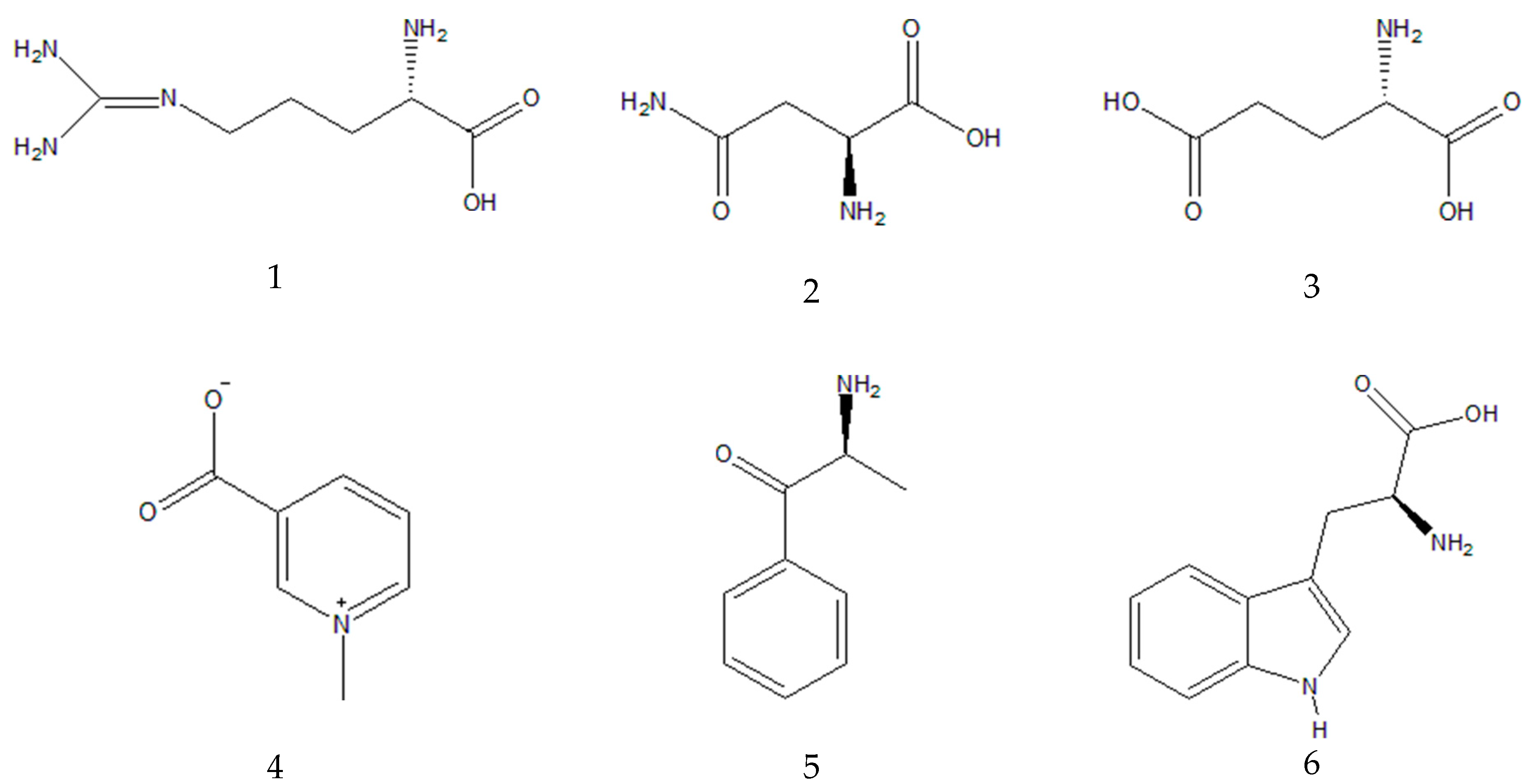
1 | 0.7 | 175.1190 | 175.1188 (100.0); 158.0922 (20.0); 130.0975 (9.7); 116.0698 (4.8) | C6H14N4O2 | Arginine a,b | |
| 2 | 0.9 | 133.0606 | 133.0607 (100.0); 116.0348 (10.1); 132.1022 (7.8); 130.0505 (5.0) | C4H8N2O3 | Asparagine a,b | ||
| 3 | 0.9 | 148.0603 | 148.0603 (33.3); 146.1177 (37.5); 130.0498 (100.0) | C5H9NO4 | Glutamic acid b | ||
| 4 | 1.0 | 138.0547 | 135.0676 (0.7); 136.0392(0.5); 136.0622 (0.5); 110.0899 | C7H7NO2 | Trigonelline a,b | ||
| 5 | 1.0 | 150.0911 | 135.0676 (24.3); 134.0600 (9.1); 132.0807 (2.0); 119.0491 (3.2); 117.0 (1.7) | C9H11NO | Cathinone b | ||
| A3 (7.51-8.00 min) |
6 | 7.6 | 205.0972 | 188.0705 (100.0); 146.0598 (43.1); 118.0650 (22.3) | C11H12N2O2 | Tryptophan a,b | |
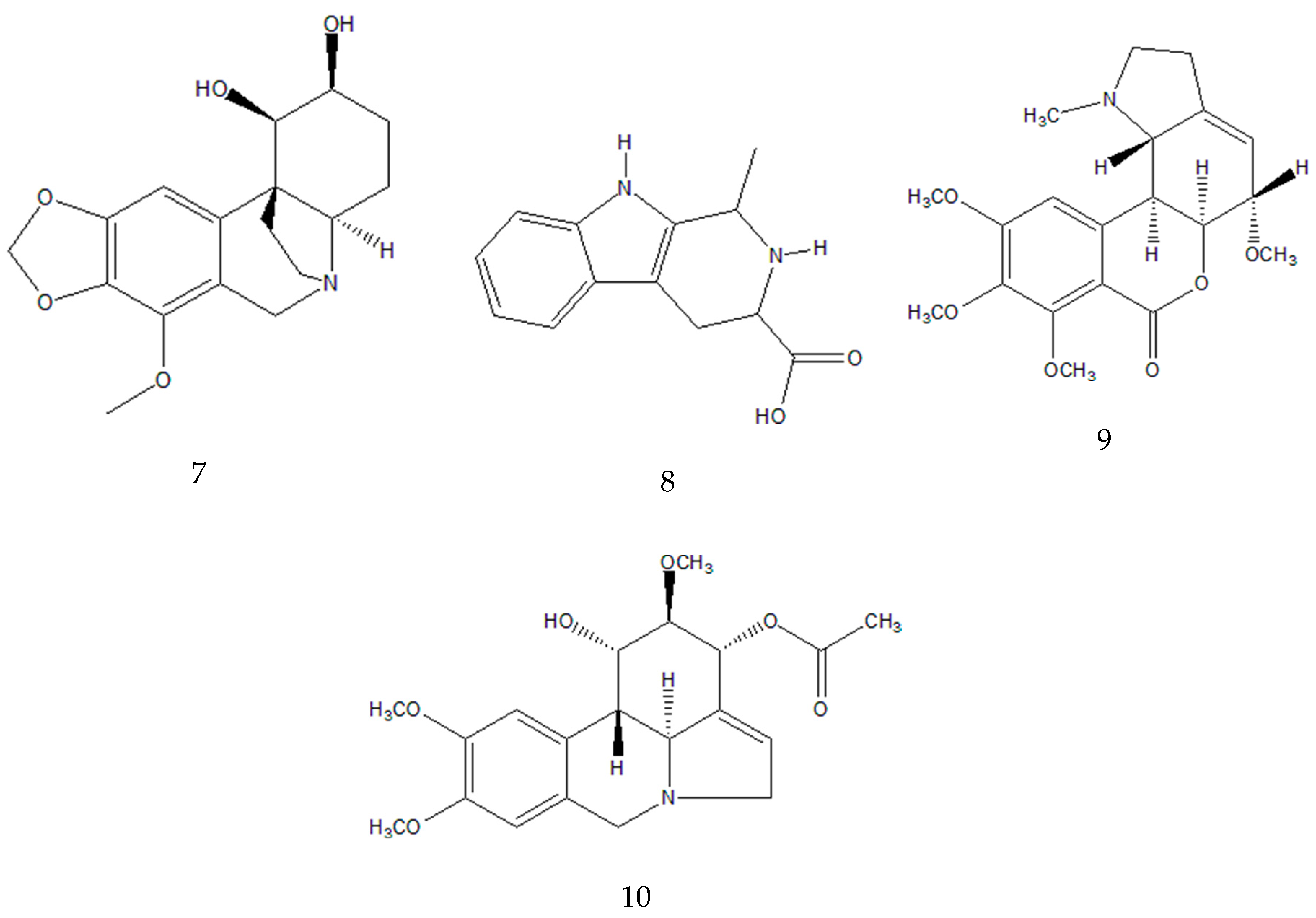
| 7 | 7.7 | 320.1490 | 147.0438 (34.6); 119.0489 (9.2); 220.0750 (5.7) | C17H21NO5 | Bulbisine b | ||
| G4 (11.01-11.50 min) |
8 | 11.1 | 231.1126 | 158.0965 (100.0); 143.0723 (96.2); 130.0650 (52.8) | C13H14N2O2 | Tetrahydroharman-3-carboxylic acid b,c | |
| 9/10 | 11.4 | 376.1755 | 376.1755 (100.0); 377.1789 (21.0); 165.0912 (16.3); 124.0754 (3.2);139.0543 (0.2) | C20H25NO6 | 3-O-acetyl narcissidine b,c |
| PARAMETERS | COMPOUNDS | INDICATORS | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Trigonelline | Cathinone | Bulbisine | Tetrahydroharman-3-carboxylic acid | 3-O-acetyl-narcissidine | 2,7-dimethoxy-homolycorine | |||||||
| MOL_WEIGHT | 137,138 | 149,193 | 319,357 | 230,267 | 375,421 | 375,421 | ||||||
| LOGP | -1.1254 | 1.2165 | 0.7652 | 1.8278 | 1.2328 | 1.9941 | Lipinski’s RO5: <5 Ideally between 1-4 | |||||
| #ROTATABLE_BONDS | 1 | 2 | 1 | 1 | 4 | 4 | ||||||
| #ACCEPTORS | 2 | 2 | 6 | 2 | 7 | 7 | ||||||
