1. Introduction
The Microbiologically Influenced Corrosion (MIC) is a global phenomenon that affects mainly metal surfaces exposed to almost all types of environments. Many definitions have been given for MIC over the years. One of the most recent ones defines MIC as “the corrosion affected by the presence or activity, or both, of microorganisms” [
1].
Since the beginning of MIC research, Sulphate-Reducing bacteria (SRB) has been associated with MIC and identified as an indicator of its presence [
2]. In addition, over the years other bacterial groups have also demonstrated their influence on this type of corrosion, such as Slime-Forming bacteria (SFB), which generates extracellular polymeric substances (EPS), or Acid-Producing bacteria (APB) [
2,
3]. Other types of micro-organisms have also been associated with MIC, such as algae, protozoa, diatoms, archaea and fungi, but bacteria stand out as the most influential, and particularly SRB [
4,
5,
6].
Different strategies have been developed to avoid MIC, including mechanical methods, as biofilm surface cleaning; or chemical, like the use of corrosion inhibitors or biocides [
1]. In particular, the use of compounds that inhibit the formation of biofilms, which adhere to the metal surface, or the use of antimicrobial agents in the system may be used [
3,
7].
In search of new materials that can efficiently, safely, economically and sustainably combat bacteria, new bionanomaterials with antimicrobial properties have been developed in the last years [
8,
9,
10,
11,
12].
In particular, a pioneer strategy of preparation of metal bionanomaterials using a protein as scaffolds and inducing agent for the formation of small metal nanoparticles, stable, monodisperse metal nanoparticles (MeNPs) with controlled size and shape, in water at room temperature have been successfully developed [
13,
14,
15,
16,
17]. Some of these bionanohybrids have been efficient against viruses and different bacteria [
18,
19,
20,
21].
Regarding to the action mechanisms of MeNPs-enzyme hybrids against bacteria, they have not been yet completely described. However, it has been proven that MeNPs serve as antimicrobial agents, and several mechanisms have been described for their antimicrobial performance. These mechanisms can be categorized based on whether they occur in the extracellular or intracellular environment of the cells.
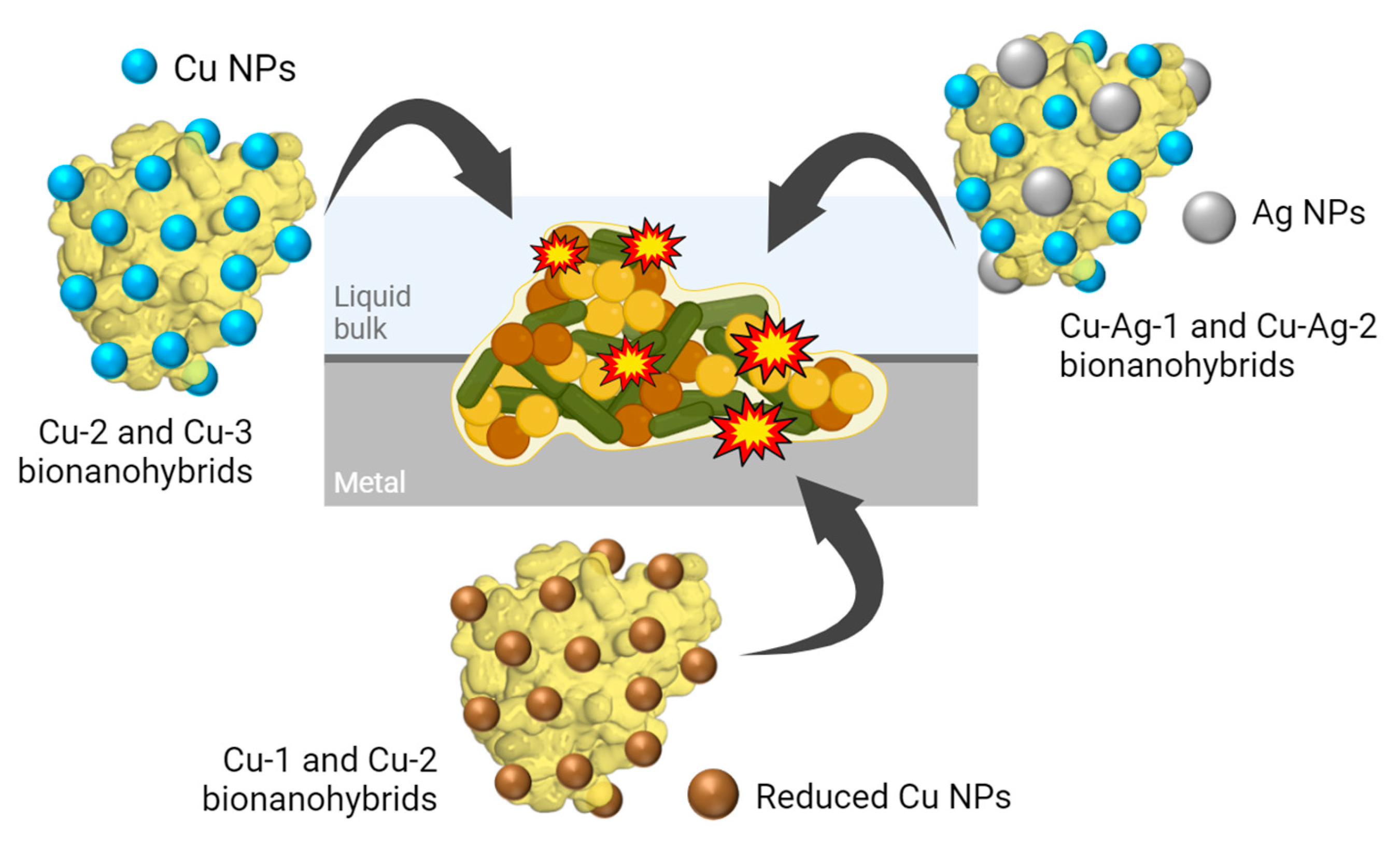
Figure 1.
Schematic illustration of bionanohybrids attack against an APB, SRB and SFB biofilm.
Figure 1.
Schematic illustration of bionanohybrids attack against an APB, SRB and SFB biofilm.
In the extracellular environment, nanoparticles can damage the cell membrane either by directly degrading the compounds that form the membrane or indirectly by generating reactive oxygen species (ROS), which cause membrane damage. In the intracellular environment, MeNPs could inhibit enzymes, induce oxidative stress and modify gene expression levels [
22,
23].
In particular, silver and copper are well-known for their antibacterial effects. Some studies suggest that copper primarily acts against bacteria by disrupting the cell membrane and inactivating enzymes [
22,
24]. In contrast, silver is believed to disrupt cellular functions, damage the cell membrane, target intracellular biomolecules, and induce oxidative stress [
25,
26,
27].
