Submitted:
02 July 2024
Posted:
02 July 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. Master Regulators of 1099 Causal Networks in Diffuse- and Intestinal-Type Gastric Cancer
2.2. HDAC1-Interacting Network and Involvement of the Regulation of EMT by Growth Factors Pathway and Coronavirus Pathogenesis Pathway
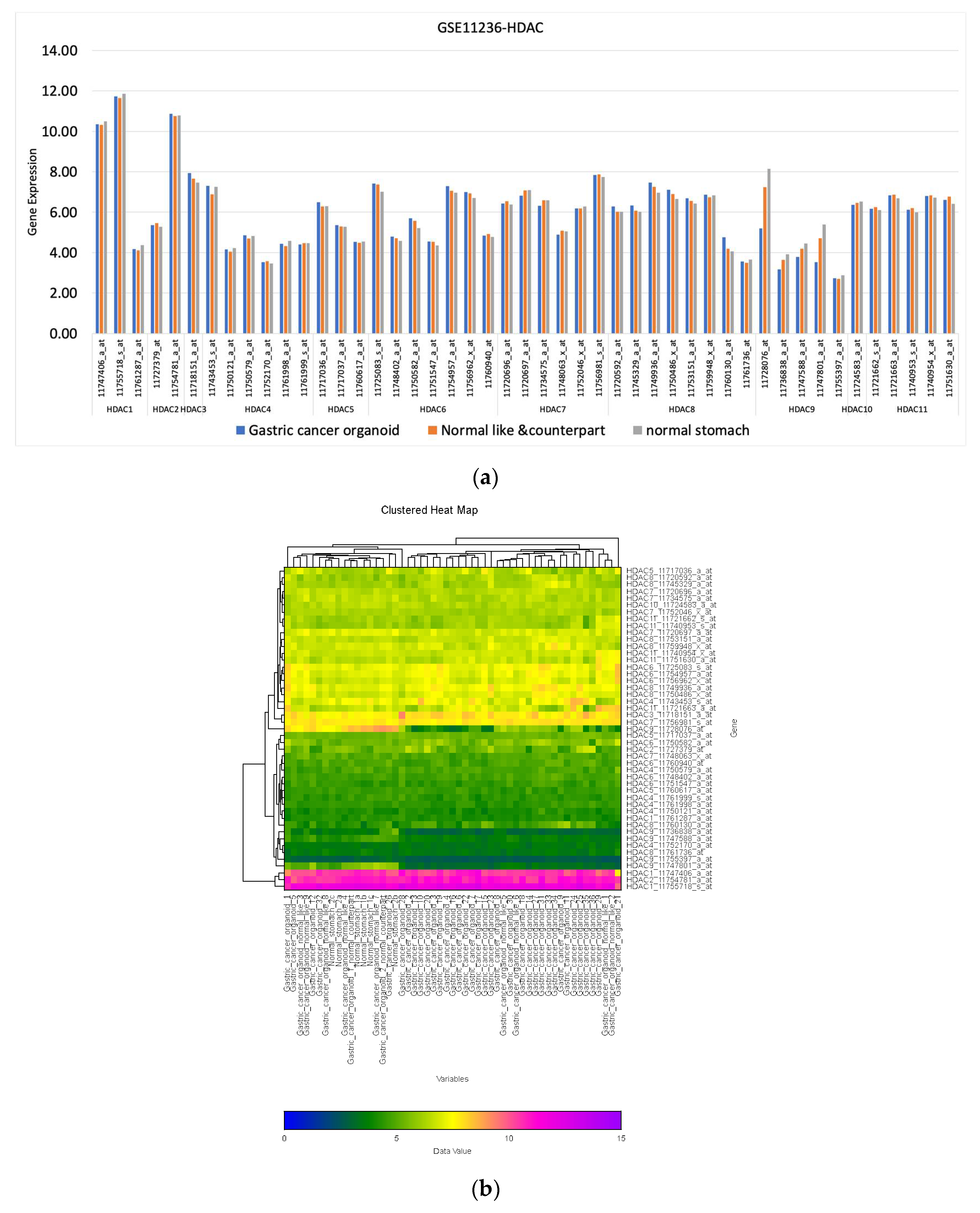
2.3. HDAC Expression Cluster Analysis
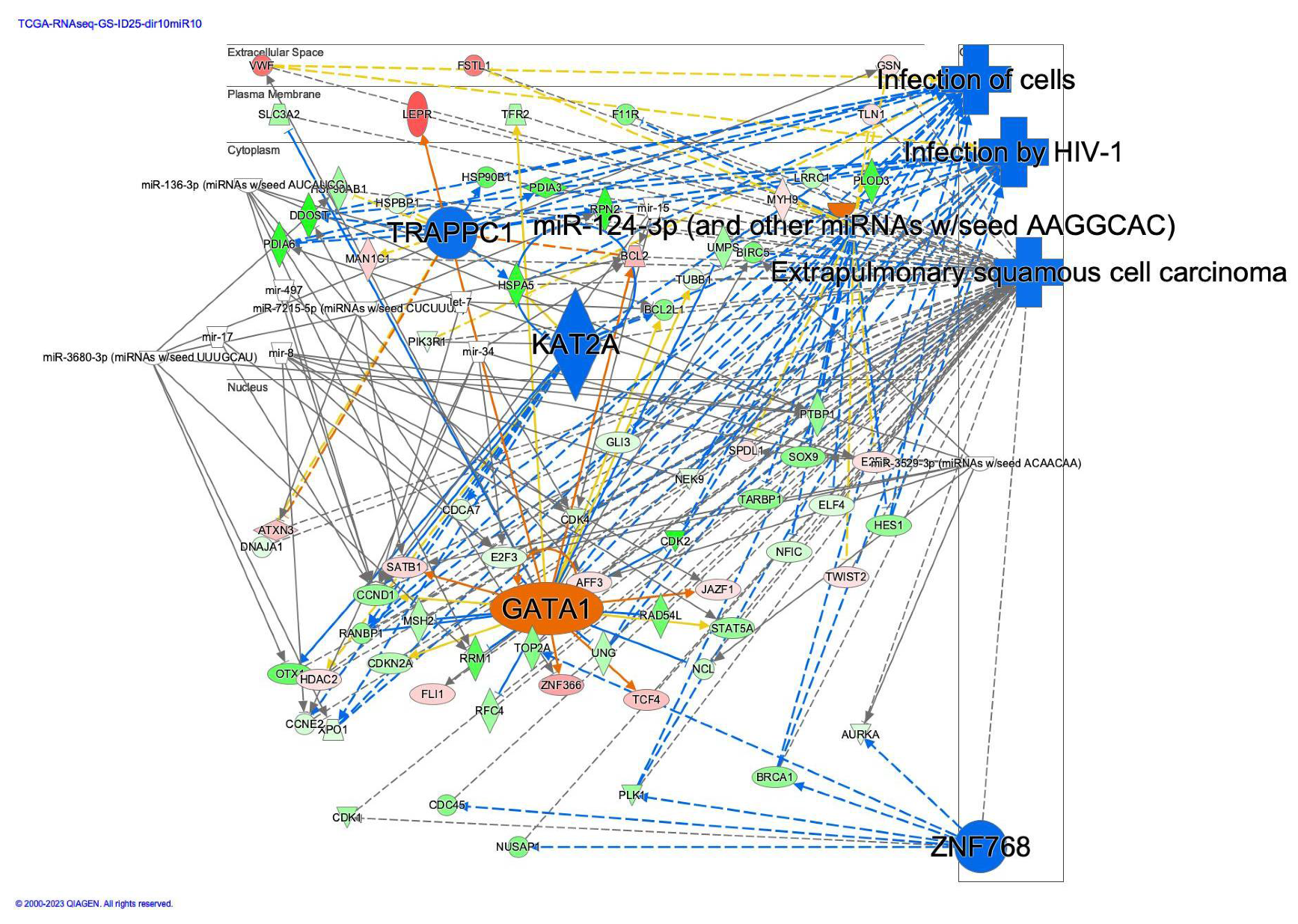
2.4. Regulator Effect Network of Diffuse-Type Gastric Cancer
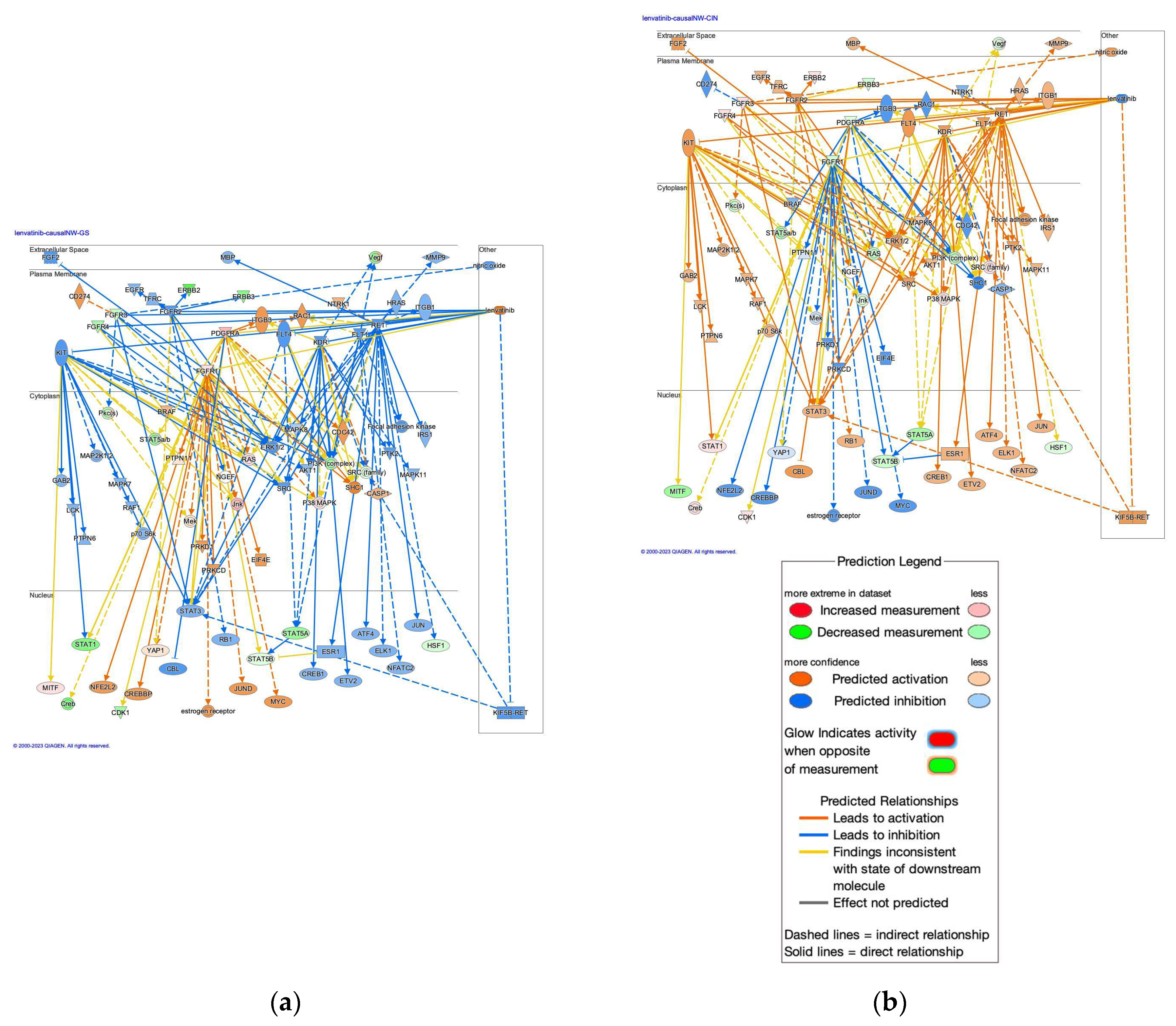
2.5. Causal Networks of Lenvatinib in Diffuse- and Intestinal-type GC
3. Discussion
4. Materials and Methods
4.1. Data Analysis of Diffuse- and Intestinal-Type GC
4.2. Network Pathway Analysis
4.3. Cluster Analysis
4.4. Data Visualization
4.5. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tanabe, S.; Quader, S.; Ono, R.; Cabral, H.; Aoyagi, K.; Hirose, A.; Perkins, E.J.; Yokozaki, H.; Sasaki, H. Regulation of Epithelial-Mesenchymal Transition Pathway and Artificial Intelligence-Based Modeling for Pathway Activity Prediction. Onco 2023, 3, 13–25. [Google Scholar] [CrossRef]
- Tanabe, S.; Quader, S.; Ono, R.; Cabral, H.; Aoyagi, K.; Hirose, A.; Yokozaki, H.; Sasaki, H. Molecular Network Profiling in Intestinal- and Diffuse-Type Gastric Cancer. Cancers (Basel) 2020, 12, 3833. [Google Scholar] [CrossRef]
- Tanabe, S.; Quader, S.; Ono, R.; Cabral, H.; Aoyagi, K.; Hirose, A.; Yokozaki, H.; Sasaki, H. Cell Cycle Regulation and DNA Damage Response Networks in Diffuse- and Intestinal-Type Gastric Cancer. Cancers 2021, 13, 5786. [Google Scholar] [CrossRef] [PubMed]
- Duan, H.; Liu, Y.; Gao, Z.; Huang, W. Recent advances in drug delivery systems for targeting cancer stem cells. Acta Pharm Sin B 2021, 11, 55–70. [Google Scholar] [CrossRef]