| #DONORS | 0 | 1 | 2 | 3 | 1 | 0 | ||||||
| SURFACE_AREA | 58,547 | 66,028 | 134,111 | 98,647 | 158,003 | 158,323 | ||||||
| Water solubility | -1.931 | -0.795 | -1.859 | -2.435 | -2.948 | -3.146 | The predicted water solubility of a compound is given as the logarithm of the molar concentration (log mol/L). | |||||
| Caco2 permeability | 1.124 | 1.237 | -0.138 | 0.832 | 0.705 | 1.472 | High CaCO-2 permeability would translate in predicted values >0.90 | |||||
| Intestinal absorption (human) | 96.44 | 76.876 | 70.972 | 94.534 | 64.054 | 97.738 | Poorly absorbed: < 30% | |||||
| Skin Permeability | -2.736 | -2.278 | -3.236 | -2.735 | -3.175 | -2.887 | Low skin permeability if it has a logKp > -2.5. | |||||
| P-glycoprotein substrate | Yes | No | Yes | Yes | Yes | No | Yes or No | |||||
| PARAMETERS | COMPOUNDS | INDICATORS | ||||||||||
| Trigonelline | Cathinone | Bulbisine | Tetrahydroharman-3-carboxylic acid | 3-O-acetylnarcissidine | 2,7-dimethoxyhomolycorine | |||||||
| P-glycoprotein I inhibitor | No | No | No | No | No | No | Yes or No |
|||||
| P-glycoprotein II inhibitor | No | No | No | No | No | No | Yes or No | |||||
| VDss (human) | -0.758 | 0.465 | 0.401 | -0.237 | 0.481 | 0.402 | Low if below 0.71 L/kg (log VDss < -0.15) and high if above 2.81 L/kg (log VDss > 0.45). | |||||
| Fraction unbound (human) | 0.857 | 0.469 | 0.436 | 0.481 | 0.413 | 0.411 | For a given compound the predicted fraction that would be unbound in plasma will be calculated. | |||||
| BBB permeability | -0.234 | -0.133 | -0.716 | 0.225 | -0.538 | -0.396 | Readily cross BBB >0.3; Poorly distributed in brain <-1 | |||||
| CNS permeability | -2.739 | -1.768 | -3.429 | -3.254 | -3.243 | -2.969 | Can penetrate, Log PS > -2 Cannot penetrate, Log PS < -3 | |||||