Thus, the present study aims to evaluate the antimicrobial efficacy of different Cu and Cu-Ag NPs-enzyme hybrids against the previously mentioned MIC-causing bacteria, as well as their potential against the formation of biofilms on susceptible surfaces
2. Materials and Methods
2.1. Materials
Copper (II) sulphate pentahydrate, hydrogen peroxide (33% v/v), sodium hydroxide, sodium dihydrogen phosphate 1-hydrate and sodium borohydride were obtained from Panreac (Barcelona, Spain). Silver nitrate was from Merck (St. Louis, MO, USA). Lipase B from Candida antarctica (CAL-B) solution was supplied by Novozymes (Copenhagen, Denmark).
All cultures tested are enrichments from Endures B.V. strain collection and their origin are a MIC damage case of an industrial process water pipe of a chemical production site in Belgium. The pipe system had a perforation and corrosion products were collected from the pipe surface and tested for presence of MIC relevant microorganisms. Three different enrichments were used from this damage case: Slime-Forming bacteria (SFB), Sulphate-Reducing bacteria (SRB) and Acid-Producing bacteria (APB). Bacteria were grown in the corresponding media and corrosion products from the failure case were used as inoculum. Enrichments were stored at 4°C in the Endures in-house strain collection.
2.2. Culture Media
SFB culture medium: 5 g of bacteriological peptone from meat and 3 g of meat extract were dissolved in 1 L of demi water and autoclaved.
APB culture medium: 5 g of D-glucose, 10 g of tryptone, 10 g of bacteriological peptone, 1.59 mL of glycerol, 0.2 g of magnesium sulphate heptahydrate (MgSO4·7H2O) and 0.01 g of phenol red were dissolved in 1 L of demi water and autoclaved. Then, 0.2 g of sodium thiosulphate pentahydrate (Na2S2O3·5H2O) and 0.2 g of dipotassium phosphate (K2HPO4) were dissolved in 50 mL and add to the previous solution by sterile filtration. APB medium changes from red to yellow when the culture is active.
SRB culture medium: first, 1 g of ammonium chloride (NH4Cl), 4.5 g of sodium sulphate anhydrous (Na2SO4), 0.04 g of calcium chloride dihydrate (CaCl2·2H2O), 0.06 g of magnesium sulphate heptahydrate (MgSO4·7H2O), 4.2 mL of DL-Na-Lactate and 1 g of yeast extract were dissolved in 990 mL of demi water and autoclaved.
Second, 4 mg of iron (II) sulfate heptahydrate (FeSO4·7H2O) was dissolved in 50 µL of H2SO4. Once completely dissolved, it was added to a third solution, composed by 0.3 g of sodium citrate dihydrate, 0.1 g of L-ascorbic acid, 0.5 g of dipotassium phosphate (K2HPO4) and 10 mL of demi water. Finally, this mixture was added to the starting solution by sterile filtration. SRB medium changes from yellow to black when the culture is active.
2.3. Synthesis of Copper Bionanohybrids
Copper bionanohybrids Cu-1 and Cu-2 were prepared adding 3.6 mL of lipase CAL-B solution (10.34 mg/mL) to 60 mL of a sodium phosphate solution (0.1 M, pH 7) in a 250 mL glass bottle containing a small magnetic bar stirrer. After that, 600 mg of Cu2SO4·5H2O (10 mg/mL) was added to the solution and it was stirred for 1 h at room temperature. Then, different quantities of NaBH4 were added to the mixture, 300 mg for Cu-1 and 30 mg for Cu-2, respectively. The reducing agent was added in a 6 mL aqueous solution in 2 times of 3 mL, and it was stirred for 30 min. After the reduction step, the mixture was centrifuged for 5 min at 3000 rpm and the solid obtained in the generated pellet was washed with distilled water. This process was repeated two times more. Finally, the solid was resuspended in 2 mL of distilled water in a cryotube, frozen with liquid nitrogen and lyophilized for 16h.
The bionanohybrid
Cu-3 was synthetized adding 1.8 mL of CAL-B to 60 mL of sodium phosphate solution (0.1 M, pH 7) and 600 mg of Cu
2SO
4·5H
2O. The mixture was stirred with a magnetic bar for 1h at room temperature. After that, a light blue emulsion with a concentration of 5000 ppm was obtained, as reported in a previous synthesis protocol for copper bionanohybrids [
18].
Cu-4 was obtained in the same way as
Cu-3 but finally adding 0.5 % (v/v) of hydrogen peroxide.
ICP-OES analyses of the metal content revealed copper percentages of 45 wt%, 27 wt%, 32 wt% and 32 wt% for Cu-1, Cu-2, Cu-3 and Cu-4, respectively.
2.4. Synthesis of Copper-Silver Bionanohybrids
Copper-silver bionanohybrids were generated adding different amounts of silver nitrate to Cu-3 hybrid using different media. First, 20 mL of Cu-3 was centrifuged and washed three times with distilled water, and the supernatant was removed. Cu-Ag-1 was prepared adding 20 mL of sodium phosphate solution (0.1 M, pH 7) to the washed Cu-3 pellet and 240 mg of silver nitrate salt. Instead, Cu-Ag-2 was synthetized adding 20 mL of distilled water and 12 mg of silver nitrate to the washed Cu-3. After that, in both cases, the mixture was stirred for 24 hours at room temperature and after that it was centrifuged for 8 min at 3000 rpm. The supernatant was removed, and the pellet was washed with distilled water, repeating this process three times. Lastly, the solid was resuspended in 2 mL of distilled water in a cryotube, frozen with liquid nitrogen and lyophilized for 16h.
ICP-OES analyses of the metal content revealed copper percentages of 25 wt% and 32 wt%, and silver percentages of 82 wt% and 4 wt% for Cu-Ag-1 and Cu-Ag-2, respectively.
2.5. Bionanohybrids Characterization Techniques
Inductively coupled plasma-optical emission spectrometry (ICP-OES) was performed using an OPTIMA 2100 DV instrument (PerkinElmer, Waltham, MA). X-ray diffraction (XRD) patterns were obtained using a Texture Analysis D8 ADVANCE Diffractometer (Bruker, Billerica, MA, USA) with Cu Kα radiation. Their analysis was performed using the X’Pert Highscore Plus programs. Transmission electron microscopy (TEM) images were obtained on a 2100F microscope (JEOL, Tokyo, Japan) equipped with an EDX detector INCA x-sight (Oxford Instruments, Abingdon, UK).
2.6. Bacteria Growth
Bacterial cultures of SFB, APB and SRB, isolated from industrial process water environments, were grown in the appropriate culture media in a final volume of 25 mL from previous cultures using inoculums of 10%. The incubation was aerobic for SFB and anaerobic for SRB and APB. In the preparation of SRB and APB cultures, anaerobic bottles closed with a Teflon rubber septum were used. After that, a current of N2 was flushed to remove the oxygen and the bottles were kept at room temperature without agitation. For the aerobic strain, SFB, tubes for the growths were used, which were kept at room temperature in an orbital shaker.
The growth curves or SRB, APB and SFB were studied using different initial concentrations of bacteria in the growths: 1·10
6, 1·10
7 and 1·10
8 cells/mL (
Figure S1, S2 and S3). After that, the of 1·10
7 cells/mL initial concentration was selected as the optimum for the following experiments.
