Submitted:
30 June 2024
Posted:
01 July 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Sequence Retrieval, Selection, Alignment, and Subtyping
2.3. Assessment of RASs in the Final HCV Dataset for the Three DAA Classes
2.4. Assessment of Clinically-Relevant RASs by DAA and HCV Genotypes/Subtypes
2.5. Data and Statistical Analysis
3. Results
3.1. HCV Dataset Characteristics
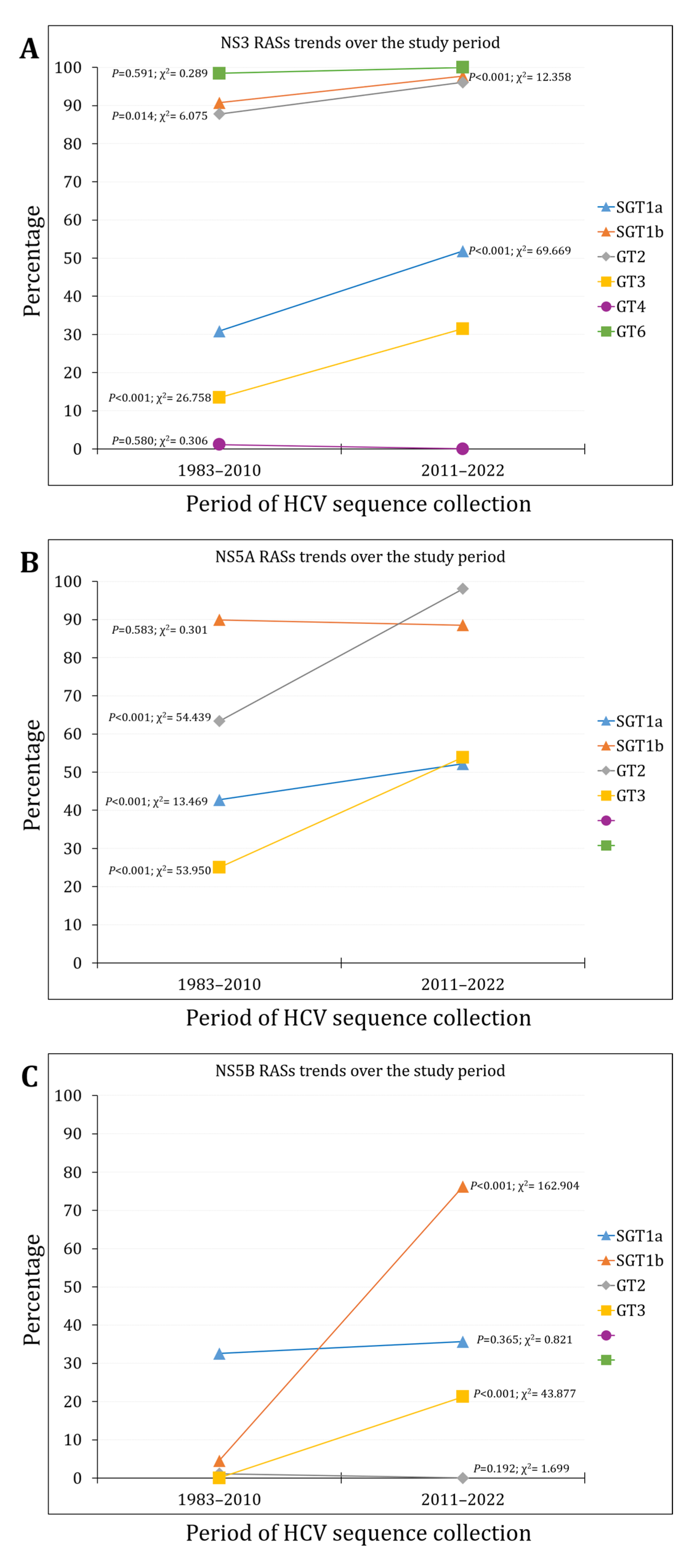
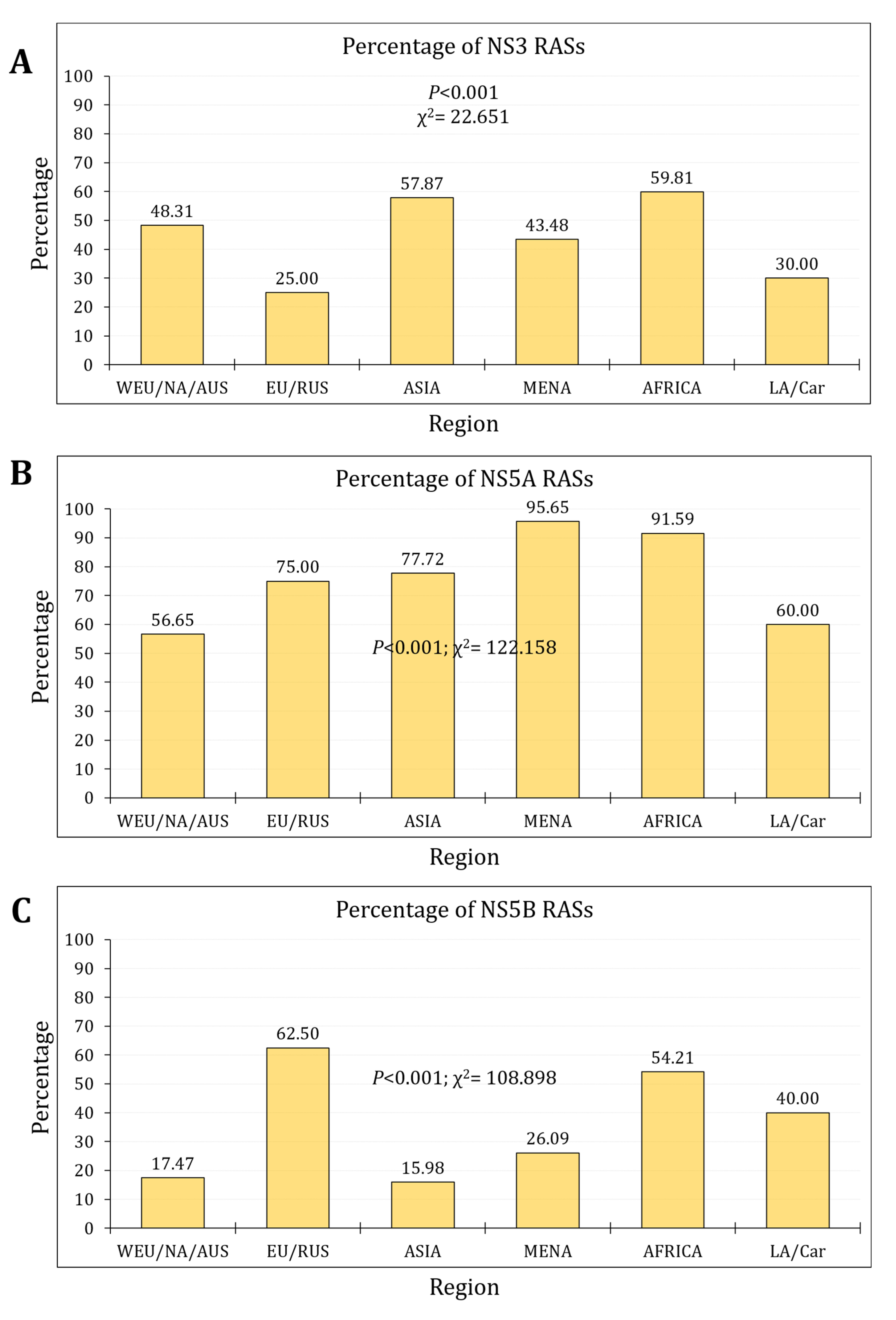
3.2. The Prevalence and Trends of NS3 RASs
3.3. The Prevalence and Trends of NS5A RASs
3.4. The Prevalence and Trends of NS5B RASs
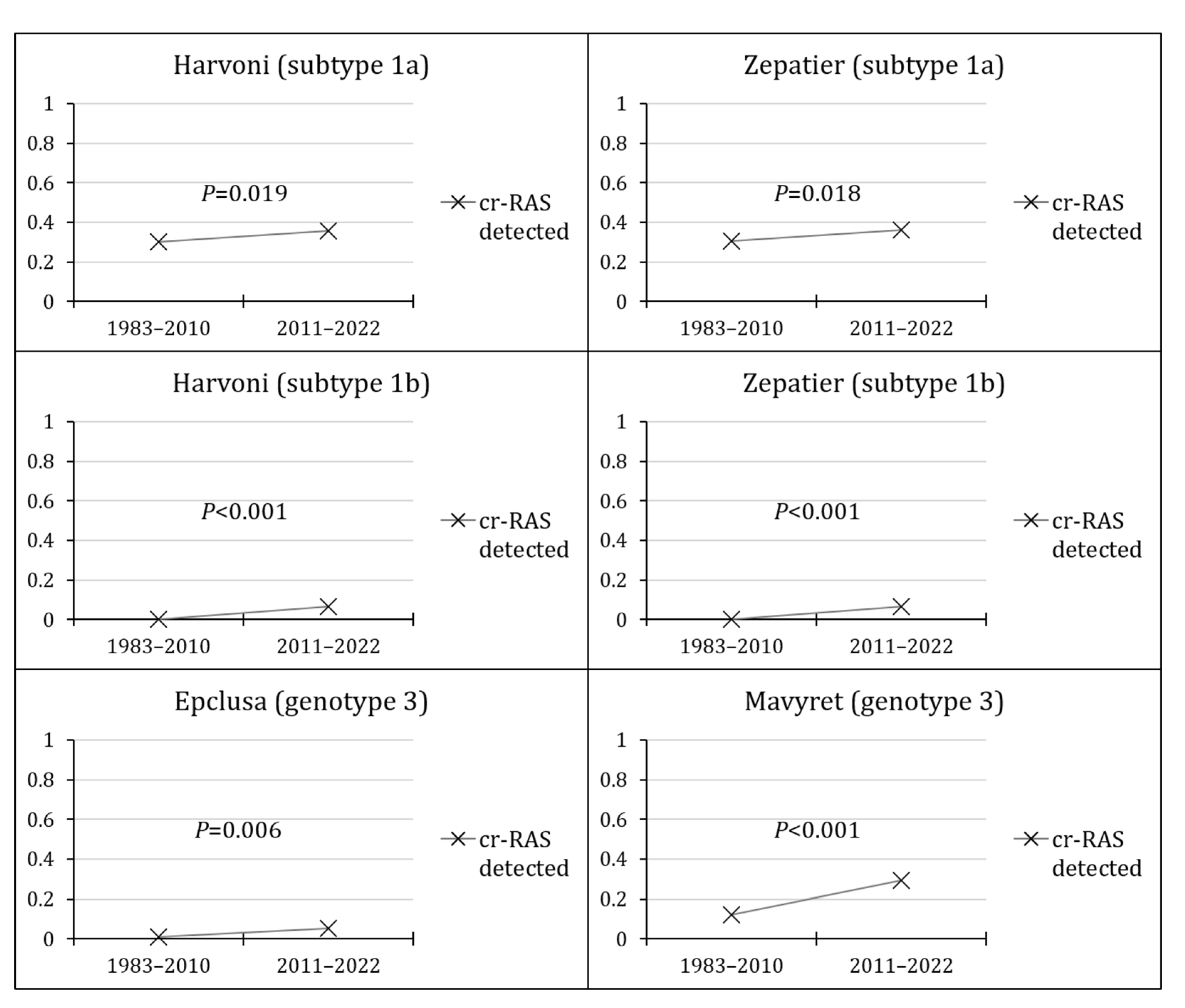
3.5. The Prevalence of cr-RASs in Four Different DAA Regimens
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization (WHO). Hepatitis C - Key Facts. Available online: https://www.who.int/news-room/fact-sheets/detail/hepatitis-c (accessed on 19 April 2024).
- Polaris Observatory HCV Collaborators. Global change in hepatitis C virus prevalence and cascade of care between 2015 and 2020: a modelling study. Lancet Gastroenterol Hepatol 2022, 7, 396–415. [Google Scholar] [CrossRef] [PubMed]
- Sallam, M.; Khalil, R. Contemporary Insights into Hepatitis C Virus: A Comprehensive Review. Microorganisms 2024, 12, 1035. [Google Scholar] [CrossRef] [PubMed]
- GBD 2017 Cirrhosis Collaborators. The global, regional, and national burden of cirrhosis by cause in 195 countries and territories, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol Hepatol 2020, 5, 245–266. [Google Scholar] [CrossRef] [PubMed]
- Moon, A.M.; Singal, A.G.; Tapper, E.B. Contemporary Epidemiology of Chronic Liver Disease and Cirrhosis. Clin Gastroenterol Hepatol 2020, 18, 2650–2666. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.Q.; Terrault, N.A.; Tacke, F.; Gluud, L.L.; Arrese, M.; Bugianesi, E.; Loomba, R. Global epidemiology of cirrhosis - aetiology, trends and predictions. Nat Rev Gastroenterol Hepatol 2023, 20, 388–398. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.; Kim, B.K.; Yon, D.K.; Lee, S.W.; Lee, H.G.; Chang, H.H.; Park, S.; Koyanagi, A.; Jacob, L.; Dragioti, E.; et al. Global burden of primary liver cancer and its association with underlying aetiologies, sociodemographic status, and sex differences from 1990-2019: A DALY-based analysis of the Global Burden of Disease 2019 study. Clin Mol Hepatol 2023, 29, 433–452. [Google Scholar] [CrossRef] [PubMed]