- Chen, L.; Yang, F.; Chen, S.; Tai, J. Mechanisms on chemotherapy resistance of colorectal cancer stem cells and research progress of reverse transformation: A mini-review. Frontiers in Medicine 2022, 9. [Google Scholar] [CrossRef]
- Dzobo, K.; Senthebane, D.A.; Ganz, C.; Thomford, N.E.; Wonkam, A.; Dandara, C. Advances in Therapeutic Targeting of Cancer Stem Cells within the Tumor Microenvironment: An Updated Review. Cells 2020, 9. [Google Scholar] [CrossRef]
- Hao, Y.; Baker, D.; Ten Dijke, P. TGF-β-Mediated Epithelial-Mesenchymal Transition and Cancer Metastasis. Int J Mol Sci 2019, 20. [Google Scholar] [CrossRef]
- Lu, W.; Kang, Y. Epithelial-Mesenchymal Plasticity in Cancer Progression and Metastasis. Dev Cell 2019, 49, 361–374. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, Y.; Ye, F.; Qian, P.; Qin, Z.; Li, D.; Ye, L.; Feng, L. DKK1 Promotes Epithelial-Mesenchymal Transition and Cisplatin Resistance in Gastric Cancer via Activation of the PI3K/AKT Pathway. Cancers (Basel) 2023, 15. [Google Scholar] [CrossRef]
- Yoon, S.J.; Park, J.; Shin, Y.; Choi, Y.; Park, S.W.; Kang, S.G.; Son, H.Y.; Huh, Y.M. Deconvolution of diffuse gastric cancer and the suppression of CD34 on the BALB/c nude mice model. BMC Cancer 2020, 20, 314. [Google Scholar] [CrossRef]
- Taja-Chayeb, L.; Vidal-Millán, S.; Trejo-Becerril, C.; Pérez-Cárdenas, E.; Chávez-Blanco, A.; Domínguez-Gómez, G.; González-Fierro, A.; Romo-Pérez, A.; Dueñas-González, A. Hereditary diffuse gastric cancer (HDGC). An overview. Clin Res Hepatol Gastroenterol 2022, 46, 101820. [Google Scholar] [CrossRef] [PubMed]
- Tanabe, S.; Boonstra, E.; Hong, T.; Quader, S.; Ono, R.; Cabral, H.; Aoyagi, K.; Yokozaki, H.; Perkins, E.J.; Sasaki, H. Molecular Networks of Platinum Drugs and Their Interaction with microRNAs in Cancer. Genes (Basel) 2023, 14. [Google Scholar] [CrossRef] [PubMed]