2.7. Cell Counts
To obtain the bacterial cell/mL concentration a Thoma Cell Counting Chamber and a Leica DM500 Microscope with a 40X objective and the equation below were used, following a standard procedure designed by Endures B.V.
2.8. Antibacterial Activity of Bionanohybrids
The bacterial viability in the presence of the bionanohybrids was performed adding 50 or 100 ppm of the bionanohybrids to bacterial solutions with a concentration of 1·107 cells/mL. The mixture was maintained in room temperature in an orbital shaker. Samples at different times were taken, from 0 to 70 hours, with 3 replicas per point, to follow the evolution of the growth curves.
To percentage of bacteria reduction was calculated as follows:
2.9. Coupons Experiments
Carbon steel coupons (2 x 2 cm) were exposed to SRB cultures for early biofilm experiments. The coupons were placed in the bottom of glass bottles with 20 mL of SRB medium, an adequate bacterial volume to obtain a concentration of order of magnitude 7 and 50 ppm of selected bionanohybrid. The set-up was performed in the anaerobic chamber, to avoid the presence of oxygen and samples were collected after 0 and 24 hours of exposure.
To analyze the bacterial attachment to the surface of the coupons and their viability, the surfaces were stained with a LIVE/DEAD® BacLight™ Bacterial Viability Kit (Thermo Fisher), for 20 minutes in the dark. After that, the coupons were carefully rinsed, dried and preserved in the dark. For the fluorescence imaging, an Olympus BX51 Fluorescence Microscope (Olympus Corporation, Tokyo, Japan) was used.
Each experiment was performed in triplicated.
3. Results and Discussion
3.1. Synthesis and Characterization of Bionanohybrids
A variety of synthesis strategies have been employed to generate the designed bionanohybrids, starting from a copper bionanohybrid and subsequently incorporating a reducing agent, an oxidant agent or a silver salt, in order to search for new sustainable antimicrobials able to combat the MIC.
Cu hybrids were synthesized by mixing an aqueous solution of lipase B from C. antarctica (CAL-B) with a buffer phosphate 0.1 M at pH 7 and a copper sulphate salt. After that, Cu-1 and Cu-2 bionanohybrids were produced by reducing the mixture using sodium borohydride as reducing agent (4 mg/mL and 0.5 mg/mL, respectively). Instead, Cu-3 was directly obtained from the original mixture, without using any reducing agent. Also, Cu-4 was generated adding 0.5% (v/v) of H2O2 to the final Cu-3.
Cu-Ag bionanohybrids were formed by the addition of different silver nitrate solutions to Cu-3, previously washed and liquid-removed. Cu-Ag-1 was produced by the addition of 12 mg/mL of silver nitrate prepared in buffer phosphate 0.1 M at pH 7, while Cu-Ag-2 was generated by adding 1 mg/mL of silver nitrate in distilled water to the generated Cu-3 pellet.
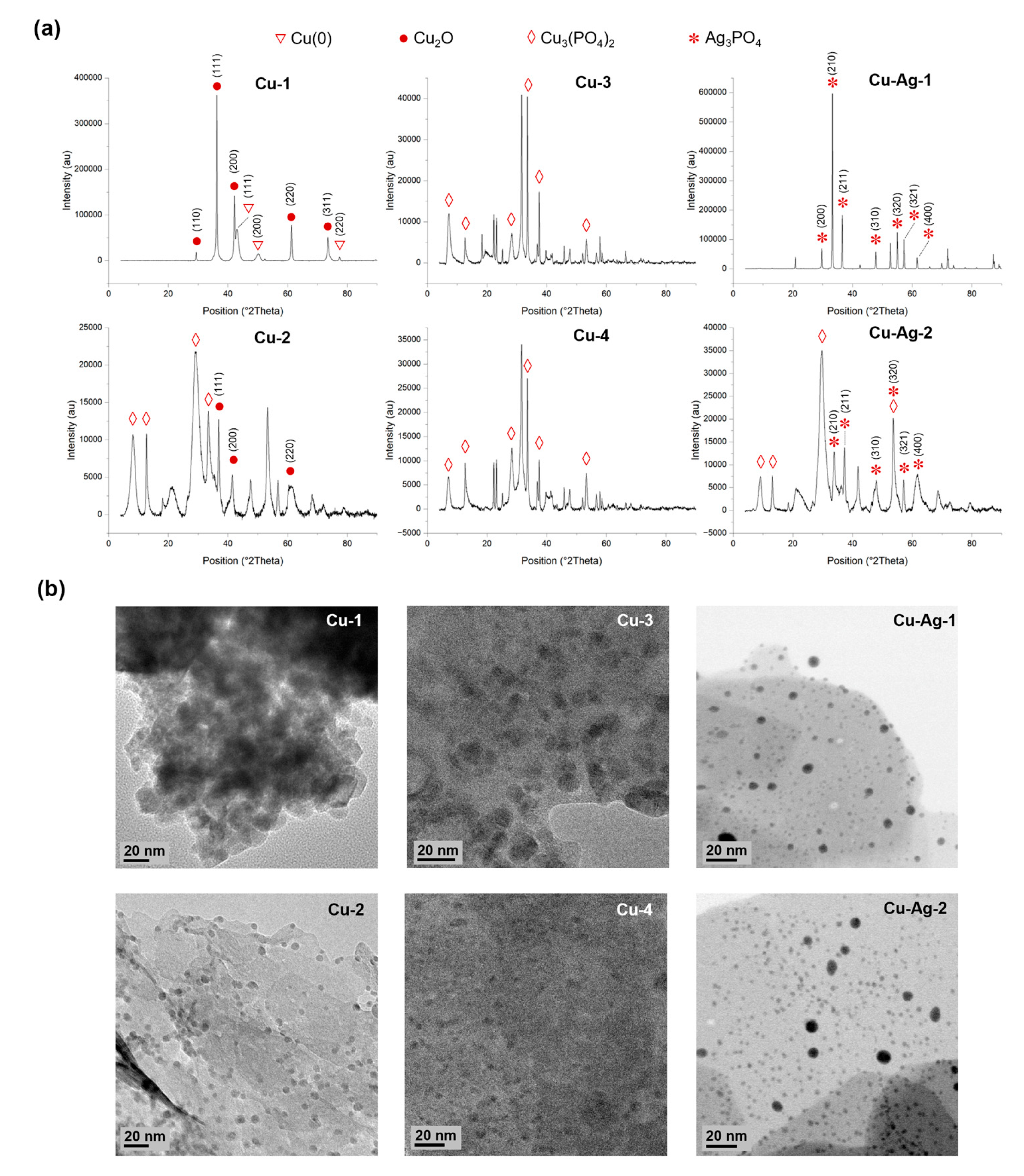
Then, XRD analyses were performed to determine the metallic species present in the different hybrids (
Figure 2a). The peaks (110), (110), (200), (220) and (311) found in
Cu-1, exhibited a high degree of correlation with Cu
2O standard data (JCPDS card 05-0667), while the peaks (111), (200) and (220) matched well with the JCPDS card 04-0836, which corresponds to Cu(0).