- Liang, T.J.; Ghany, M.G. Current and future therapies for hepatitis C virus infection. N Engl J Med 2013, 368, 1907–1917. [Google Scholar] [CrossRef] [PubMed]
- Geddawy, A.; Ibrahim, Y.F.; Elbahie, N.M.; Ibrahim, M.A. Direct Acting Anti-hepatitis C Virus Drugs: Clinical Pharmacology and Future Direction. J Transl Int Med 2017, 5, 8–17. [Google Scholar] [CrossRef]
- Calleja, J.L.; Crespo, J.; Rincón, D.; Ruiz-Antorán, B.; Fernandez, I.; Perelló, C.; Gea, F.; Lens, S.; García-Samaniego, J.; Sacristán, B.; et al. Effectiveness, safety and clinical outcomes of direct-acting antiviral therapy in HCV genotype 1 infection: Results from a Spanish real-world cohort. J Hepatol 2017, 66, 1138–1148. [Google Scholar] [CrossRef] [PubMed]
- Crespo, J.; Calleja, J.L.; Fernández, I.; Sacristan, B.; Ruiz-Antorán, B.; Ampuero, J.; Hernández-Conde, M.; García-Samaniego, J.; Gea, F.; Buti, M.; et al. Real-World Effectiveness and Safety of Oral Combination Antiviral Therapy for Hepatitis C Virus Genotype 4 Infection. Clin Gastroenterol Hepatol 2017, 15, 945–949. [Google Scholar] [CrossRef] [PubMed]
- Chan, A.; Patel, K.; Naggie, S. Genotype 3 Infection: The Last Stand of Hepatitis C Virus. Drugs 2017, 77, 131–144. [Google Scholar] [CrossRef] [PubMed]
- Goossens, N.; Negro, F. Is genotype 3 of the hepatitis C virus the new villain? Hepatology 2014, 59, 2403–2412. [Google Scholar] [CrossRef] [PubMed]
- Wei, B.; Ji, F.; Yeo, Y.H.; Ogawa, E.; Stave, C.D.; Dang, S.; Li, Z.; Furusyo, N.; Cheung, R.C.; Nguyen, M.H. Systematic review and meta-analysis: real-world effectiveness of direct-acting antiviral therapies in chronic hepatitis C genotype 3 in Asia. BMJ Open Gastroenterol 2018, 5, e000209. [Google Scholar] [CrossRef]
- Isfordink, C.J.; van de Laar, T.J.W.; Rebers, S.P.H.; Wessels, E.; Molenkamp, R.; Knoester, M.; Baak, B.C.; van Nieuwkoop, C.; van Hoek, B.; Brakenhoff, S.M.; et al. Direct-Acting Antiviral Treatment for Hepatitis C Genotypes Uncommon in High-Income Countries: A Dutch Nationwide Cohort Study. Open Forum Infect Dis 2021, 8, ofab006. [Google Scholar] [CrossRef] [PubMed]
- Burgess, S.V.; Hussaini, T.; Yoshida, E.M. Concordance of sustained virologic response at weeks 4, 12 and 24 post-treatment of hepatitis c in the era of new oral direct-acting antivirals: A concise review. Ann Hepatol 2016, 15, 154–159. [Google Scholar] [PubMed]
