Submitted:
05 July 2024
Posted:
08 July 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. Tissues Characteristics
| Types of breast tissues | Sample size (total 130) | Age (SD+) | Age range | < 50 years | > 50 years |
|---|---|---|---|---|---|
| Mammoplasty reduction | 28 | 36 (SD+14.5) | 19-69 | 23 | 5 |
| Fibrocystic | 31 | 49 (SD+10.9) | 29-83 | 18 | 13 |
| Fibroadenoma | 34 | 31 (SD+14.6) | 13-76 | 31 | 3 |
| ER-positive BCa | 15 | 60.8 (SD+13) | 39-82 | 4 | 11 |
| ER-negative BCa | 22 | 54.3 (SD+11.2) | 31-74 | 9 | 13 |
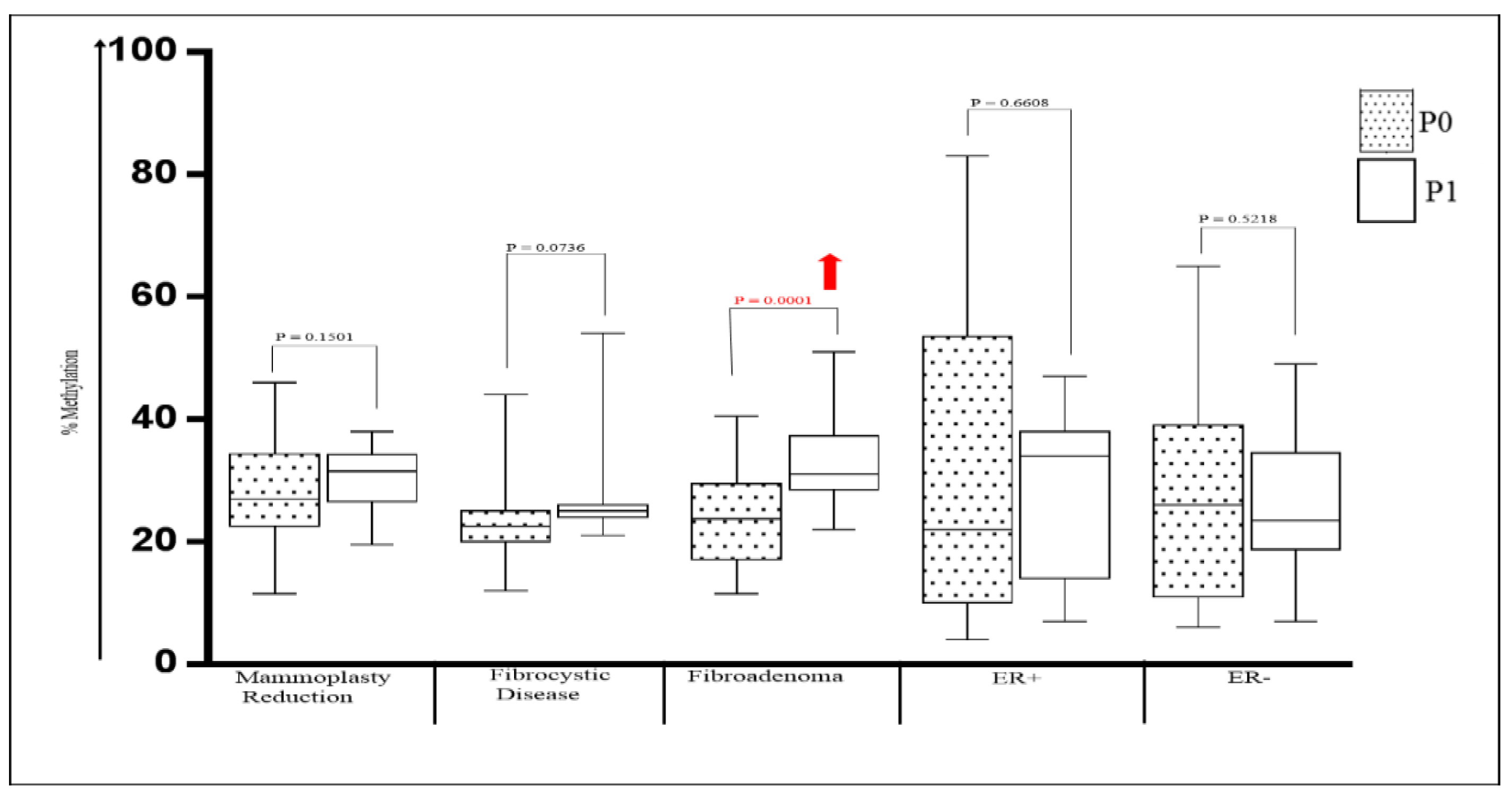
2.2. Methylation of P0 and P1 Promoters of ESRα in Human Breast Tissues
2.3. Methylation in the P0 and P1 Promoters of ESRα in Breast Cancer Cell Lines
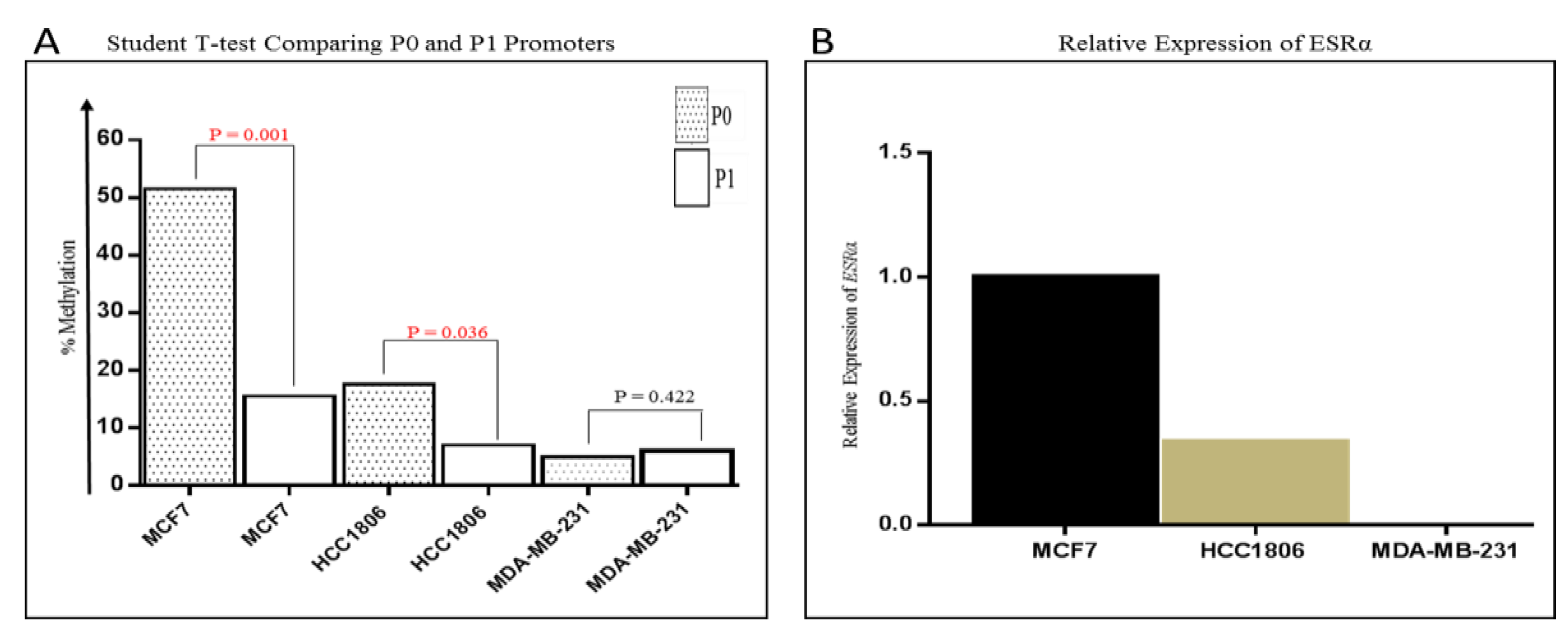
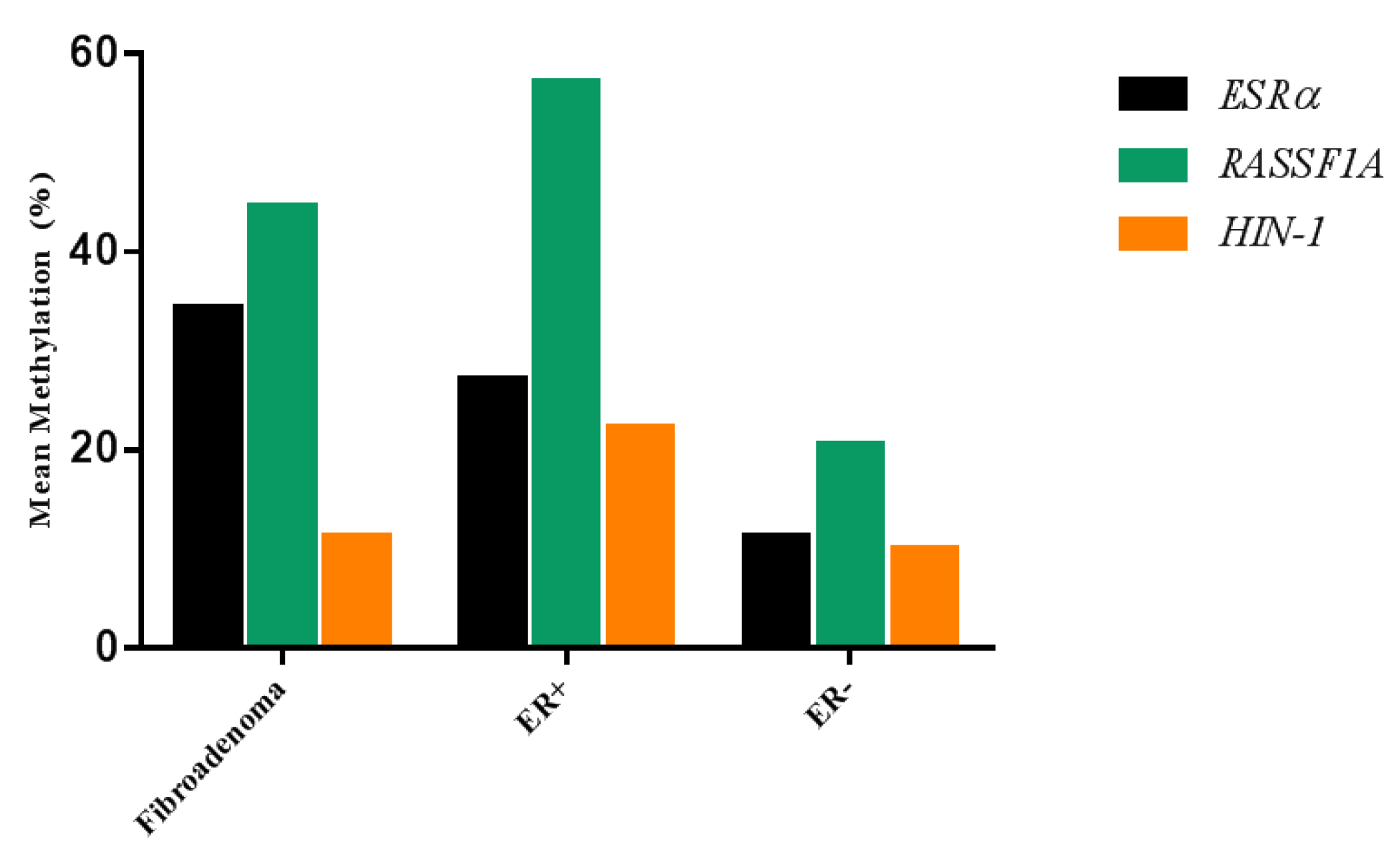
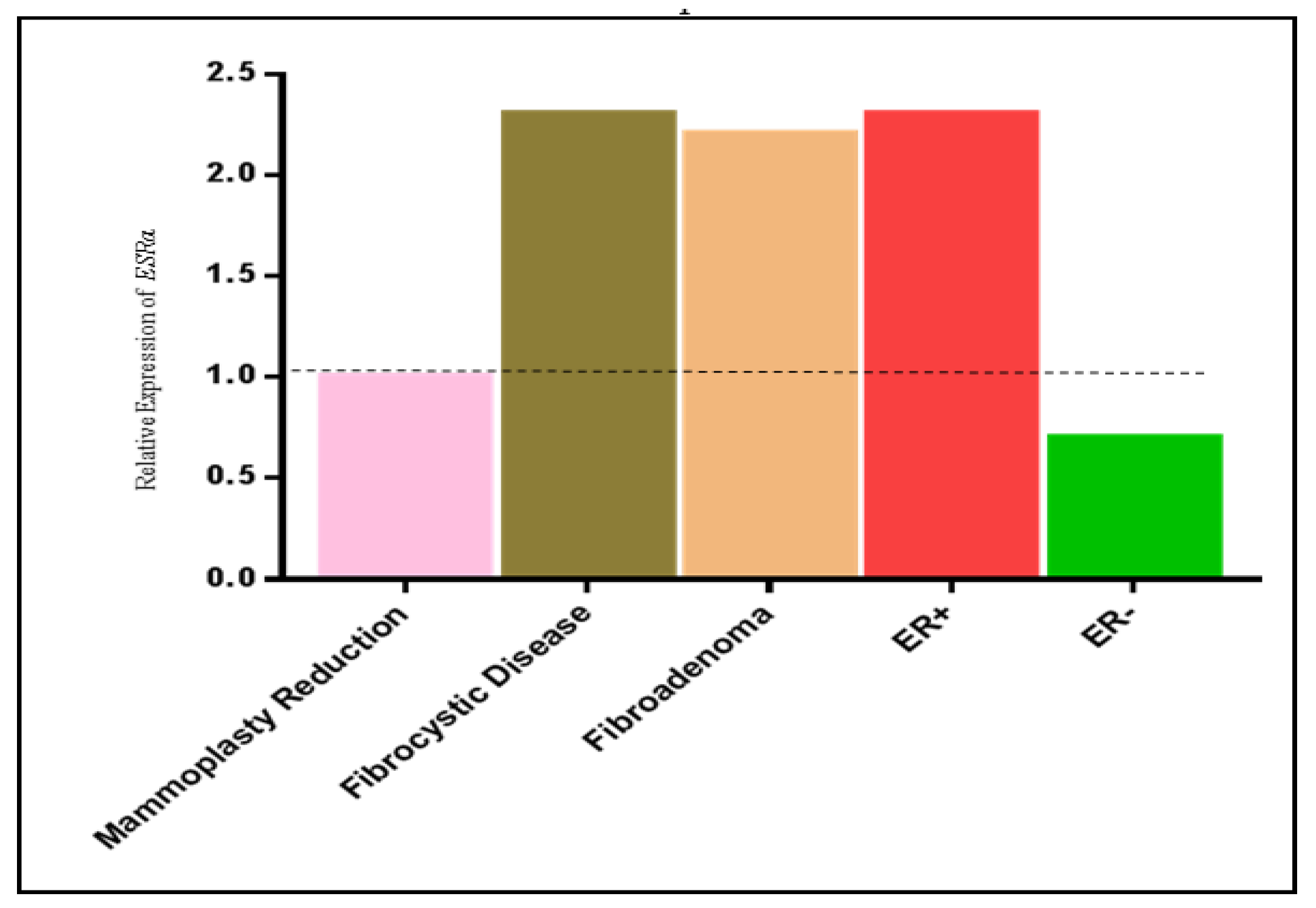
2.4. Correlation between ESRα Promoter Methylation and ESRα, RASSF1A and HIN-1 Expression
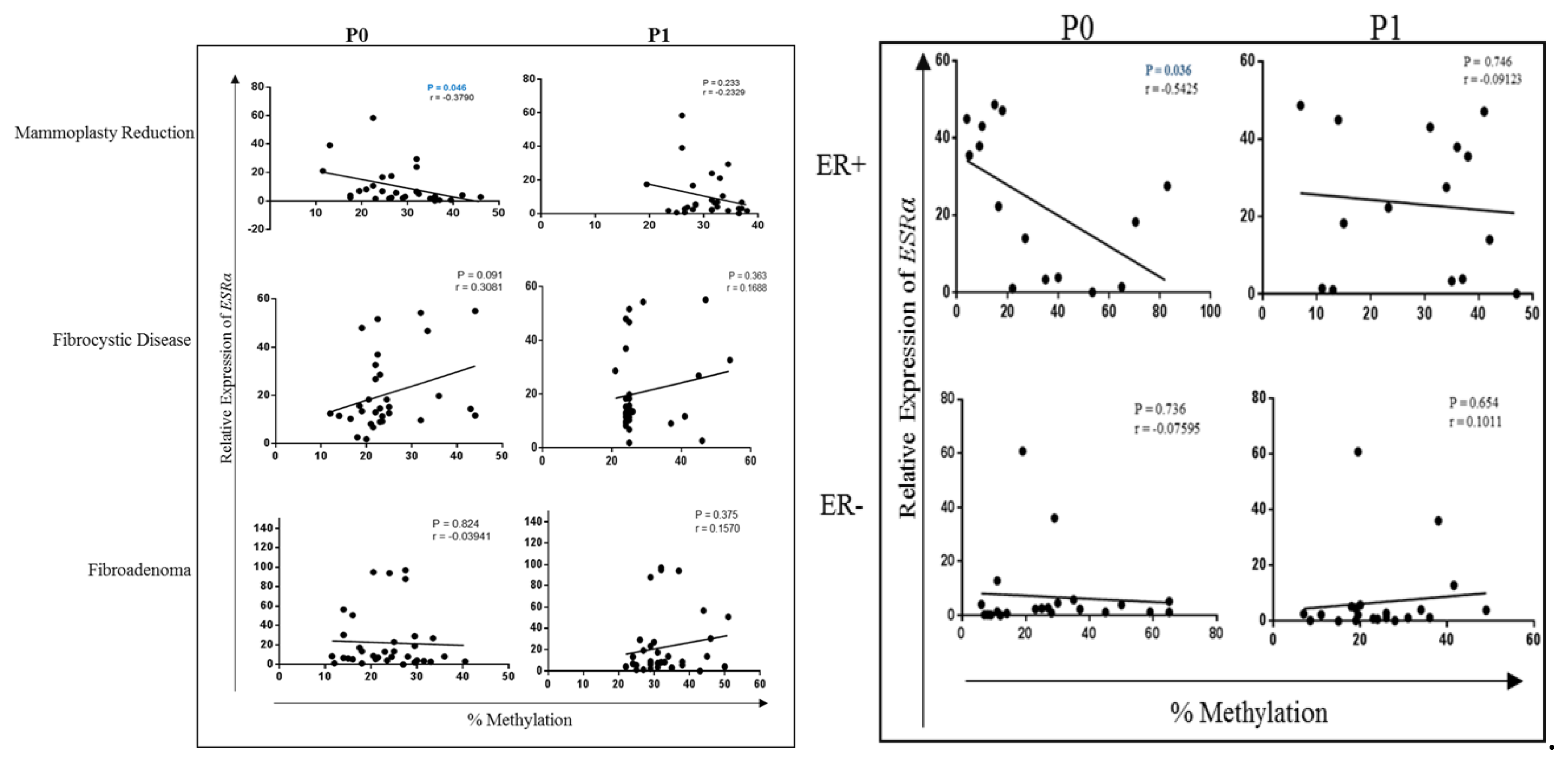
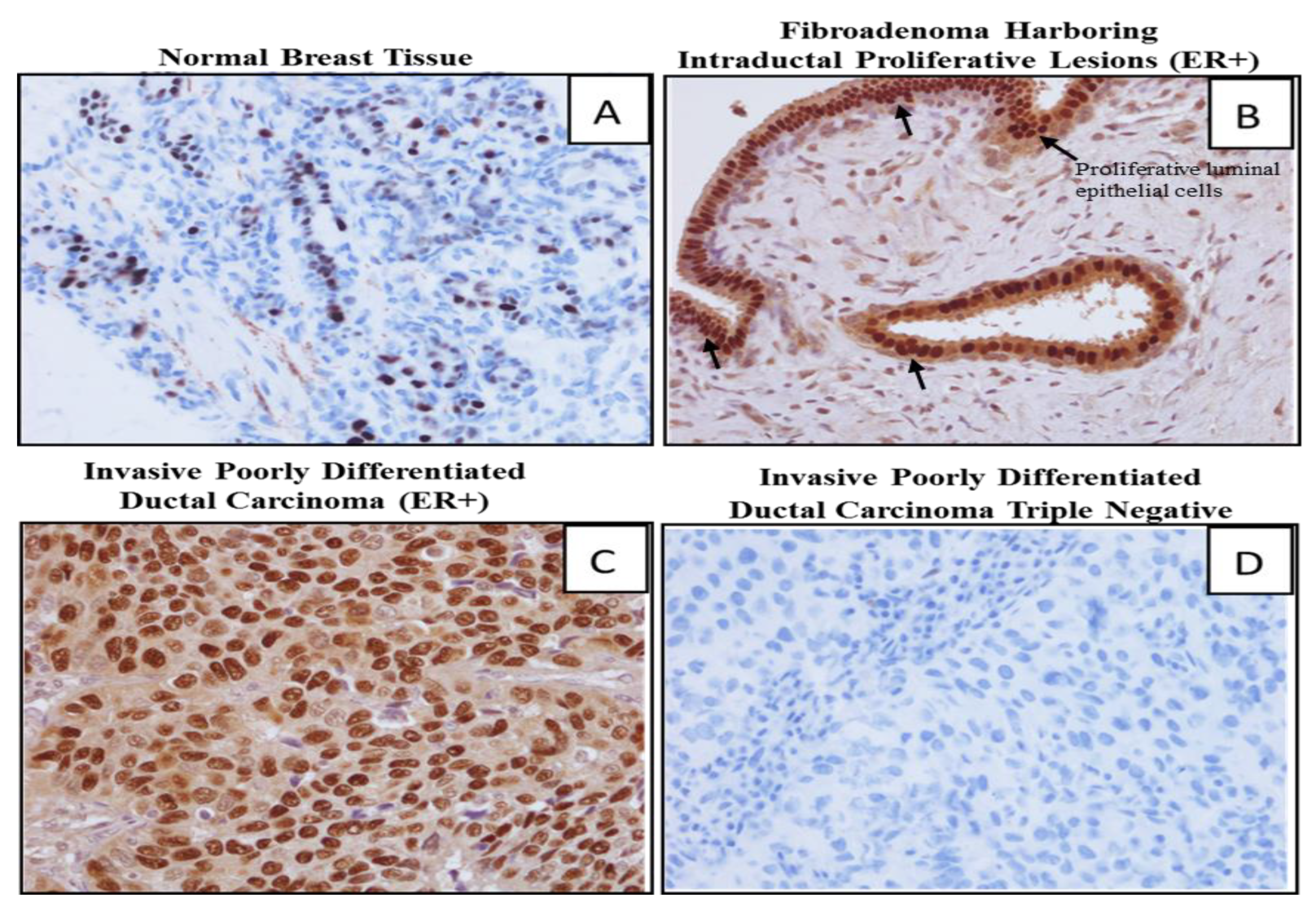
2.5. Comparing Methylation of P1 Promoters of ESRα in Fibroadenoma with ER+ve and ER-ve BCa
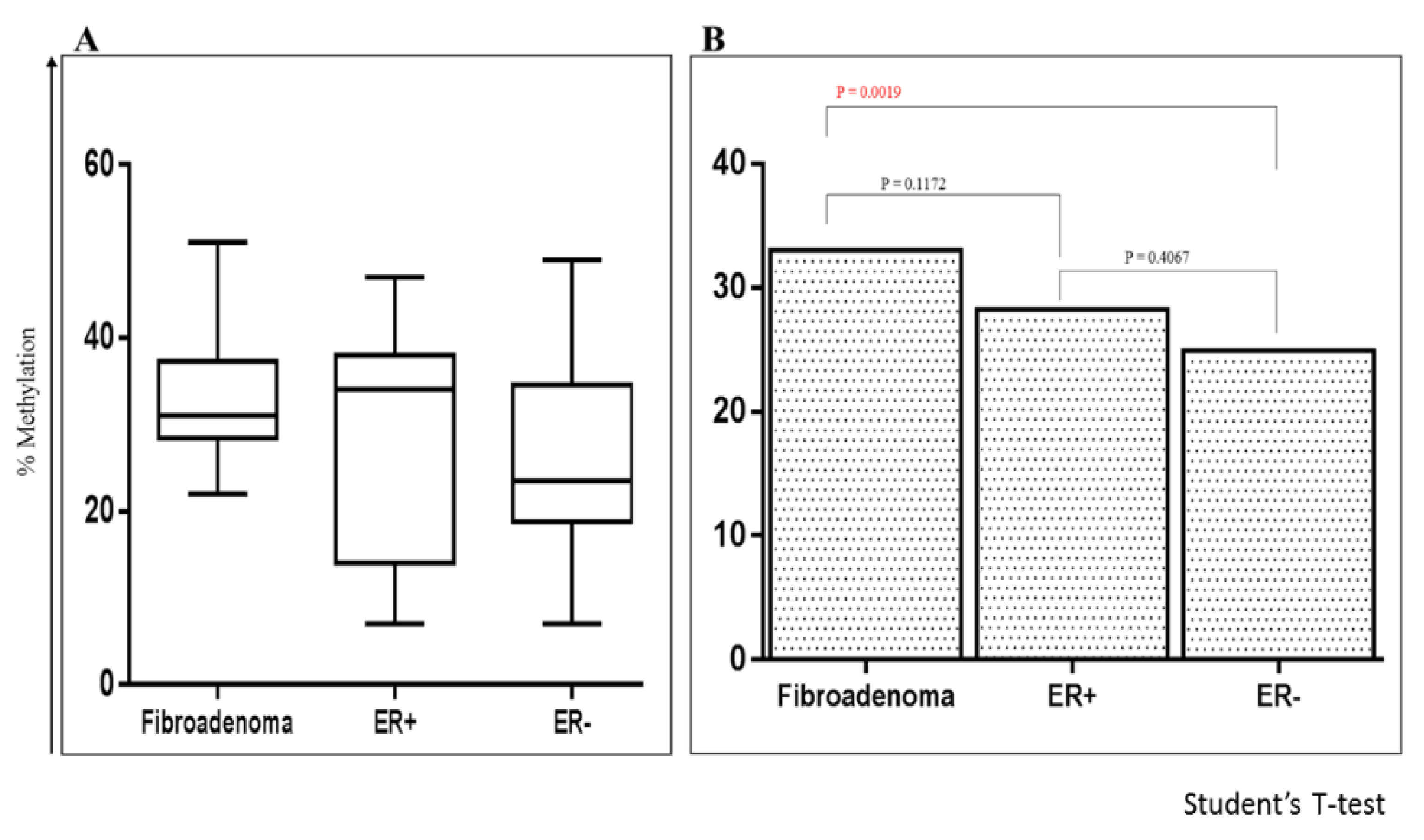
2.6. Methylation of RASSF1A and HIN1 in the Benign Fibroadenoma and BCa Tissues