Cu-2, present peaks that correspond to Cu
2O standard, located in 2 theta positions 36.8, 41.4 and 60.3. Also, peaks that match well to Cu
3(PO
4)
2 standard data (JCPDS card 00-022-0548) were observed. In
Cu-3 and
Cu-4, the predominant peaks correspond to Cu
3(PO
4)
2 standard. Furthermore, a higher crystallinity was observed for
Cu-1, in comparison to
Cu-2,
Cu-3 and
Cu-4, which present a more amorphous structure due to the low peak intensities encountered (
Figure 2).
On the other hand, (200), (210), (211), (310), (320), (321) and (400) peaks that match well with Ag
3PO
4 standard (JCPDS 06-0505) were identified in Cu-Ag bionanohybrids, with a higher prevalence of this species observed in
Cu-Ag-1 than in
Cu-Ag-2. In addition, peaks corresponding to Cu
3(PO
4)
2 were observed in
Cu-Ag-2 (
Figure 2a).
TEM characterization showed the formation of small, crystalline nanoparticles on the surface of the different bionanohybrids (
Figure 2b). The larger NPs were found on
Cu-1, which showed an average particle diameter size of 11.5 nm, follow by
Cu-2, with 6.4 nm.
Cu-3 and
Cu-4 showed smaller NPs, with average diameter sizes of 2.9 and 3.0 nm, respectively. In the NPs distribution graphs of Cu-Ag hybrids, larger NPs about 15 to 20 nm were found (
Figure 2b). However, the average particle sizes were 3.2 and 3.5 nm for
Cu-Ag-1 and
Cu-Ag-2 (
Figure S4).
Therefore, hybrids with larger NPs are obtained when using a reducing agent, as in the synthesis of
Cu-1 and
Cu-2, which present the higher average particles sizes. Also, when incorporating silver in the synthesis process, larger NPs can be obtained, as is observed in
Cu-Ag-1 and
Cu-Ag-2. But in contrast to
Cu-1 and
Cu-2, Cu-Ag hybrids maintain a high proportion of smaller particles, and both have an average particle size of 3 nm, as
Cu-3 and
Cu-4 [
18,
28]. On the other hand, in TEM images of Cu-Ag-1, we can also observe bigger structures (>50 nm) (
Figure S5) which correspond to silver phosphate crystals, which corroborates the XRD spectra obtained. This was confirmed by EDX analyses (
Figure S6) of Cu-Ag-1, which also corroborates that smaller NPs (<20 nm) correspond to CuNPs.
3.2. Antibacterial Activity of Bionanohybrids
The antibacterial activity of the bionanohybrids was tested against three different MIC-relevant enrichments isolated from industrial process water environments, all from the Endures B.V. strain collection: Slime-Forming bacteria, Sulphate-Reducing bacteria and Acid-Producing bacteria.
3.2.1. Slime-Forming Bacteria
Slime-Forming bacteria (SFB) are characterized by the production of a variety of extracellular polymeric substances, or slime, under aerobic conditions. One of their main roles in MIC is to create a favorable environment for anaerobic bacteria, such as SRB, and to protect them from the external environment [
3,
29].
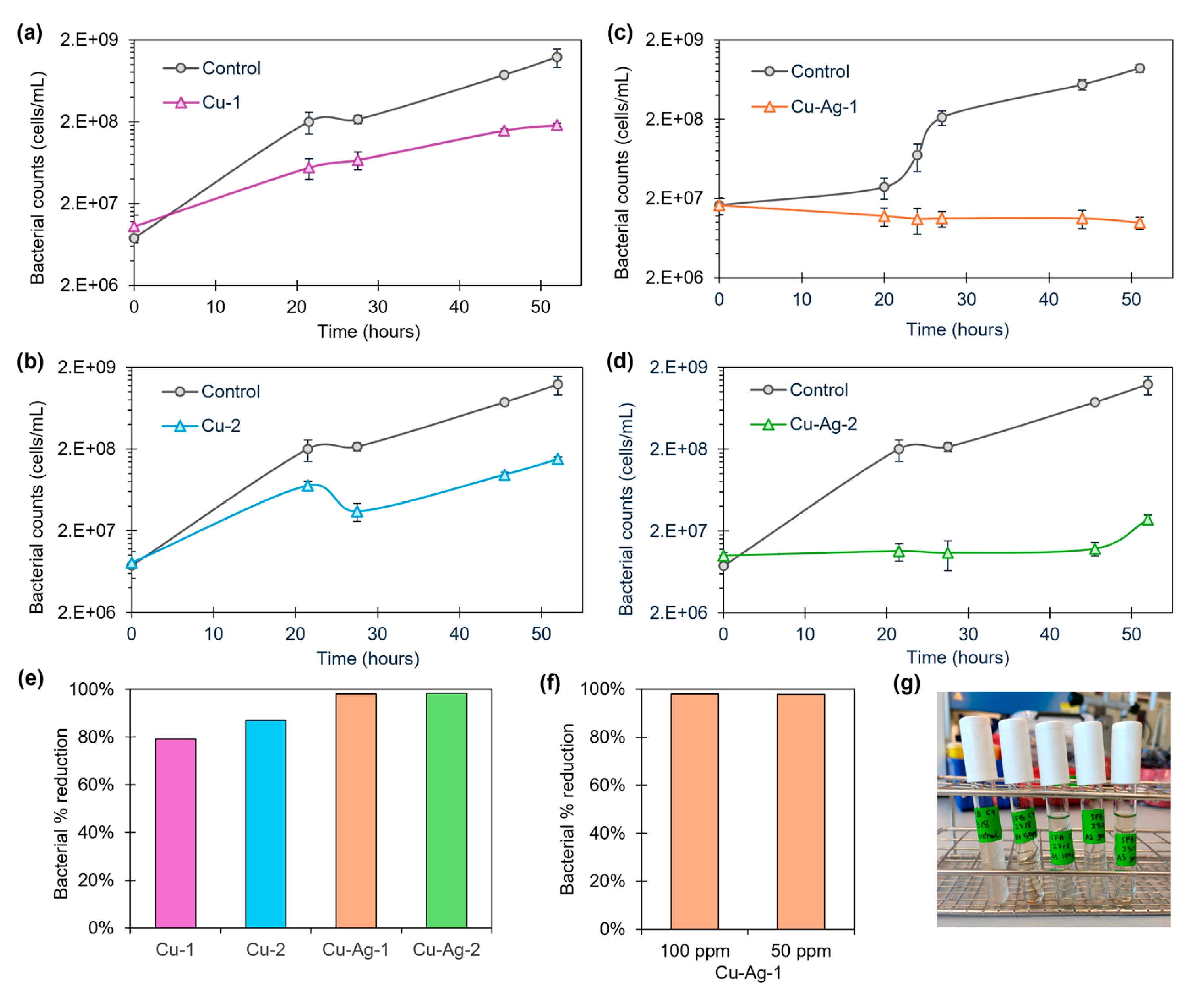
The Cu and Cu-Ag bionanohybrids were tested in a 100-ppm concentration against a starting SFB solution of 1·10
7 cells/mL. The growth of SFB was followed taking different sampling points between 0 and 52 hours. A decrease in the bacterial population was found on the SFB growth, especially after 24 hours, compared to the controls (
Figure 3). Percentages reductions of the cell count of 80%, 87%, 98% and 98% were found for
Cu-1,
Cu-2,
Cu-Ag-1 and
Cu-Ag-2, respectively, after 45 hours of exposition (
Figure 3e).