- Chen, J.; Florian, J.; Carter, W.; Fleischer, R.D.; Hammerstrom, T.S.; Jadhav, P.R.; Zeng, W.; Murray, J.; Birnkrant, D. Earlier sustained virologic response end points for regulatory approval and dose selection of hepatitis C therapies. Gastroenterology 2013, 144, 1450–1455. [Google Scholar] [CrossRef] [PubMed]
- Fraser, H.; Martin, N.K.; Brummer-Korvenkontio, H.; Carrieri, P.; Dalgard, O.; Dillon, J.; Goldberg, D.; Hutchinson, S.; Jauffret-Roustide, M.; Kåberg, M.; et al. Model projections on the impact of HCV treatment in the prevention of HCV transmission among people who inject drugs in Europe. J Hepatol 2018, 68, 402–411. [Google Scholar] [CrossRef] [PubMed]
- Ayoub, H.H.; Mahmud, S.; Chemaitelly, H.; Abu-Raddad, L.J. Treatment as prevention for hepatitis C virus in the Middle East and North Africa: a modeling study. Front Public Health 2023, 11, 1187786. [Google Scholar] [CrossRef] [PubMed]
- van Santen, D.K.; Sacks-Davis, R.; Stewart, A.; Boyd, A.; Young, J.; van der Valk, M.; Smit, C.; Rauch, A.; Braun, D.L.; Jarrin, I.; et al. Treatment as prevention effect of direct-acting antivirals on primary hepatitis C virus incidence: Findings from a multinational cohort between 2010 and 2019. EClinicalMedicine 2023, 56, 101810. [Google Scholar] [CrossRef] [PubMed]
- Hellard, M.; Schroeder, S.E.; Pedrana, A.; Doyle, J.; Aitken, C. The Elimination of Hepatitis C as a Public Health Threat. Cold Spring Harb Perspect Med 2020, 10. [Google Scholar] [CrossRef] [PubMed]
- Cox, A.L. Challenges and Promise of a Hepatitis C Virus Vaccine. Cold Spring Harb Perspect Med 2020, 10. [Google Scholar] [CrossRef] [PubMed]
- Duncan, J.D.; Urbanowicz, R.A.; Tarr, A.W.; Ball, J.K. Hepatitis C Virus Vaccine: Challenges and Prospects. Vaccines (Basel) 2020, 8, 90. [Google Scholar] [CrossRef] [PubMed]
- Izhari, M.A. Molecular Mechanisms of Resistance to Direct-Acting Antiviral (DAA) Drugs for the Treatment of Hepatitis C Virus Infections. Diagnostics (Basel) 2023, 13, 3102. [Google Scholar] [CrossRef] [PubMed]
- European Association for the Study of the Liver. EASL recommendations on treatment of hepatitis C: Final update of the series(☆). J Hepatol 2020, 73, 1170–1218. [Google Scholar] [CrossRef] [PubMed]
- AASLD-IDSA. Recommendations for testing, managing, and treating hepatitis C. Available online: https://www.hcvguidelines.org/evaluate/resistance (accessed on 3 May 2024).
- Bhattacharya, D.; Aronsohn, A.; Price, J.; Lo Re, V. Hepatitis C Guidance 2023 Update: AASLD-IDSA Recommendations for Testing, Managing, and Treating Hepatitis C Virus Infection. Clin Infect Dis 2023. [Google Scholar] [CrossRef] [PubMed]