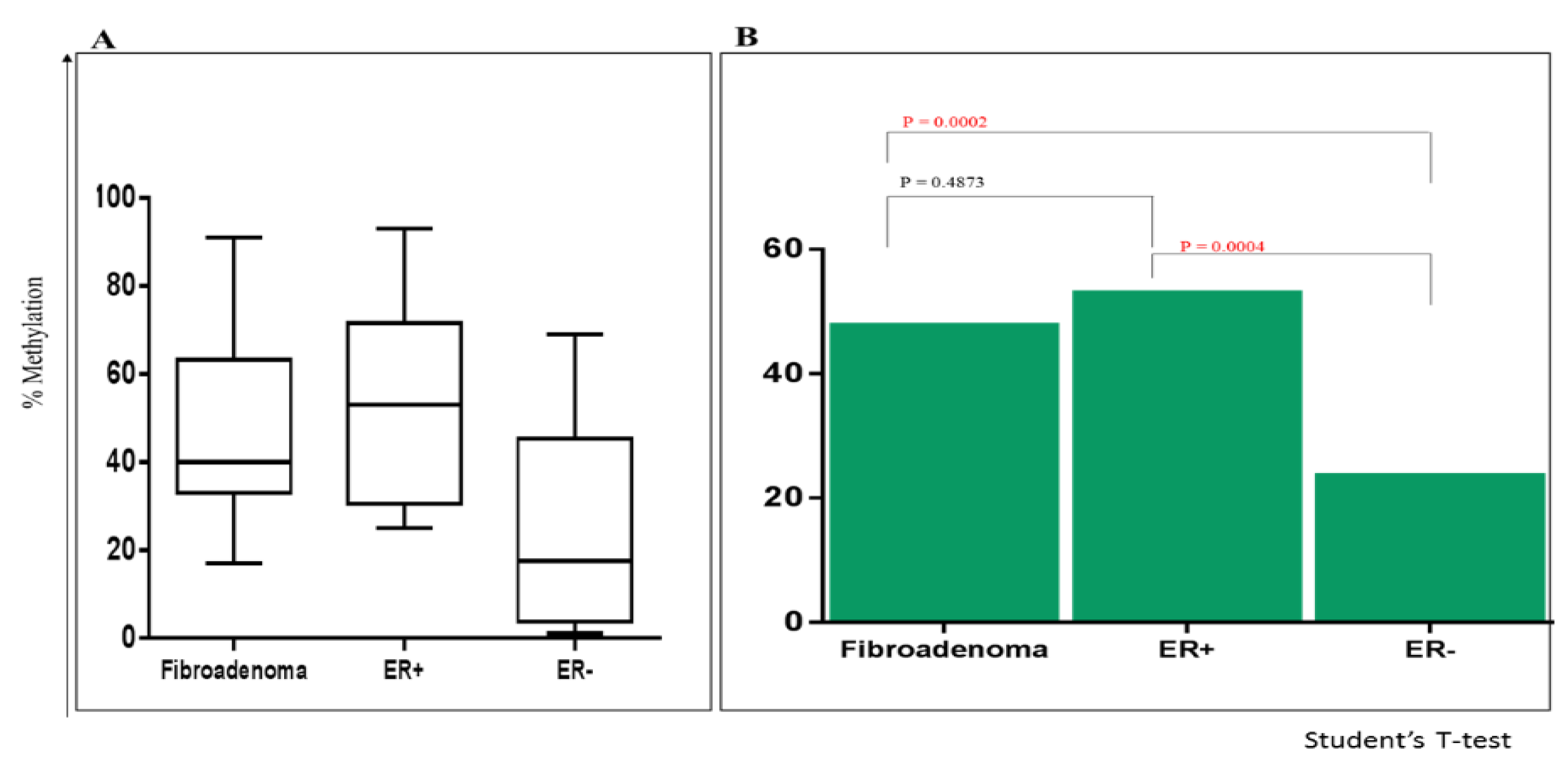
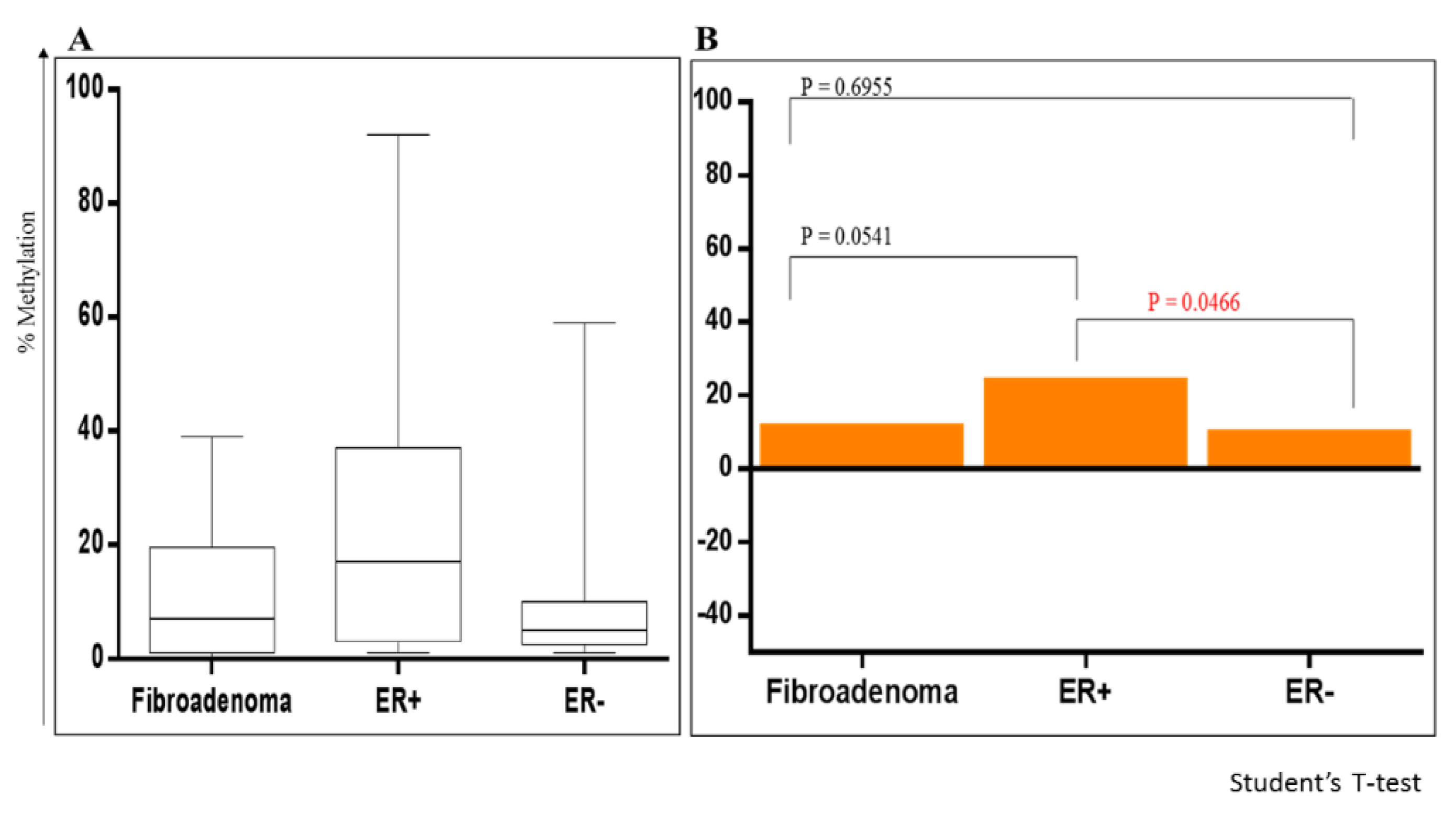
2.7. Logistic Regression Analysis of Fibroadenoma with ER+ve and ER-ve BCa
| Genes | ESRα | RASSF1A | HIN-1 | ESRα +HIN1 | RASSF1A + HIN1 | ESRα + RASSF1A | ESRα + RASSF1A + HIN1 |
|---|---|---|---|---|---|---|---|
| ESRα | 0.098 | –––– | –––– | 0.164 | –––– | 0.066 | 0.129 |
| RASSF1A | –––– | 0.114 | –––– | –––– | 0.052 | 0.078 | 0.044 |
| HIN-1 | –––– | 0.173 | 0.321 | 0.069 | –––– | 0.155 |
| ESRα | 0.009 | –––– | –––– | 0.009 | –––– | 0.024 | 0.025 |
| RASSF1A | –––– | 0.022 | –––– | –––– | 0.005 | 0.015 | 0.016 |
| HIN-1 | –––– | –––– | .091 | 0.595 | 0.949 | –––– | 0.865 |
2.8. Correlation of DNA Methylation in the P0 and P1 Promoters of ESRα with the Clinicopathological Characteristics
| Clinicopathological characteristics | ER-positive | ER-negative | ||
| N | p-value | N | p-value | |
| Histology of Tumor | 0.156 | 0.768 | ||
| Adenocarcinoma | 2 | |||
| Ductal carcinoma | 2 | |||
| Infiltrating ductal carcinoma | 11 | 19 | ||
| Medullary Adenocarcinoma | 1 | |||
| Medullary carcinoma | 1 | |||
| Metaplastic carcinoma | 1 | |||
| Age | 15 | 0.072 | 22 | 0.149 |