After that,
Cu-Ag-1, one of the hybrids with the best antibacterial effect in the previous test, was selected to perform an extra experiment reducing the concentration to 50 ppm. (
Figure S7). The antimicrobial capacity was found to be maintained, reaching a percentage reduction of 98% after 45 hours of contact (
Figure 3f). This showed that reducing to 50 ppm the concentration of the hybrid had no effect on the antibacterial performance.
On the other hand, the visual observation of the test tubes where the experiment was conducted indicated a notable reduction in the slime content, being visually undetectable after two days of exposure, compared to the control (
Figure 3g).
The results demonstrate that copper-silver nanohybrids, with a reduction of SFB cells of 98% in both cases after 45 hours, are more effective against SFB than copper nanohybrids. Furthermore, the differences in the SFB inhibition between Cu-Ag-1 and Cu-Ag-2 were minimal. This suggests that the incorporation of silver into the copper bionanohybrid enhances the antibacterial effect against SFB. On the other hand, Cu-Ag-1 have a considerable silver content than Cu-Ag-2 (20% higher). This indicates that elevated silver quantities in the hybrid composition do not improve its antibacterial efficacy.
3.2.2. Sulphate-Reducing Bacteria
Sulphate-Reducing bacteria (SRB) do not require oxygen for growth and activity. They consume sulphates during the respiration process and produce sulphides, which can lead to corrosion and environmental issues [
3]. As a result, they are of particular concern in the battle against MIC.
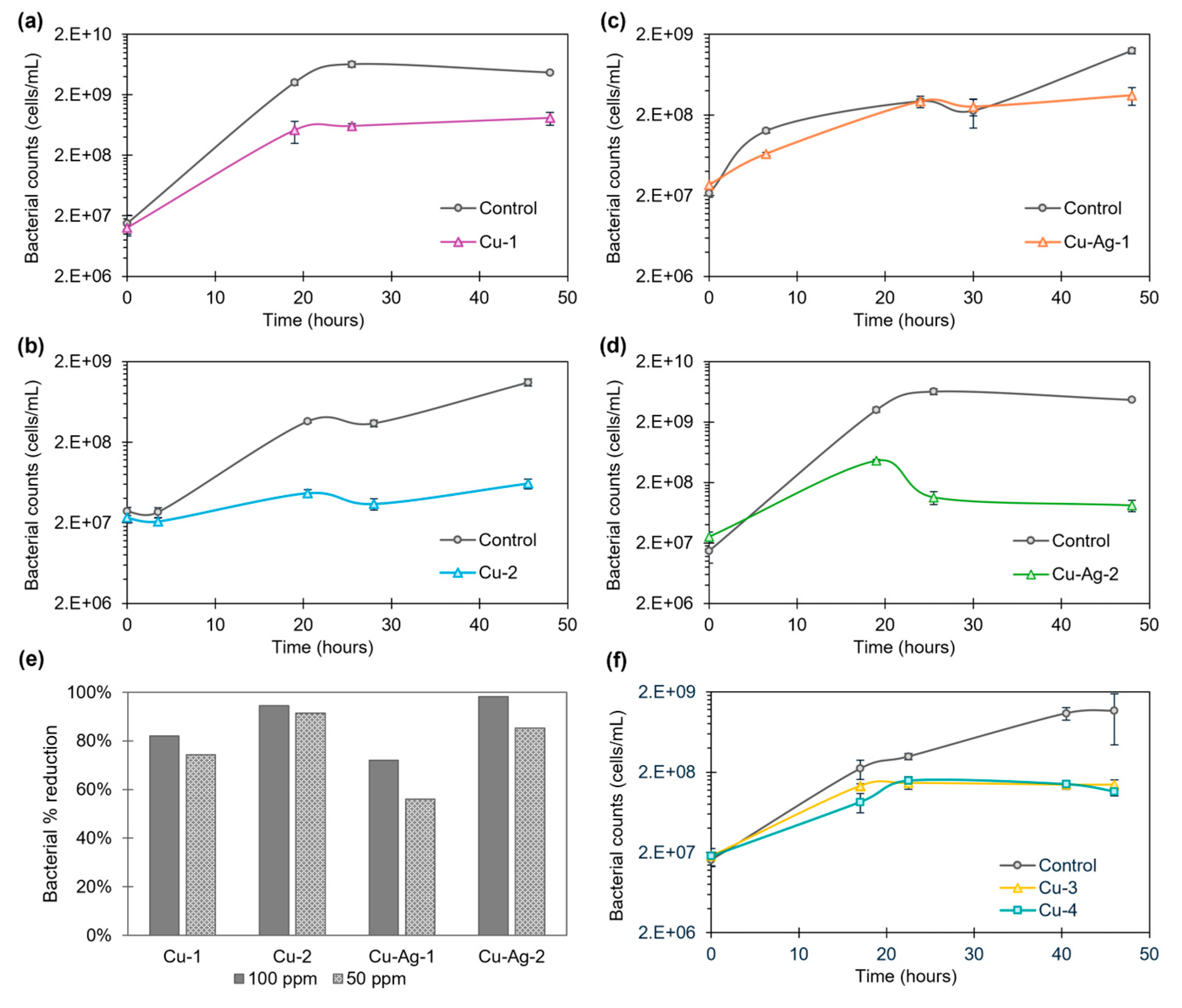
In a first step,
Cu-1,
Cu-2,
Cu-Ag-1 and
Cu-Ag-2 hybrids were used in a concentration of 100 ppm against SRB (1∙10
7 cells/mL), following the cell growing from 0 to 48 hours (
Figure 4 a-d). In all cases, an inhibition in SFB growth was demonstrated, but it was more pronounced with the hybrids
Cu-1,
Cu-2 and
Cu-Ag-2, particularly after 24 hours. Following a 48-hour period, percentages of bacterial reduction of the cell counts of 82%, 94%, 72% and 98% for
Cu-1,
Cu-2,
Cu-Ag-1 and
Cu-Ag-2 were achieved.
Then, the concentration of the hybrids was reduced to 50 ppm, and the growth curves were obtained (
Figure S8). In this instance, the growth of SRB was slightly less inhibited than at 100 ppm. Nevertheless, the values reached with the hybrids were lower than those observed in the controls, with the best inhibition values being exhibited by
Cu-2 and
Cu-Ag-2. Percentages of bacterial reduction of 74%, 92%, 56% and 85% were obtained respectively for
Cu-1,
Cu-2,
Cu-Ag-1 and
Cu-Ag-2 after 48 hours of exposure.