- Korivi, B.R.; Faria, S.; Aly, A.; Sun, J.; Patnana, M.; Jensen, C.T.; Wagner-Bartak, N.; Bhosale, P.R. Intestinal and diffuse gastric cancer: a retrospective study comparing primary sites. Clin Imaging 2019, 56, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Mariette, C.; Carneiro, F.; Grabsch, H.I.; van der Post, R.S.; Allum, W.; de Manzoni, G. Consensus on the pathological definition and classification of poorly cohesive gastric carcinoma. Gastric Cancer 2019, 22, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.C.; Sohn, B.H.; Cheong, J.H.; Kim, S.B.; Lee, J.E.; Park, K.C.; Lee, S.H.; Park, J.L.; Park, Y.Y.; Lee, H.S.; et al. Clinical and genomic landscape of gastric cancer with a mesenchymal phenotype. Nat Commun 2018, 9, 1777. [Google Scholar] [CrossRef] [PubMed]
- Cristescu, R.; Lee, J.; Nebozhyn, M.; Kim, K.M.; Ting, J.C.; Wong, S.S.; Liu, J.; Yue, Y.G.; Wang, J.; Yu, K.; et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med 2015, 21, 449–456. [Google Scholar] [CrossRef] [PubMed]
- Peng, K.; Zhang, F.; Wang, Y.; Sahgal, P.; Li, T.; Zhou, J.; Liang, X.; Zhang, Y.; Sethi, N.; Liu, T.; et al. Development of Combination Strategies for Focal Adhesion Kinase Inhibition in Diffuse Gastric Cancer. Clin Cancer Res 2023, 29, 197–208. [Google Scholar] [CrossRef]
- Schlumberger, M.; Tahara, M.; Wirth, L.J.; Robinson, B.; Brose, M.S.; Elisei, R.; Habra, M.A.; Newbold, K.; Shah, M.H.; Hoff, A.O.; et al. Lenvatinib versus placebo in radioiodine-refractory thyroid cancer. N Engl J Med 2015, 372, 621–630. [Google Scholar] [CrossRef] [PubMed]
- Roskoski, R., Jr. Small molecule protein kinase inhibitors approved by regulatory agencies outside of the United States. Pharmacol Res 2023, 194, 106847. [Google Scholar] [CrossRef]