| Molecular Subtypes | 0.014 | 0.795 | ||
| Luminal A | 1 | |||
| Luminal B | 6 | |||
| Triple Negative | 15 | |||
| HER2 Type | 4 | |||
| ER+ve, PR-ve, HER2-ve | 5 | |||
| ER+ve, PR+ve, Unknown | 2 | |||
| ER+ve, PR-ve, Unknown | 1 | |||
| ER-ve, PR+ve, HER2-ve | 3 | |||
| Tumor Size | 0.922 | 0.018 | ||
| ≤ 2 cm | 5 | 8 | ||
| > 2 cm | 6 | 12 | ||
| Unknowns | 4 | 2 | ||
| Grade | 0.022 | 0.774 | ||
| Grade I | ||||
| Grade II | 5 | 1 | ||
| Grade III | 8 | 20 | ||
| Unknown | 1 | |||
| Stage | 0.186 | 0.77 | ||
| 0 | 1 | |||
| 1 | 2 | 6 | ||
| 2 | 4 | 5 | ||
| 3 | 4 | 7 | ||
| 4 | 1 | 1 | ||
| Unknowns | 3 | 3 | ||
| Lymph Node | 0.3 | 0.853 | ||
| Lymph Node positive | 8 | 6 | ||
| Lymph Node negative | 2 | 12 | ||
| Unknowns | 4 | 4 | ||
| Survival Status | ||||
| Dead | 3 | 12 | ||
| Alive | 11 | 10 | ||
| unknown | 1 | |||
2.9. Correlation of DNA Methylation in Promoters of RASSF1 and HIN-1 with the Clinicopathological Characteristics
| Clinicopathological characteristics | RASSF1A | HIN-1 | ||
| N | p-value | N | p-value | |
| Histology of tumor | 0.713 | 0.203 | ||
| Infiltrating ductal carcinoma | 29 | 29 | ||
| Others | 4 | 6 | ||
| Age | 0.377 | 0.403 | ||