- Bagaglio, S.; Uberti-Foppa, C.; Morsica, G. Resistance Mechanisms in Hepatitis C Virus: implications for Direct-Acting Antiviral Use. Drugs 2017, 77, 1043–1055. [Google Scholar] [CrossRef] [PubMed]
- Mushtaq, S.; Hashmi, A.H.; Khan, A.; Asad Raza Kazmi, S.M.; Manzoor, S. Emergence and Persistence of Resistance-Associated Substitutions in HCV GT3 Patients Failing Direct-Acting Antivirals. Front Pharmacol 2022, 13, 894460. [Google Scholar] [CrossRef] [PubMed]
- Malandris, K.; Kalopitas, G.; Theocharidou, E.; Germanidis, G. The Role of RASs /RVs in the Current Management of HCV. Viruses 2021, 13, 2096. [Google Scholar] [CrossRef] [PubMed]
- Eltahla, A.A.; Leung, P.; Pirozyan, M.R.; Rodrigo, C.; Grebely, J.; Applegate, T.; Maher, L.; Luciani, F.; Lloyd, A.R.; Bull, R.A. Dynamic evolution of hepatitis C virus resistance-associated substitutions in the absence of antiviral treatment. Sci Rep 2017, 7, 41719. [Google Scholar] [CrossRef] [PubMed]
- Preciado, M.V.; Valva, P.; Escobar-Gutierrez, A.; Rahal, P.; Ruiz-Tovar, K.; Yamasaki, L.; Vazquez-Chacon, C.; Martinez-Guarneros, A.; Carpio-Pedroza, J.C.; Fonseca-Coronado, S.; et al. Hepatitis C virus molecular evolution: transmission, disease progression and antiviral therapy. World J Gastroenterol 2014, 20, 15992–16013. [Google Scholar] [CrossRef] [PubMed]
- Fourati, S.; Pawlotsky, J.M. Virologic Tools for HCV Drug Resistance Testing. Viruses 2015, 7, 6346–6359. [Google Scholar] [CrossRef] [PubMed]
- Soria, M.E.; García-Crespo, C.; Martínez-González, B.; Vázquez-Sirvent, L.; Lobo-Vega, R.; de Ávila, A.I.; Gallego, I.; Chen, Q.; García-Cehic, D.; Llorens-Revull, M.; et al. Amino Acid Substitutions Associated with Treatment Failure for Hepatitis C Virus Infection. J Clin Microbiol 2020, 58. [Google Scholar] [CrossRef] [PubMed]
- Kalaghatgi, P.; Sikorski, A.M.; Knops, E.; Rupp, D.; Sierra, S.; Heger, E.; Neumann-Fraune, M.; Beggel, B.; Walker, A.; Timm, J.; et al. Geno2pheno[HCV] - A Web-based Interpretation System to Support Hepatitis C Treatment Decisions in the Era of Direct-Acting Antiviral Agents. PLoS One 2016, 11, e0155869. [Google Scholar] [CrossRef] [PubMed]
- Lontok, E.; Harrington, P.; Howe, A.; Kieffer, T.; Lennerstrand, J.; Lenz, O.; McPhee, F.; Mo, H.; Parkin, N.; Pilot-Matias, T.; et al. Hepatitis C virus drug resistance-associated substitutions: State of the art summary. Hepatology 2015, 62, 1623–1632. [Google Scholar] [CrossRef] [PubMed]