- Zou, M.; Li, Y.; Dai, Y.; Sun, L.; Huang, T.; Yuan, X.; Qiu, H. AFP-producing hepatoid adenocarcinoma (HAC) of peritoneum and omentum: a case report and literature review. Onco Targets Ther 2019, 12, 7649–7654. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, X.; Shi, Y.; Ni, Y.; Fei, J.; Jin, Z.; Li, W.; Wang, X.; Wu, N. Role and potential therapeutic value of histone methyltransferases in drug resistance mechanisms in lung cancer. Front Oncol 2024, 14, 1376916. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Wu, E.; Wang, D.; Niu, Y.; Yue, H.; Zhang, D.; Luo, J.; Chen, R. LncRNA HRCEG, regulated by HDAC1, inhibits cells proliferation and epithelial-mesenchymal-transition in gastric cancer. Cancer Genet 2020, 241, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Zeng, J.; Liu, H.; Wang, T.; Yu, Z.; Chen, J. Role of HDAC1 in the progression of gastric cancer and the correlation with lncRNAs. Oncol Lett 2019, 17, 3296–3304. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Jing, X.; Chen, Z.; Pan, X.; Xu, D.; Yu, X.; Zhong, F.; Zhao, L.; Yang, C.; Wang, B.; et al. Histone deacetylase-mediated tumor microenvironment characteristics and synergistic immunotherapy in gastric cancer. Theranostics 2023, 13, 4574–4600. [Google Scholar] [CrossRef] [PubMed]
- Jenke, R.; Oliinyk, D.; Zenz, T.; Körfer, J.; Schäker-Hübner, L.; Hansen, F.K.; Lordick, F.; Meier-Rosar, F.; Aigner, A.; Büch, T. HDAC inhibitors activate lipid peroxidation and ferroptosis in gastric cancer. Biochem Pharmacol 2024, 225, 116257. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Das, K.; Wu, J.; Lee, M.H.; Tan, P. RNH1 regulation of reactive oxygen species contributes to histone deacetylase inhibitor resistance in gastric cancer cells. Oncogene 2014, 33, 1527–1537. [Google Scholar] [CrossRef] [PubMed]
- Robert, C.; Rassool, F.V. Chapter Three - HDAC Inhibitors: Roles of DNA Damage and Repair. In Advances in Cancer Research, Grant, S., Ed.; Academic Press: 2012; Volume 116, pp. 87-129.