| ER-status | 0.0003 | 0.032 | ||
| ER-positive | 12 | 14 | ||
| ER-negative | 22 | 21 | ||
| Tumor size | 0.499 | 0.89 | ||
| < 2 cm | 12 | 15 | ||
| > 2 cm | 17 | 18 | ||
| Grade | 0.599 | 0.157 | ||
| Grade II | 4 | 5 | ||
| Grade III | 29 | 29 | ||
| Stage | 0.995 | 0.287 | ||
| 1 | 7 | 8 | ||
| 2 | 8 | 8 | ||
| 3 | 11 | 10 | ||
| 4 | 4 | 2 | ||
| Lymph node | 0.307 | 0.927 | ||
| Positive lymph node | 13 | 13 | ||
| Negative lymph node | 16 | 16 | ||
3. Discussion
Comparing Methylation in the P0 and P1 Promoters of ESRα in Breast Tissues
The Influence of Menstrual Cycle on ESRα Transcription
Significance of P1 Promoter Methylation in Fibroadenoma and BCa
Logistic Regression Analysis
4. Materials and Methods
4.1. Human Breast Tissues
4.2. Immunohistochemistry (IHC) and Hematoxylin and Eosin (H&E) Staining
4.3. Breast Cancer Cell Lines
4.4. DNA and RNA Isolation
4.5. Bisulfite Modification
4.6. Designing Polymerase Chain Reaction (PCR) and Pyrosequencing Primers
| PyroMark PCR and sequencing primers for ESRα P0 and P1 promoters | |
| P0 Promoter primers | Forward: 5' GGGAAGTAGTTAGTAGGTAGGGTATTTG 3' Reverse: 5' Biotin-TCACTCCCCACTACCATTCAT 3' Sequencing: 5' AGGGTATTTGGTAGTTTTT 3' Sequence analyzed: 5’TTYGGTAGATAYGTAGTTGGGTTATTGTAT AGYGTTGGATGAATGGTAGTGGGGAGTG 3’ |