- Feld, J.J. Resistance testing: Interpretation and incorporation into HCV treatment algorithms. Clin Liver Dis (Hoboken) 2017, 9, 115–120. [Google Scholar] [CrossRef] [PubMed]
- Kliemann, D.A.; Tovo, C.V.; Gorini da Veiga, A.B.; Machado, A.L.; West, J. Genetic Barrier to Direct Acting Antivirals in HCV Sequences Deposited in the European Databank. PLoS One 2016, 11, e0159924. [Google Scholar] [CrossRef] [PubMed]
- Cento, V.; Mirabelli, C.; Salpini, R.; Dimonte, S.; Artese, A.; Costa, G.; Mercurio, F.; Svicher, V.; Parrotta, L.; Bertoli, A.; et al. HCV genotypes are differently prone to the development of resistance to linear and macrocyclic protease inhibitors. PLoS One 2012, 7, e39652. [Google Scholar] [CrossRef]
- Vidal, L.L.; Soares, M.A.; Santos, A.F. NS3 protease polymorphisms and genetic barrier to drug resistance of distinct hepatitis C virus genotypes from worldwide treatment-naïve subjects. J Viral Hepat 2016, 23, 840–849. [Google Scholar] [CrossRef]
- de Salazar, A.; Dietz, J.; di Maio, V.C.; Vermehren, J.; Paolucci, S.; Müllhaupt, B.; Coppola, N.; Cabezas, J.; Stauber, R.E.; Puoti, M.; et al. Prevalence of resistance-associated substitutions and retreatment of patients failing a glecaprevir/pibrentasvir regimen. J Antimicrob Chemother 2020, 75, 3349–3358. [Google Scholar] [CrossRef] [PubMed]
- Pisaturo, M.; Starace, M.; Minichini, C.; De Pascalis, S.; Occhiello, L.; Fraia, A.D.; Messina, V.; Sangiovanni, V.; Claar, E.; Coppola, N. Virological patterns of hepatitis C virus patients with failure to the current-generation direct-acting antivirals. Int J Antimicrob Agents 2020, 56, 106067. [Google Scholar] [CrossRef]
- Sarrazin, C.; Cooper, C.L.; Manns, M.P.; Reddy, K.R.; Kowdley, K.V.; Roberts, S.K.; Dvory-Sobol, H.; Svarovskia, E.; Martin, R.; Camus, G.; et al. No impact of resistance-associated substitutions on the efficacy of sofosbuvir, velpatasvir, and voxilaprevir for 12 weeks in HCV DAA-experienced patients. J Hepatol 2018, 69, 1221–1230. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.D.; Maitra, S.; Singh, N.; Tyagi, P.; Ashraf, A.; Kumar, R.; Shalimar. Systematic review with meta-analysis: impact of baseline resistance-associated substitutions on the efficacy of glecaprevir/pibrentasvir among chronic hepatitis C patients. Aliment Pharmacol Ther 2020, 51, 490–504. [Google Scholar] [CrossRef] [PubMed]