- Zheng, H.C.; Xue, H.; Jiang, H.M. The roles of ING5 in cancer: A tumor suppressor. Front Cell Dev Biol 2022, 10, 1012179. [Google Scholar] [CrossRef] [PubMed]
- González-Sánchez, G.D.; Granados-López, A.J.; López-Hernández, Y.; Robles, M.J.G.; López, J.A. miRNAs as Interconnectors between Obesity and Cancer. Noncoding RNA 2024, 10. [Google Scholar] [CrossRef]
- Santaolalla, R.; Abreu, M.T. Innate immunity in the small intestine. Curr Opin Gastroenterol 2012, 28, 124–129. [Google Scholar] [CrossRef]
- Prinz, C.; Fehring, L.; Frese, R. MicroRNAs as Indicators of Malignancy in Pancreatic Ductal Adenocarcinoma (PDAC) and Cystic Pancreatic Lesions. Cells 2022, 11. [Google Scholar] [CrossRef]
- Kanno, S.; Nosho, K.; Ishigami, K.; Yamamoto, I.; Koide, H.; Kurihara, H.; Mitsuhashi, K.; Shitani, M.; Motoya, M.; Sasaki, S.; et al. MicroRNA-196b is an independent prognostic biomarker in patients with pancreatic cancer. Carcinogenesis 2017, 38, 425–431. [Google Scholar] [CrossRef] [PubMed]