After that,
Cu-3 and
Cu-4 were tested in a concentration of 50 ppm against SRB, due to the good results that the copper hybrids have been demonstrated in the previous experiment. Samples were collected at different intervals between 0 and 46 hours and the SRB growth profiles shown in
Figure 4f were obtained. They showed growth inhibition compared to the control, with bacterial reduction percentages of 88% and 90% for
Cu-3 and
Cu-4 at 46 hours.
Results indicate that bionanohybrids tested showed antibacterial activity against SRB in concentration of 100 and 50 ppm. In addition, decreasing the concentration to 50 ppm generated bacterial reduction percentages of up to 92% after a 2-days exposure for the best of them, Cu-2. Among the copper-silver hybrids, Cu-Ag-2 showed the best results, with bacterial reductions of 98% and 85% after 48 hours with a concentration of 100 ppm and 50 ppm. In this way, modifications in the synthesis processes have led to differences in the structure of the bionanohybrids designed that play a key role in their efficiency.
3.2.3. Acid-Producing Bacteria
Acid-Producing bacteria (APB) generate organic acids when growing under certain conditions, including the absence of oxygen. These low pH conditions facilitate the corrosion of metal surfaces. This group of bacteria is normally associated with SRB [
2,
3].
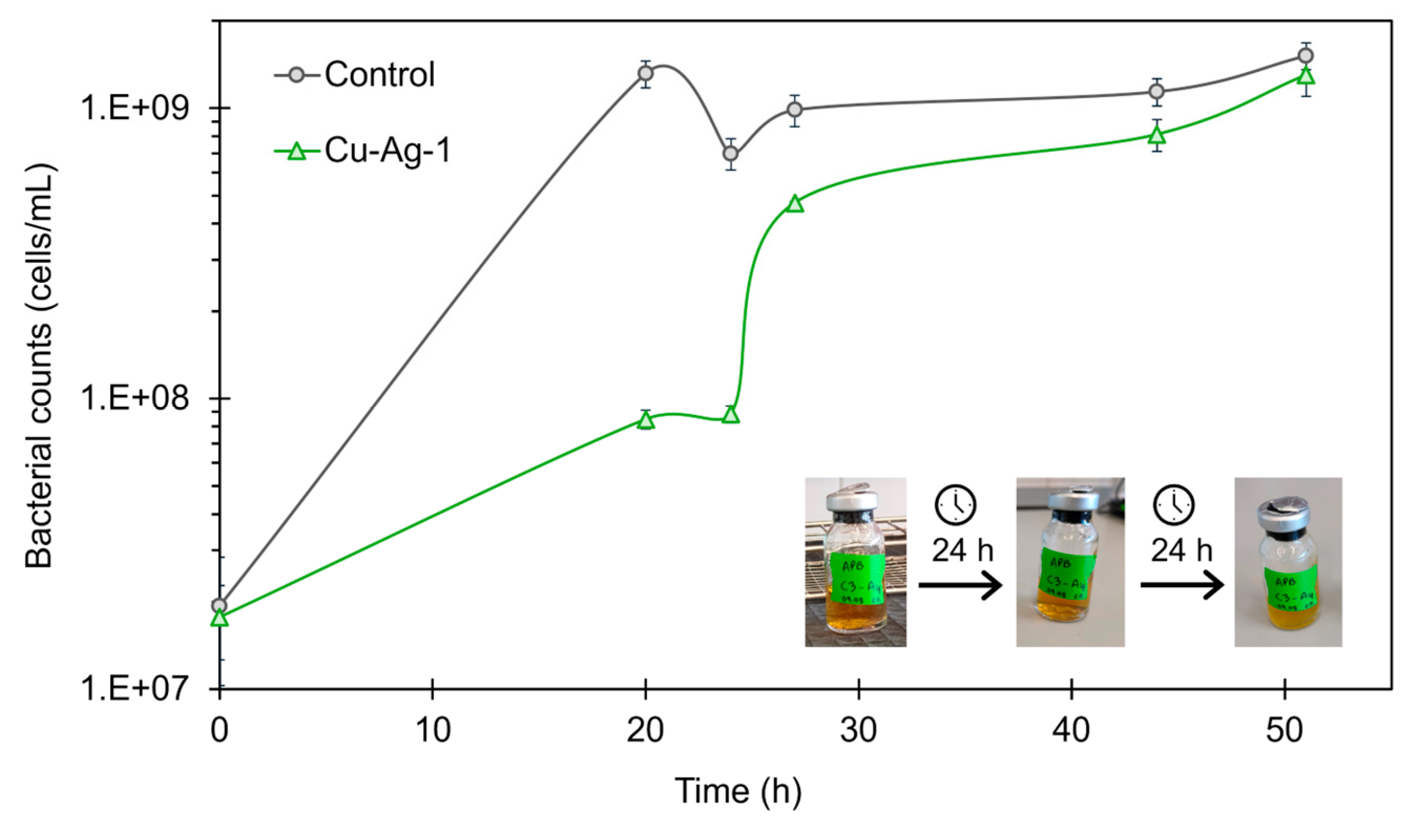
Cu-Ag-1 bionanohybrid was selected to be used against APB, in a concentration of 100 ppm. According to the growth curves obtained, as is shown in
Figure 5, the growth of APB was inhibited by
Cu-Ag-1 in the first stage of the curve, with a percentage of cells reduction of 94% at 20 hours. However, after 24 hours of exposition, the concentration of APB cells grew strongly and 48 hours later it almost reached the control concentration.
During the experiment, pH measurements were taken at 24 and 48 hours. After 24 h, the control showed a pH of 4, while the sample exposed to Cu-Ag-1 had a pH of 6. The bacterial reduction percentage obtained for the hybrid at that point was 87%. However, after 48 hours, the pH values were 4 for both solutions, and the bacterial growth curve with Cu-Ag-1 indicated that the bacterial community had returned to control levels.
Moreover, the color of the APB culture medium, which changes from red to yellow when the bacteria are active, also indicated that the bacterial activity persisted after the addition of the bionanohybrid at 48 hours, as is observed in
Figure 5. Thus, the decrease in the pH value and the color of the culture suggested that the acids and the low pH generated by APB made the bionanohybrids unstable, so their effect disappeared, and the bacteria grew back to control levels.
Therefore, the remaining hybrids were not used for testing against APB, as they are not suitable for controlling this type of bacteria, and a new design would be necessary so that they can withstand the low pH generated by APB.
3.3. Coupons Experiments
In the final step of this work, the capacity of the bionanohybrids to inhibit SRB adhesion to a metal surface and prevent the resulting corrosion was evaluated. Carbon steel coupons were used as the metal surface, and Cu-2 and Cu-Ag-2 were selected for this experiment at a concentration of 50 ppm, as they had demonstrated the best performance against SRB.