- Sayan, M.; Yıldırım, F.S.; Akhan, S.; Yıldırım, A.A.; Şirin, G.; Cabalak, M.; Demir, M.; Can, S.; Ersöz, G.; Altıntaş, E.; et al. NS5A resistance - associated substitutions in chronic hepatitis C patients with direct acting antiviral treatment failure in Turkey. Int J Infect Dis 2020, 95, 84–89. [Google Scholar] [CrossRef] [PubMed]
- Ridruejo, E.; Pereson, M.J.; Flichman, D.M.; Di Lello, F.A. Hepatitis C virus treatment failure: Clinical utility for testing resistance-associated substitutions. World J Hepatol 2021, 13, 1069–1078. [Google Scholar] [CrossRef] [PubMed]
- Wyles, D.L.; Luetkemeyer, A.F. Understanding Hepatitis C Virus Drug Resistance: Clinical Implications for Current and Future Regimens. Top Antivir Med 2017, 25, 103–109. [Google Scholar] [PubMed]
- Onorato, L.; Pisaturo, M.; Starace, M.; Minichini, C.; Di Fraia, A.; Astorri, R.; Coppola, N. Virological Factors Associated with Failure to the Latest Generation of Direct Acting Agents (DAA) and Re-Treatment Strategy: A Narrative Review. Viruses 2021, 13, 432. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharjee, C.; Singh, M.; Das, D.; Chaudhuri, S.; Mukhopadhyay, A. Current therapeutics against HCV. Virusdisease 2021, 32, 228–243. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M. Era of direct acting anti-viral agents for the treatment of hepatitis C. World J Hepatol 2018, 10, 670–684. [Google Scholar] [CrossRef]
- Sayers, E.W.; Cavanaugh, M.; Clark, K.; Ostell, J.; Pruitt, K.D.; Karsch-Mizrachi, I. GenBank. Nucleic Acids Res 2020, 48, D84–d86. [Google Scholar] [CrossRef]
- Kuiken, C.; Hraber, P.; Thurmond, J.; Yusim, K. The hepatitis C sequence database in Los Alamos. Nucleic Acids Res 2008, 36, D512–516. [Google Scholar] [CrossRef]
- Yusim, K.; Richardson, R.; Tao, N.; Dalwani, A.; Agrawal, A.; Szinger, J.; Funkhouser, R.; Korber, B.; Kuiken, C. Los alamos hepatitis C immunology database. Appl Bioinformatics 2005, 4, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Villesen, P. FaBox: an online toolbox for fasta sequences. Molecular Ecology Notes 2007, 7, 965–968. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Struck, D.; Lawyer, G.; Ternes, A.M.; Schmit, J.C.; Bercoff, D.P. COMET: adaptive context-based modeling for ultrafast HIV-1 subtype identification. Nucleic Acids Res 2014, 42, e144. [Google Scholar] [CrossRef] [PubMed]
- AASLD-IDSA HCV Guidance Panel. Hepatitis C Guidance 2018 Update: AASLD-IDSA Recommendations for Testing, Managing, and Treating Hepatitis C Virus Infection. Clin Infect Dis 2018, 67, 1477–1492. [Google Scholar] [CrossRef] [PubMed]