| P1 Promoter primers | Qiagen Inc. Valencia, California Catalog Number: PM00024619 |
| PyroMark PCR and sequencing primers for RASSF1 | |
| RASSFIA | Forward: 5' GGGGGAGTTTGAGTTTATTGA 3' Reverse: 5' Biotin- CTACCCCTTAACTACCCCTTCC 3' Sequencing: 5' GGGTAGTATTAGGTTGGAG 3' |
| PyroMark PCR and sequencing primers for HIN1 | |
4.7. Quantitative Analysis of DNA Methylation by Pyrosequencing
4.8. Gene Expression Analysis
4.9. Statistical Analyses
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, Z., R. Zhang, and D. Li, Molecular Biology Mechanisms and Emerging Therapeutics of Triple-Negative Breast Cancer. Biologics, 2023. 17: p. 113-128. [CrossRef]
- Chen, P., B. Li, and L. Ou-Yang, Role of estrogen receptors in health and disease. Front Endocrinol (Lausanne), 2022. 13: p. 839005. [CrossRef]
- Jia, M., K. Dahlman-Wright, and J.A. Gustafsson, Estrogen receptor alpha and beta in health and disease. Best Pract Res Clin Endocrinol Metab, 2015. 29(4): p. 557-68. [CrossRef]
- Gao, L., et al., Estrogen receptor beta promoter methylation: a potential indicator of malignant changes in breast cancer. Arch Med Sci, 2016. 12(1): p. 129-36. [CrossRef]