- Letafati, A.; Najafi, S.; Mottahedi, M.; Karimzadeh, M.; Shahini, A.; Garousi, S.; Abbasi-Kolli, M.; Sadri Nahand, J.; Tamehri Zadeh, S.S.; Hamblin, M.R.; et al. MicroRNA let-7 and viral infections: focus on mechanisms of action. Cell Mol Biol Lett 2022, 27, 14. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Yao, H.; Zhu, P.; Zhang, X.; Pan, Q.; Gong, C.; Huang, Y.; Hu, X.; Su, F.; Lieberman, J.; et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell 2007, 131, 1109–1123. [Google Scholar] [CrossRef] [PubMed]
- Gilles, M.E.; Slack, F.J. Let-7 microRNA as a potential therapeutic target with implications for immunotherapy. Expert Opin Ther Targets 2018, 22, 929–939. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Vega, F.; Mina, M.; Armenia, J.; Chatila, W.K.; Luna, A.; La, K.C.; Dimitriadoy, S.; Liu, D.L.; Kantheti, H.S.; Saghafinia, S.; et al. Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell 2018, 173, 321–337.e310. [Google Scholar] [CrossRef] [PubMed]
- Bass, A.J.; Thorsson, V.; Shmulevich, I.; Reynolds, S.M.; Miller, M.; Bernard, B.; Hinoue, T.; Laird, P.W.; Curtis, C.; Shen, H.; et al. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014, 513, 202–209. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013, 6, pl1. [Google Scholar] [CrossRef]
- Grossman, R.L.; Heath, A.P.; Ferretti, V.; Varmus, H.E.; Lowy, D.R.; Kibbe, W.A.; Staudt, L.M. Toward a Shared Vision for Cancer Genomic Data. N Engl J Med 2016, 375, 1109–1112. [Google Scholar] [CrossRef]
- Krämer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef]
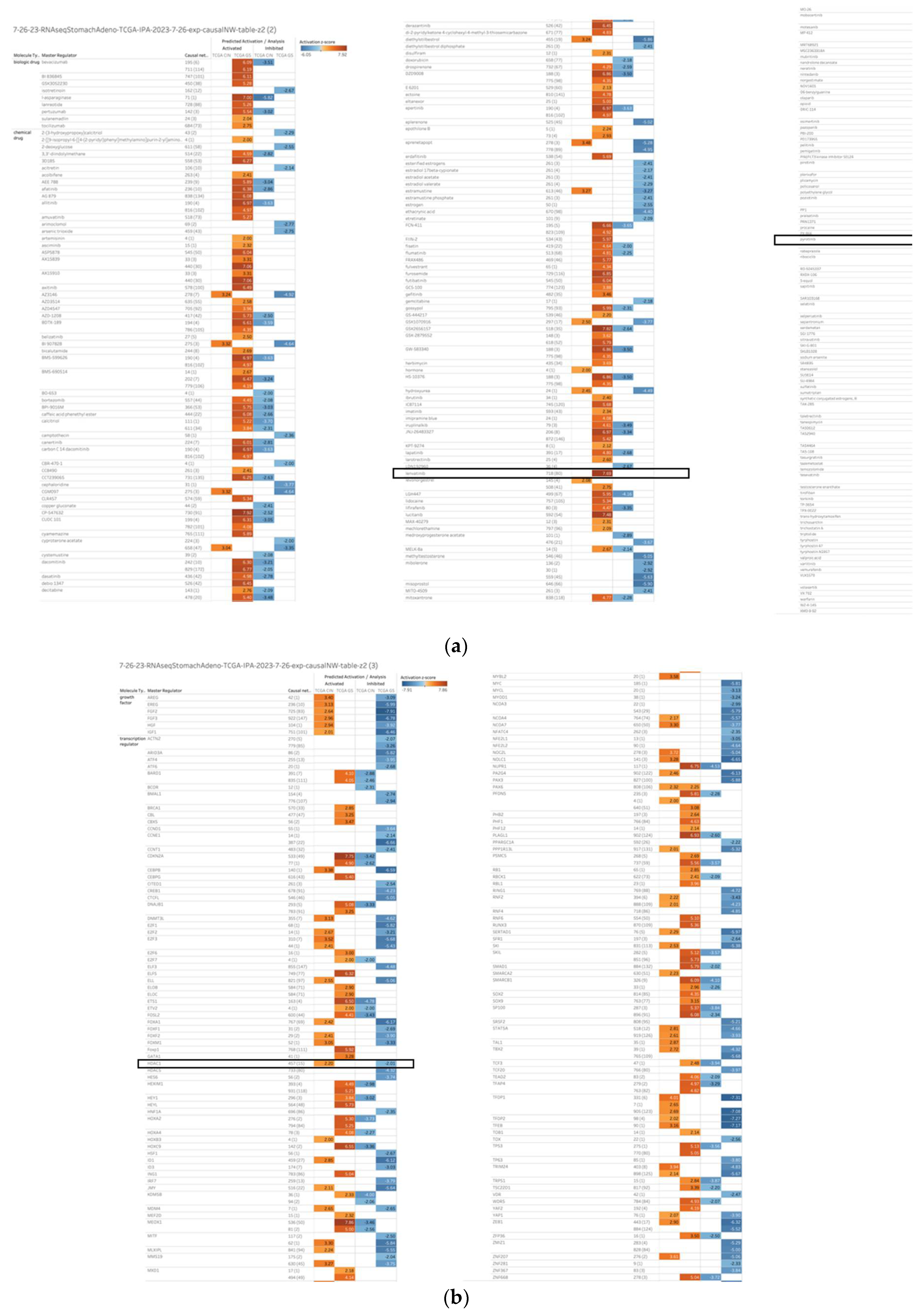
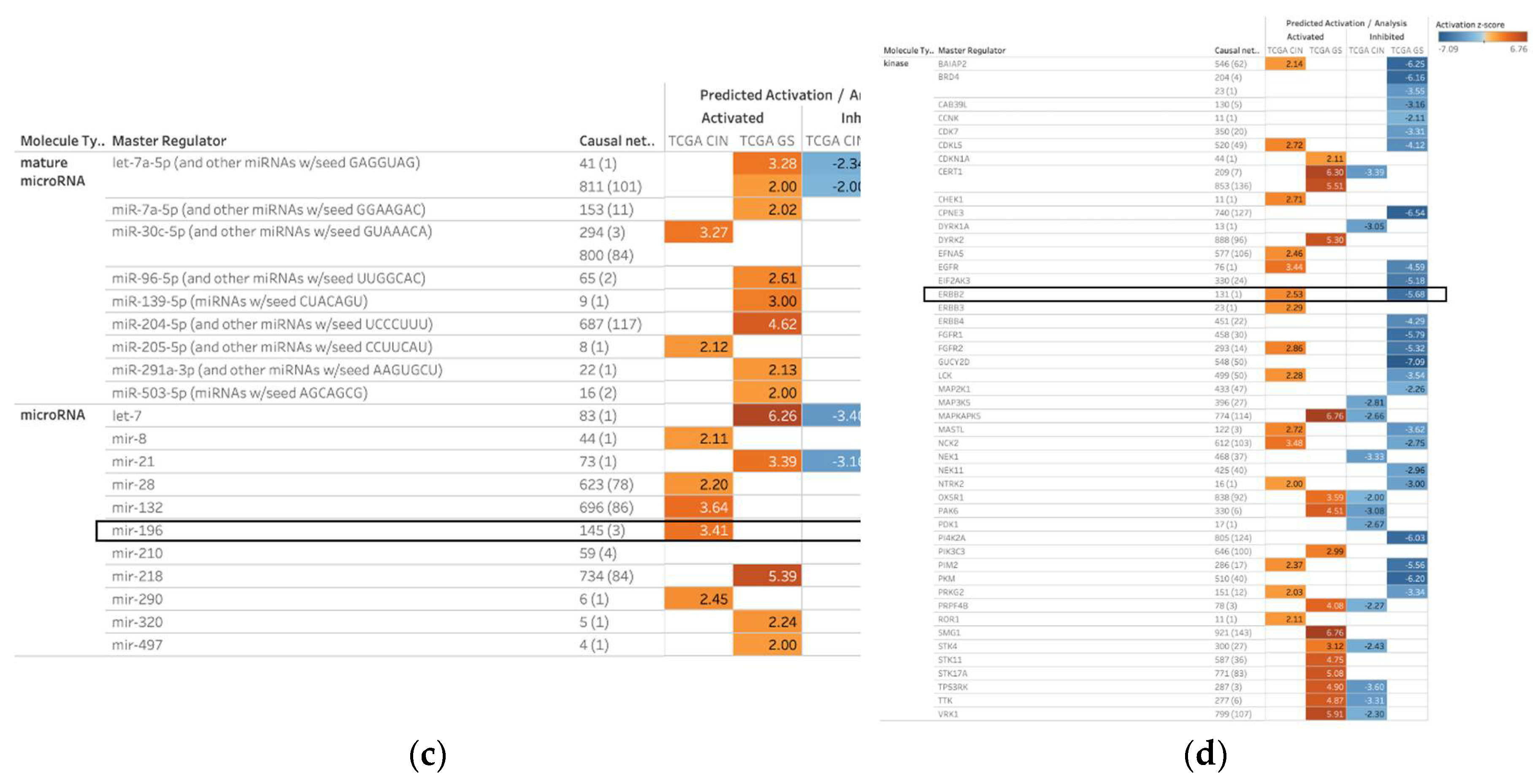
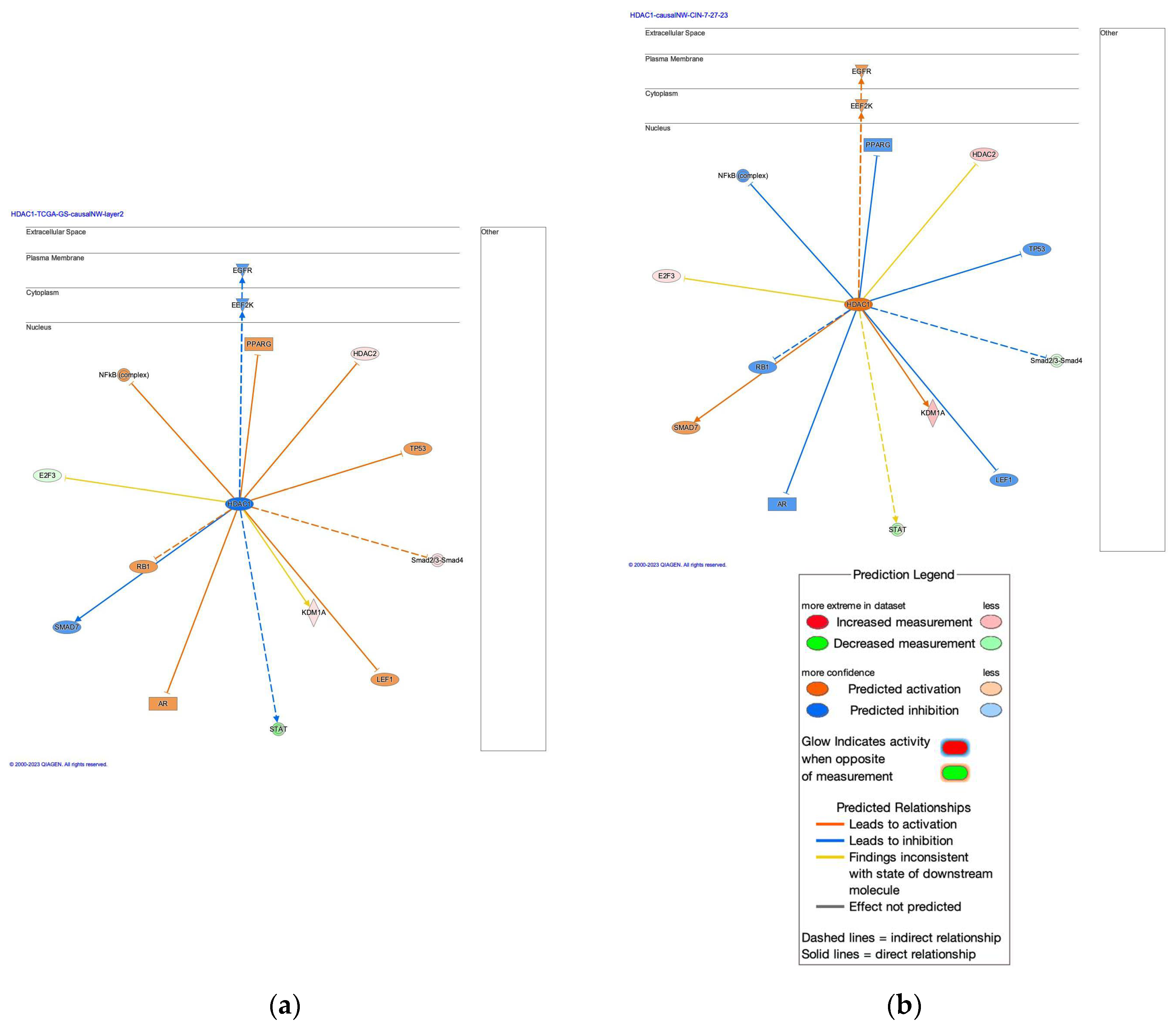
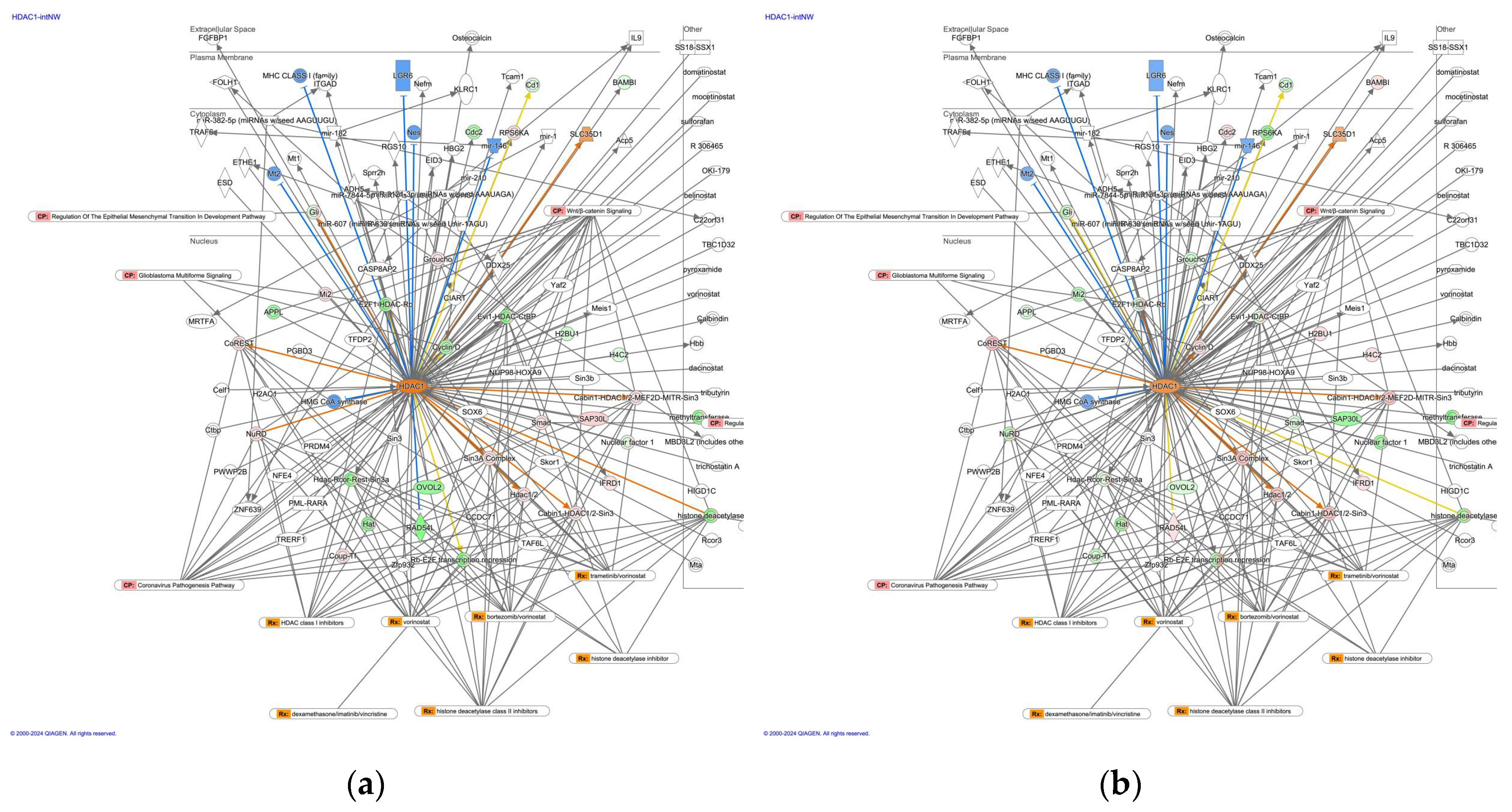

| Master Regulator |
Analysis | Depth | Predicted Activation |
Activation z-score |
p-value of overlap |
Network bias- corrected p-value |
Causal network |
Target-connected regulators |
| HDAC1 | TCGA GS | 3 | 0.405 | 2.87E-26 | 2.00E-04 | 878 (117) | 115 | |
| TCGA CIN | 3 | 1.08 | 2.87E-26 | 2.00E-04 | 878 (117) | 115 | ||