- Svarovskaia, E.S.; Dvory-Sobol, H.; Parkin, N.; Hebner, C.; Gontcharova, V.; Martin, R.; Ouyang, W.; Han, B.; Xu, S.; Ku, K.; et al. Infrequent development of resistance in genotype 1-6 hepatitis C virus-infected subjects treated with sofosbuvir in phase 2 and 3 clinical trials. Clin Infect Dis 2014, 59, 1666–1674. [Google Scholar] [CrossRef] [PubMed]
- Eltahla, A.A.; Leung, P.; Pirozyan, M.R.; Rodrigo, C.; Grebely, J.; Applegate, T.; Maher, L.; Luciani, F.; Lloyd, A.R.; Bull, R.A. Dynamic evolution of hepatitis C virus resistance-associated substitutions in the absence of antiviral treatment. Scientific Reports 2017, 7, 41719. [Google Scholar] [CrossRef] [PubMed]
- Bertoli, A.; Sorbo, M.C.; Aragri, M.; Lenci, I.; Teti, E.; Polilli, E.; Di Maio, V.C.; Gianserra, L.; Biliotti, E.; Masetti, C.; et al. Prevalence of Single and Multiple Natural NS3, NS5A and NS5B Resistance-Associated Substitutions in Hepatitis C Virus Genotypes 1-4 in Italy. Sci Rep 2018, 8, 8988. [Google Scholar] [CrossRef] [PubMed]
- Paolucci, S.; Fiorina, L.; Mariani, B.; Gulminetti, R.; Novati, S.; Barbarini, G.; Bruno, R.; Baldanti, F. Naturally occurring resistance mutations to inhibitors of HCV NS5A region and NS5B polymerase in DAA treatment-naïve patients. Virol J 2013, 10, 355. [Google Scholar] [CrossRef] [PubMed]
- Martínez, A.P.; García, G.; Ridruejo, E.; Culasso, A.C.; Pérez, P.S.; Pereson, M.J.; Neukam, K.; Flichman, D.; Di Lello, F.A. Hepatitis C virus genotype 1 infection: Prevalence of NS5A and NS5B resistance-associated substitutions in naïve patients from Argentina. J Med Virol 2019, 91, 1970–1978. [Google Scholar] [CrossRef] [PubMed]
- Valutite, D.; Ostankova, Y.; Semenov, A.; Lyalina, L.; Totolian, A. Distribution of Primary Resistance Mutations in Saint Petersburg in Patients with Chronic Hepatitis C. Diagnostics (Basel) 2022, 12, 1054. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.W.; Li, H.; Ren, H.; Hu, P. Global prevalence of pre-existing HCV variants resistant to direct-acting antiviral agents (DAAs): mining the GenBank HCV genome data. Sci Rep 2016, 6, 20310. [Google Scholar] [CrossRef] [PubMed]