- Paruthiyil, S., et al., Estrogen receptor beta causes a G2 cell cycle arrest by inhibiting CDK1 activity through the regulation of cyclin B1, GADD45A, and BTG2. Breast Cancer Res Treat, 2011. 129(3): p. 777-84. [CrossRef]
- Stingl, J., Estrogen and progesterone in normal mammary gland development and in cancer. Horm Cancer, 2011. 2(2): p. 85-90. [CrossRef]
- Saji, S., et al., Estrogen receptors alpha and beta in the rodent mammary gland. Proc Natl Acad Sci U S A, 2000. 97(1): p. 337-42. [CrossRef]
- Kerdivel, G., G. Flouriot, and F. Pakdel, Modulation of estrogen receptor alpha activity and expression during breast cancer progression. Vitam Horm, 2013. 93: p. 135-60. [CrossRef]
- Allred, D.C., S.K. Mohsin, and S.A. Fuqua, Histological and biological evolution of human premalignant breast disease. Endocr Relat Cancer, 2001. 8(1): p. 47-61. [CrossRef]
- Simpson, J.F., Update on atypical epithelial hyperplasia and ductal carcinoma in situ. Pathology, 2009. 41(1): p. 36-9. [CrossRef]
- Kurebayashi, J., et al., Expression levels of estrogen receptor-alpha, estrogen receptor-beta, coactivators, and corepressors in breast cancer. Clin Cancer Res, 2000. 6(2): p. 512-8.