The metal coupons were submerged in a solution of SRB with an average concentration of 1∙10
7 cells/mL and the selected hybrids under anaerobic condition. Samples were taken after 0 and 24 hours. Then, coupons were removed from the media and stained for fluorescent imaging. The analysis of fluorescence images after 24 hours revealed a minor bacterial growth on the surface of the coupons which were in contact with the bionanohybrids, compared to the control (
Figure 6a). The percentage of fluorescence area of the control samples indicated a bacterial occupation of 6.3% after 24 hours. However, the fluorescent area percentage for
Cu-2 and
Cu-Ag-2 samples was 0.1% and 1.8%, respectively (
Figure 6b).
These data indicates that the presence of the bionanohybrids effectively inhibits the growth of SRB on the surface of the coupons after 24 hours. This approach has demonstrated the efficacy of Cu-2 and Cu-Ag-2 bionanohybrids in the reduction of bacteria concentration on the surface of metal coupons with the use of small concentrations of the hybrids, of only 50 ppm. The most efficient results were observed with Cu-2, where it was demonstrated that the percentage of bacterial occupancy on the coupon was reduced by 98%, and the number of bacteria in the aqueous medium by a 98%, after 24 hours.
4. Conclusions
Different synthetic methods have been used to produce Cu and Cu-Ag bionanohybrids, all based in a “green” approach, using water and room temperature, in the search for new sustainable antimicrobials.
For the preparation of copper hybrids, different concentrations of reducer agent were used, resulting in hybrids with different NPs diameters and copper metallic species. Comparing Cu-1 with Cu-2, with eight times less of reducing agent in the synthesis, Cu-2 demonstrated better performance against SRB and SFB. This suggests that the presence of Cu(0) and larger NPs in Cu-1 does not improve the antibacterial effect. However, Cu-2 showed better activity against SRB than Cu-3 and Cu-4, indicating that the presence of Cu(I) is favorable for the enhancement of the antibacterial effect in copper hybrids.
The incorporation of silver into the synthesis protocol produced hybrids with superior performance against SFB, compared to copper-only hybrids. Moreover, Cu-Ag-1 and Cu-Ag-2 presented similar efficiencies against SFB, but not against SRB, where Cu-Ag-2 demonstrated greater efficacy compared to Cu-Ag-1. This suggests that an increment in the silver content of the bimetallic hybrid does not necessarily enhance the antibacterial effect against SRB and SFB. In addition, Cu-2 exhibited higher effectiveness against SRB in comparison to Cu-Ag-2, as was also evidenced by the coupons experiment, suggesting that its composition is more adept at combating this particular bacterial strain.
It should be noted that the reduction in concentration from 100 ppm to 50 ppm did not drastically reduce the activity of the hybrids, but in some cases, it was maintained, as in the case of Cu-Ag-1 against SFB or Cu-2 against SRB.
Bionanohybrids were also shown to inhibit the adhesion of SRB on metal surfaces, as it was showed in the coupons experiment, where a maximum reduction in the bacterial adhesion to the surface was obtained with Cu-2 of 98%, after 24 hours of exposure. On the other hand, hybrids have demonstrated to not be effective against APB, due to the damaging effects that low pH levels generate on the bionanohybrid structure.
Overall, the results of this study suggest that these nanoparticles-enzyme hybrids may be a promising alternative in the fight against MIC. These results demonstrate that both copper and copper-silver hybrids could be effective against MIC, so one or the other could be chosen depending on the microbiota of the particular study situation.
Additionally, key areas for future research and comprehensive treatment against MIC include developing more robust hybrid systems that can resist the acidic pH generated by APB, creating attachment systems for using these hybrids as coatings on various surfaces, studying the specific mechanisms involved in the process and exploring longer exposure times against MIC-causing bacteria.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org, Figure S1. Growth curves of Slime-Forming bacteria starting from different concentrations; Figure S2. Growth curves of Acid-Producing bacteria starting from different concentrations; Figure S3. Growth curves of Sulphate-Reducing bacteria starting from different concentrations; Figure S4. Nanoparticles size distribution of Cu and Cu-Ag bionanohybrids; Figure S5. TEM images of Cu-Ag-1 hybrid; Figure S6. HAADF-STEM-EDX characterization of Cu-Ag-1 hybrid; Figure S7. SFB cell growth with Cu-Ag-1; Figure S8. SRB cell growth with Cu-1, Cu-2, Cu-3, Cu-4, Cu-Ag-1 and Cu-Ag-2 bionanohybrids
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript. C. O.-N. contributed to writing-original draft, methodology, investigation, formal analysis, data curation. M. S. and N. N. H. contributed to conceptualization, methodology, formal analysis, resources, supervision, writing-review & editing. J. M. P. contributed to conceptualization, methodology, project administration, resources, supervision, writing-review & editing.
Funding
This work was funded by the Spanish National Research Council (CSIC) (CSIC PTI-Global Health SGL2103036 (J.M.P) and project PIE 202480E088.
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article.
Acknowledgments
This work was supported by the Spanish National Research Council (CSIC) (CSIC PTI-Global Health SGL2103036 (J.M.P) and project PIE 202480E088).The authors would like to acknowledge the networking support from the COST Action-European MIC Network—New paths for science, sustainability and standards (Euro-MIC), CA20130, supported by COST (European Cooperation in Science and Technology). Authors thank Dr. Martinez from Novozymes.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Knisz, J.; Eckert, R.; Gieg, L.M.; Koerdt, A.; Lee, J.S.; Silva, E.R.; Skovhus, T.L.; An Stepec, B.A.; Wade, S.A. Microbiologically Influenced Corrosion—More than Just Microorganisms. FEMS Microbiol Rev 2023, 47, fuad041. [Google Scholar] [CrossRef] [PubMed]
- Eckert, R.B.; Skovhus, T.L. Failure Analysis of Microbiologically Influenced Corrosion; Eckert, R.B., Skovhus, T.L., Eds.; CRC Press, 2021; ISBN 9780367356804.