| HDAC5 | TCGA GS | 3 | Inhibited | -4.321 | 2.01E-25 | 1.00E-04 | 733 (80) | 78 |
| TCGA CIN | 3 | 0.85 | 2.01E-25 | 1.00E-04 | 733 (80) | 78 | ||
| Hdac1/2 | TCGA GS | 3 | Inhibited | -4.33 | 2.53E-25 | 1.00E-03 | 820 (95) | 95 |
| TCGA CIN | 3 | 1.606 | 2.53E-25 | 1.00E-03 | 820 (95) | 95 | ||
| HDAC1 | TCGA GS | 2 | Inhibited | -2.011 | 7.62E-19 | 1.00E-04 | 457 (15) | 15 |
| TCGA CIN | 2 | Activated | 2.199 | 7.62E-19 | 1.00E-04 | 457 (15) | 15 | |
| HDAC10 | TCGA GS | 3 | -0.089 | 1.43E-15 | 1.70E-02 | 508 (37) | 37 | |
| TCGA CIN | 3 | -1.509 | 1.43E-15 | 1.70E-02 | 508 (37) | 37 | ||
| HDAC2 | TCGA GS | 1 | 1.528 | 3.55E-03 | 2.24E-02 | 21 (1) | 1 | |
| TCGA CIN | 1 | -0.655 | 3.55E-03 | 2.24E-02 | 21 (1) | 1 |
| let-7 | miR-136-3p (miRNAs w/seed AUCAUCG) |
| mir-15 | miR-3529-3p (miRNAs w/seed ACAACAA) |
| mir-17 | miR-3680-3p (miRNAs w/seed UUUGCAU) |
| mir-34 | miR-7215-5p (miRNAs w/seed CUCUUUA) |
| mir-8 | mir-497 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