- Brinkman, J.A. and D. El-Ashry, ER re-expression and re-sensitization to endocrine therapies in ER-negative breast cancers. J Mammary Gland Biol Neoplasia, 2009. 14(1): p. 67-78. [CrossRef]
- Lachner, M., R.J. O'Sullivan, and T. Jenuwein, An epigenetic road map for histone lysine methylation. J Cell Sci, 2003. 116(Pt 11): p. 2117-24. [CrossRef]
- Macaluso, M., et al., Epigenetic modulation of estrogen receptor-alpha by pRb family proteins: a novel mechanism in breast cancer. Cancer Res, 2007. 67(16): p. 7731-7. [CrossRef]
- Zhu, K., et al., Methyl-group dietary intake and risk of breast cancer among African-American women: a case-control study by methylation status of the estrogen receptor alpha genes. Cancer Causes Control, 2003. 14(9): p. 827-36. [CrossRef]
- Stirzaker, C., E. Zotenko, and S.J. Clark, Genome-wide DNA methylation profiling in triple-negative breast cancer reveals epigenetic signatures with important clinical value. Mol Cell Oncol, 2016. 3(1): p. e1038424. [CrossRef]
- Jovanovic, J., et al., The epigenetics of breast cancer. Mol Oncol, 2010. 4(3): p. 242-54. [CrossRef]
- Fatemi, M., et al., Footprinting of mammalian promoters: use of a CpG DNA methyltransferase revealing nucleosome positions at a single molecule level. Nucleic Acids Res, 2005. 33(20): p. e176. [CrossRef]
- Illingworth, R.S. and A.P. Bird, CpG islands--'a rough guide'. FEBS Lett, 2009. 583(11): p. 1713-20. [CrossRef]
- Takai, D. and P.A. Jones, Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc Natl Acad Sci U S A, 2002. 99(6): p. 3740-5. [CrossRef]
- Lapidus, R.G., et al., Mapping of ER gene CpG island methylation-specific polymerase chain reaction. Cancer Res, 1998. 58(12): p. 2515-9.
- Lapidus, R.G., et al., Methylation of estrogen and progesterone receptor gene 5' CpG islands correlates with lack of estrogen and progesterone receptor gene expression in breast tumors. Clin Cancer Res, 1996. 2(5): p. 805-10.
- Weigel, R.J., et al., Quantitative analysis of the transcriptional start sites of estrogen receptor in breast carcinoma. Cell Growth Differ, 1995. 6(6): p. 707-11.
- Grandien, K., et al., Estrogen target tissue determines alternative promoter utilization of the human estrogen receptor gene in osteoblasts and tumor cell lines. Endocrinology, 1995. 136(5): p. 2223-9. [CrossRef]
- Herman, J.G. and S.B. Baylin, Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med, 2003. 349(21): p. 2042-54. [CrossRef]
- O'Doherty, A.M., et al., Methylation status of oestrogen receptor-alpha gene promoter sequences in human ovarian epithelial cell lines. Br J Cancer, 2002. 86(2): p. 282-4. [CrossRef]
- Ottaviano, Y.L., et al., Methylation of the estrogen receptor gene CpG island marks loss of estrogen receptor expression in human breast cancer cells. Cancer Res, 1994. 54(10): p. 2552-5.
- Falette, N.S., et al., Estrogen receptor gene methylation in human breast tumors. Cancer Res, 1990. 50(13): p. 3974-8.
- Holst, C.R., et al., Methylation of p16(INK4a) promoters occurs in vivo in histologically normal human mammary epithelia. Cancer Res, 2003. 63(7): p. 1596-601.
- Laird, P.W., The power and the promise of DNA methylation markers. Nat Rev Cancer, 2003. 3(4): p. 253-66. [CrossRef]
- Wittenberger, T., et al., DNA methylation markers for early detection of women's cancer: promise and challenges. Epigenomics, 2014. 6(3): p. 311-27. [CrossRef]
- Sharma, G., et al., Clinical significance of promoter hypermethylation of DNA repair genes in tumor and serum DNA in invasive ductal breast carcinoma patients. Life Sci, 2010. 87(3-4): p. 83-91. [CrossRef]
- Jing, M.X., et al., Estrogen receptor-alpha promoter methylation in sporadic basal-like breast cancer of Chinese women. Tumour Biol, 2011. 32(4): p. 713-9. [CrossRef]
- Lee, J.S., et al., A comparative study of Korean with Caucasian breast cancer reveals frequency of methylation in multiple genes correlates with breast cancer in young, ER, PR-negative breast cancer in Korean women. Cancer Biol Ther, 2007. 6(7): p. 1114-20. [CrossRef]
- Mehrotra, J., et al., Estrogen receptor/progesterone receptor-negative breast cancers of young African-American women have a higher frequency of methylation of multiple genes than those of Caucasian women. Clin Cancer Res, 2004. 10(6): p. 2052-7. [CrossRef]
- Bulut, N., et al., Demographic and clinico-pathological characteristics in patients with triple-negative and non-triple-negative breast cancer. Med Oncol, 2011. 28 Suppl 1: p. S75-9. [CrossRef]
- Lee, J.S., et al., Basal-like breast cancer displays distinct patterns of promoter methylation. Cancer Biol Ther, 2010. 9(12): p. 1017-24. [CrossRef]
- Porras, L., H. Ismail, and S. Mader, Positive Regulation of Estrogen Receptor Alpha in Breast Tumorigenesis. Cells, 2021. 10(11). [CrossRef]
- Raos, D., et al., Epigenetically inactivated RASSF1A as a tumor biomarker. Bosn J Basic Med Sci, 2021. 21(4): p. 386-397. [CrossRef]
- Liu, L., et al., Frequent hypermethylation of the RASSF1A gene in prostate cancer. Oncogene, 2002. 21(44): p. 6835-40. [CrossRef]
- Yoon, J.H., R. Dammann, and G.P. Pfeifer, Hypermethylation of the CpG island of the RASSF1A gene in ovarian and renal cell carcinomas. Int J Cancer, 2001. 94(2): p. 212-7. [CrossRef]
- Trivers, K.F., et al., Oral contraceptives and survival in breast cancer patients aged 20 to 54 years. Cancer Epidemiol Biomarkers Prev, 2007. 16(9): p. 1822-7. [CrossRef]
- Moorman, P.G., R.C. Millikan, and B. Newman, Oral contraceptives and breast cancer among African-american women and white women. J Natl Med Assoc, 2001. 93(9): p. 329-34.
- Tisserand, P., et al., Lack of HIN-1 methylation defines specific breast tumor subtypes including medullary carcinoma of the breast and BRCA1-linked tumors. Cancer Biol Ther, 2003. 2(5): p. 559-63. [CrossRef]
- Pang, J.M., A. Dobrovic, and S.B. Fox, DNA methylation in ductal carcinoma in situ of the breast. Breast Cancer Res, 2013. 15(3): p. 206. [CrossRef]
- Krop, I., et al., HIN-1, an inhibitor of cell growth, invasion, and AKT activation. Cancer Res, 2005. 65(21): p. 9659-69. [CrossRef]
- Gupta, V., P. Agarwal, and P. Deshpande, Impact of RASSF1A gene methylation on clinico-pathological features of tumor and non-tumor tissue of breast cancer. Ann Diagn Pathol, 2021. 52: p. 151722. [CrossRef]
- Fackler, M.J., et al., DNA methylation of RASSF1A, HIN-1, RAR-beta, Cyclin D2 and Twist in in situ and invasive lobular breast carcinoma. Int J Cancer, 2003. 107(6): p. 970-5.