- Javaherdashti, R. Microbiologically Influenced Corrosion (MIC). In Microbiologically Influenced Corrosion: An Engineering Insight; Javaherdashti, R., Ed.; Springer International Publishing: Cham, 2017; pp. 29–79. [Google Scholar]
- Amendola, R.; Acharjee, A. Microbiologically Influenced Corrosion of Copper and Its Alloys in Anaerobic Aqueous Environments: A Review. Front Microbiol 2022, 13. [Google Scholar] [CrossRef] [PubMed]
- Blackwood, D. An Electrochemist Perspective of Microbiologically Influenced Corrosion. Corros Mater Degrad 2018, 1, 59–76. [Google Scholar] [CrossRef]
- Yuan, S.; Liang, B.; Zhao, Y.; Pehkonen, S.O. Surface Chemistry and Corrosion Behaviour of 304 Stainless Steel in Simulated Seawater Containing Inorganic Sulphide and Sulphate-Reducing Bacteria. Corros Sci 2013, 74, 353–366. [Google Scholar] [CrossRef]
- Byrnes, T. Pipeline Coatings. Trends in Oil and Gas Corrosion Research and Technologies: Production and Transmission 2017, 563–591. [CrossRef]
- Rawashdeh, R.; Haik, Y. Antibacterial Mechanisms of Metallic Nanoparticles: A Review. Dyn Biochem Process Biotechnol Mol Biol 2009, 3, 12–20. [Google Scholar]
- Pormohammad, A.; Turner, R.J. Silver Antibacterial Synergism Activities with Eight Other Metal(Loid)-Based Antimicrobials against Escherichia Coli, Pseudomonas Aeruginosa, and Staphylococcus Aureus. Antibiotics 2020, 9, 853. [Google Scholar] [CrossRef] [PubMed]
- Lemire, J.A.; Harrison, J.J.; Turner, R.J. Antimicrobial Activity of Metals: Mechanisms, Molecular Targets and Applications. Nat Rev Microbiol 2013, 11, 371–384. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-López, E.; Gomes, D.; Esteruelas, G.; Bonilla, L.; Lopez-Machado, A.L.; Galindo, R.; Cano, A.; Espina, M.; Ettcheto, M.; Camins, A.; et al. Metal-Based Nanoparticles as Antimicrobial Agents: An Overview. Nanomaterials 2020, 10, 292. [Google Scholar] [CrossRef] [PubMed]
- Gawande, M.B.; Goswami, A.; Felpin, F.-X.; Asefa, T.; Huang, X.; Silva, R.; Zou, X.; Zboril, R.; Varma, R.S. Cu and Cu-Based Nanoparticles: Synthesis and Applications in Catalysis. Chem Rev 2016, 116, 3722–3811. [Google Scholar] [CrossRef] [PubMed]
- Benavente, R.; Lopez-Tejedor, D.; Palomo, J.M. Synthesis of a Superparamagnetic Ultrathin FeCO3 Nanorods–Enzyme Bionanohybrid as a Novel Heterogeneous Catalyst. Chem. Commun. 2018, 54, 6256–6259. [Google Scholar] [CrossRef]
- Benavente, R.; Lopez-Tejedor, D.; del Puerto Morales, M.; Perez-Rizquez, C.; Palomo, J.M. The Enzyme-Induced Formation of Iron Hybrid Nanostructures with Different Morphologies. Nanoscale 2020, 12, 12917–12927. [Google Scholar] [CrossRef] [PubMed]
- Filice, M.; Marciello, M.; Morales, M. del P.; Palomo, J.M. Synthesis of Heterogeneous Enzyme–Metal Nanoparticle Biohybrids in Aqueous Media and Their Applications in C–C Bond Formation and Tandem Catalysis. Chem Comm 2013, 49, 6876. [Google Scholar] [CrossRef] [PubMed]
- Naapuri, J.M.; Losada-Garcia, N.; Deska, J.; Palomo, J.M. Synthesis of Silver and Gold Nanoparticles–Enzyme–Polymer Conjugate Hybrids as Dual-Activity Catalysts for Chemoenzymatic Cascade Reactions. Nanoscale 2022, 14, 5701–5715. [Google Scholar] [CrossRef] [PubMed]
- Palomo, J.M. Nanobiohybrids: A New Concept for Metal Nanoparticles Synthesis. Chem. Commun. 2019, 55, 9583–9589. [Google Scholar] [CrossRef] [PubMed]
- Losada-Garcia, N.; Vazquez-Calvo, A.; Ortega-Alarcon, D.; Abian, O.; Velazquez-Campoy, A.; Domingo-Calap, P.; Alcami, A.; Palomo, J.M. Nanostructured Biohybrid Material with Wide-Ranging Antiviral Action. Nano Res 2023, 16, 11455–11463. [Google Scholar] [CrossRef]
- Ortega-Nieto, C.; Losada-Garcia, N.; Pessela, B.C.; Domingo-Calap, P.; Palomo, J.M. Design and Synthesis of Copper Nanobiomaterials with Antimicrobial Properties. ACS Bio & Med Chem Au 2023, 3, 349–358. [Google Scholar] [CrossRef]
- Rodríguez-Otero, A.; Losada-García, N.; Guerra-Rodríguez, S.; Palomo, J.M.; Rodríguez-Chueca, J. Antibacterial Effect of Metal-Enzyme Hybrid Nanomaterials. J Environ Chem Eng 2023, 11, 110499. [Google Scholar] [CrossRef]
- Losada-Garcia, N.; Vazquez-Calvo, A.; Alcami, A.; Palomo, J.M. Preparation of Highly Stable and Cost-Efficient Antiviral Materials for Reducing Infections and Avoiding the Transmission of Viruses Such as SARS-CoV-2. ACS Appl Mater Interfaces 2023, 15, 22580–22589. [Google Scholar] [CrossRef] [PubMed]
- Godoy-Gallardo, M.; Eckhard, U.; Delgado, L.M.; de Roo Puente, Y.J.D.; Hoyos-Nogués, M.; Gil, F.J.; Perez, R.A. Antibacterial Approaches in Tissue Engineering Using Metal Ions and Nanoparticles: From Mechanisms to Applications. Bioact Mater 2021, 6, 4470–4490. [Google Scholar] [CrossRef] [PubMed]
- Shaikh, S.; Nazam, N.; Rizvi, S.M.D.; Ahmad, K.; Baig, M.H.; Lee, E.J.; Choi, I. Mechanistic Insights into the Antimicrobial Actions of Metallic Nanoparticles and Their Implications for Multidrug Resistance. Int J Mol Sci 2019, 20, 2468. [Google Scholar] [CrossRef] [PubMed]
- Calvano, C.D.; Picca, R.A.; Bonerba, E.; Tantillo, G.; Cioffi, N.; Palmisano, F. MALDI-TOF Mass Spectrometry Analysis of Proteins and Lipids in Escherichia Coli Exposed to Copper Ions and Nanoparticles. J Mass Spectrom 2016, 51, 828–840. [Google Scholar] [CrossRef]
- Wang, L.; Hu, C.; Shao, L. The Antimicrobial Activity of Nanoparticles: Present Situation and Prospects for the Future. Int J Nanomedicine 2017, Volume 12, 1227–1249. [Google Scholar] [CrossRef]
- Kędziora, A.; Speruda, M.; Krzyżewska, E.; Rybka, J.; Łukowiak, A.; Bugla-Płoskońska, G. Similarities and Differences between Silver Ions and Silver in Nanoforms as Antibacterial Agents. Int J Mol Sci 2018, 19, 444. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.; Bulut, O.; Some, S.; Mandal, A.K.; Yilmaz, M.D. Green Synthesis of Silver Nanoparticles: Biomolecule-Nanoparticle Organizations Targeting Antimicrobial Activity. RSC Adv 2019, 9, 2673–2702. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, C.; Losada-Garcia, N.; Palomo, J.M. Direct Synthesis of Phenols from Phenylboronic Acids in Aqueous Media Catalyzed by a Cu(0)-Nanoparticles Biohybrid. ChemistrySelect 2020, 5, 7492–7496. [Google Scholar] [CrossRef]
- El-Sherik, A.M. Trends in Oil and Gas Corrosion Research and Technologies: Production and Transmission; El-Sherik, A.M., Ed.; 2017; ISBN 978-0-08-101105-8.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).