| CYP2D6 substrate | No | No | No | Yes | No | No | Yes or No | |||||
| CYP3A4 substrate | No | No | Yes | No | Yes | Yes | Yes or No | |||||
| CYP1A2 inhibitor | No | Yes | No | No | No | No | Yes or No | |||||
| CYP2C19 inhibitor | No | No | No | No | No | No | Yes or No | |||||
| CYP2C9 inhibitor | No | No | No | No | No | No | Yes or No | |||||
| CYP2D6 inhibitor | No | No | No | No | No | No | Yes or No | |||||
| CYP3A4 inhibitor | No | No | No | No | No | No | Yes or No | |||||
| Total Clearance | 0.378 | 0.811 | 1.137 | 0.694 | 0.902 | 0.636 | - | |||||
| Renal OCT2 substrate | No | No | No | No | No | No | Yes or No | |||||
| AMES toxicity | No | No | No | No | No | No | Yes or No | |||||
| Max. tolerated dose (human) | 0.743 | 0.779 | 0.025 | 0.323 | -0.558 | -0.099 | High is greater than 0.477 | |||||
| hERG I inhibitor | No | No | No | No | No | No | Yes or No | |||||
| hERG II inhibitor | No | No | No | No | No | No | Yes or No | |||||
| PARAMETERS | COMPOUNDS | INDICATORS | ||||||||||
| Trigonelline | Cathinone | Bulbisine | Tetrahydroharman-3-carboxylic acid | 3-O-acetylnarcissidine | 2,7-dimethoxyhomolycorine | |||||||
| Oral Rat Acute Toxicity (LD50) | 1.878 | 2.131 | 3.124 | 2.412 | 3.109 | 2.739 | The LD50 is the amount of a compound given all at once that causes the death of 50% of a group of test animals. | |||||
| Oral Rat Chronic Toxicity (LOAEL) | 0.454 | 1.542 | 2.614 | 1.115 | 1.610 | 2.689 | The LOAEL results need to be interpreted relative to the bioactive concentration and treatment lengths required. | |||||
| Hepatotoxicity | No | No | Yes | No | Yes | Yes | Yes or No | |||||
| Skin Sensitisation | No | No | No | No | No | No | Yes or No | |||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