- Ajmal, M., M. Khan, and K. Van Fossen, Breast Fibroadenoma, in StatPearls. 2024: Treasure Island (FL).
- Xu, L., et al., Breast carcinoma arising in a fibroadenoma: A case series of 16 patients and review of the literature. Oncol Lett, 2024. 27(1): p. 39. [CrossRef]
- Kirn, V., et al., ESR1-promoter-methylation status in primary breast cancer and its corresponding metastases. Clin Exp Metastasis, 2018. 35(7): p. 707-712. [CrossRef]
- Tanimoto, K., et al., Regulation of estrogen receptor alpha gene mediated by promoter B responsible for its enhanced expressionin human breast cancer. Nucleic Acids Res, 1999. 27(3): p. 903-9.. [CrossRef]
- Treilleux, et al., Human estrogen receptor (ER) gene promoter-P1: estradiol-independent activity and estradiol inducibility in ER+ and ER- cells. Mol Endocrinol, 1997. 11(9): p. 1319-31. [CrossRef]
- Yoshida, T., et al., Distinct mechanisms of loss of estrogen receptor alpha gene expression in human breast cancer: methylation of the gene and alteration of trans-acting factors. Carcinogenesis, 2000. 21(12): p. 2193-201. [CrossRef]
- Jones, P.A. and G. Liang, Rethinking how DNA methylation patterns are maintained. Nat Rev Genet, 2009. 10(11): p. 805-11. [CrossRef]
- Kondo, Y., et al., Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nat Genet, 2008. 40(6): p. 741-50. [CrossRef]
- Koster, F., et al., Triple-negative breast cancers express receptors for growth hormone-releasing hormone (GHRH) and respond to GHRH antagonists with growth inhibition. Breast Cancer Res Treat, 2009. 116(2): p. 273-9. [CrossRef]
- Oh, A.S., et al., Hyperactivation of MAPK induces loss of ERalpha expression in breast cancer cells. Mol Endocrinol, 2001. 15(8): p. 1344-59. [CrossRef]
- Ferguson, A.T., et al., Demethylation of the estrogen receptor gene in estrogen receptor-negative breast cancer cells can reactivate estrogen receptor gene expression. Cancer Res, 1995. 55(11): p. 2279-83.
- deConinck, E.C., L.A. McPherson, and R.J. Weigel, Transcriptional regulation of estrogen receptor in breast carcinomas. Mol Cell Biol, 1995. 15(4): p. 2191-6. [CrossRef]
- Kajabova, V., et al., RASSF1A Promoter Methylation Levels Positively Correlate with Estrogen Receptor Expression in Breast Cancer Patients. Transl Oncol, 2013. 6(3): p. 297-304. [CrossRef]
- Buhmeida, A., et al., RASSF1A methylation is predictive of poor prognosis in female breast cancer in a background of overall low methylation frequency. Anticancer Res, 2011. 31(9): p. 2975-81.
- Gaudet, M.M., et al., DNA hypermethylation of ESR1 and PGR in breast cancer: pathologic and epidemiologic associations. Cancer Epidemiol Biomarkers Prev, 2009. 18(11): p. 3036-43. [CrossRef]
- Marquardt, R.M., et al., Progesterone and Estrogen Signaling in the Endometrium: What Goes Wrong in Endometriosis? Int J Mol Sci, 2019. 20(15).. [CrossRef]
- Hori, M., et al., Assessment of hypermethylated DNA in two promoter regions of the estrogen receptor alpha gene in human endometrial diseases. Gynecol Oncol, 2000. 76(1): p. 89-96. [CrossRef]
- Atalay, C., M. Kanlioz, and M. Altinok, Menstrual cycle and hormone receptor status in breast cancer patients. Neoplasma, 2002. 49(4): p. 278.
- Hayashi, S., et al., Two promoters in expression of estrogen receptor messenger RNA in human breast cancer. Carcinogenesis, 1997. 18(3): p. 459-64. [CrossRef]
- Quintas-Granados, L.I., et al., The high methylation level of a novel 151-bp CpG island in the ESR1 gene promoter is associated with a poor breast cancer prognosis. Cancer Cell Int, 2021. 21(1): p. 649. [CrossRef]
- Chintamani, et al., Carcinoma developing in a fibroadenoma in a woman with a family history of breast cancer: a case report and review of literature. Cases J, 2009. 2: p. 9348.
- Hayes, B.D. and C.M. Quinn, Microinvasive lobular carcinoma arising in a fibroadenoma. Int J Surg Pathol, 2013. 21(4): p. 419-21. [CrossRef]
- Monsefi, N., et al., Mucinous subtype of invasive ductal carcinoma arising within a fibroadenoma. Arch Iran Med, 2013. 16(6): p. 366-8.
- Worsham, M.J., et al., Risk factors for breast cancer from benign breast disease in a diverse population. Breast Cancer Res Treat, 2009. 118(1): p. 1-7. [CrossRef]
- Kuijper, A., et al., Multiple fibroadenomas harbouring carcinoma in situ in a woman with a family history of breast/ovarian cancer. J Clin Pathol, 2002. 55(10): p. 795-7. [CrossRef]
- Zmetakova, I., et al., Evaluation of protein expression and DNA methylation profiles detected by pyrosequencing in invasive breast cancer. Neoplasma, 2013. 60(6): p. 635-46. [CrossRef]
- Wang, S., et al., Relationship between tumor DNA methylation status and patient characteristics in African-American and European-American women with breast cancer. PLoS One, 2012. 7(5): p. e37928. [CrossRef]
- Thaler, S., et al., RASSF1A inhibits estrogen receptor alpha expression and estrogen-independent signalling: implications for breast cancer development. Oncogene, 2012. 31(47): p. 4912-22. [CrossRef]
- Feng, W., et al., Correlation between CpG methylation profiles and hormone receptor status in breast cancers. Breast Cancer Res, 2007. 9(4): p. R57. [CrossRef]
- Cho, Y.H., et al., Prognostic significance of gene-specific promoter hypermethylation in breast cancer patients. Breast Cancer Res Treat, 2012. 131(1): p. 197-205. [CrossRef]
- Marzese, D.M., et al., Aberrant DNA methylation of cancer-related genes in giant breast fibroadenoma: a case report. J Med Case Rep, 2011. 5: p. 516. [CrossRef]
- Sturgeon, S.R., et al., Detection of promoter methylation of tumor suppressor genes in serum DNA of breast cancer cases and benign breast disease controls. Epigenetics, 2012. 7(11): p. 1258-67. [CrossRef]
- Xu, J., et al., Estrogen receptor-alpha promoter methylation is a biomarker for outcome prediction of cisplatin resistance in triple-negative breast cancer. Oncol Lett, 2018. 15(3): p. 2855-2862. [CrossRef]
- Beyene, D., et al., Cyclin A2 and Ki-67 proliferation markers could be used to identify tumors with poor prognosis in African American women with breast cancer. J Cancer Biol, 2023. 4(1): p. 3-16. [CrossRef]
- Iwase, H., et al., DNA methylation analysis at distal and proximal promoter regions of the oestrogen receptor gene in breast cancers. Br J Cancer, 1999. 80(12): p. 1982-6. [CrossRef]
- Kwabi-Addo, B., et al., Identification of differentially methylated genes in normal prostate tissues from African American and Caucasian men. Clin Cancer Res, 2010. 16(14): p. 3539-47. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