- Kitrinos, K.M.; Corsa, A.C.; Worth, A.; Hedskog, C.; Brainard, D.M.; Miller, M.D.; Mo, H. Nonstructural protein 5A resistance profile in patients with chronic hepatitis C treated with ledipasvir-containing regimens without sofosbuvir. J Viral Hepat 2018, 25, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Howe, A.Y.M.; Rodrigo, C.; Cunningham, E.B.; Douglas, M.W.; Dietz, J.; Grebely, J.; Popping, S.; Sfalcin, J.A.; Parczewski, M.; Sarrazin, C.; et al. Characteristics of hepatitis C virus resistance in an international cohort after a decade of direct-acting antivirals. JHEP Rep 2022, 4, 100462. [Google Scholar] [CrossRef] [PubMed]
- Wyles, D.; Dvory-Sobol, H.; Svarovskaia, E.S.; Doehle, B.P.; Martin, R.; Afdhal, N.H.; Kowdley, K.V.; Lawitz, E.; Brainard, D.M.; Miller, M.D.; et al. Post-treatment resistance analysis of hepatitis C virus from phase II and III clinical trials of ledipasvir/sofosbuvir. J Hepatol 2017, 66, 703–710. [Google Scholar] [CrossRef] [PubMed]
- Lahser, F.; Galloway, A.; Hwang, P.; Palcza, J.; Brunhofer, J.; Wahl, J.; Robertson, M.; Barr, E.; Black, T.; Asante-Appiah, E.; et al. Interim analysis of a 3-year follow-up study of NS5A and NS3 resistance-associated substitutions after treatment with grazoprevir-containing regimens in participants with chronic HCV infection. Antivir Ther 2018, 23, 593–603. [Google Scholar] [CrossRef] [PubMed]
- Kjellin, M.; Kileng, H.; Akaberi, D.; Palanisamy, N.; Duberg, A.S.; Danielsson, A.; Kristiansen, M.G.; Nöjd, J.; Aleman, S.; Gutteberg, T.; et al. Effect of the baseline Y93H resistance-associated substitution in HCV genotype 3 for direct-acting antiviral treatment: real-life experience from a multicenter study in Sweden and Norway. Scand J Gastroenterol 2019, 54, 1042–1050. [Google Scholar] [CrossRef] [PubMed]
- Perales, C.; Quer, J.; Gregori, J.; Esteban, J.I.; Domingo, E. Resistance of Hepatitis C Virus to Inhibitors: Complexity and Clinical Implications. Viruses 2015, 7, 5746–5766. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, M.; Gupta, E. Emerging resistance to directly-acting antiviral therapy in treatment of chronic Hepatitis C infection-A brief review of literature. J Family Med Prim Care 2020, 9, 531–538. [Google Scholar] [CrossRef] [PubMed]
- Sharafi, H.; Alavian, S.M. Hepatitis C resistance to NS5A inhibitors: Is it going to be a problem? World J Hepatol 2018, 10, 543–548. [Google Scholar] [CrossRef] [PubMed]
- Sarrazin, C. Treatment failure with DAA therapy: Importance of resistance. J Hepatol 2021, 74, 1472–1482. [Google Scholar] [CrossRef]
- Li, H.C.; Yang, C.H.; Lo, S.Y. Hepatitis C Viral Replication Complex. Viruses 2021, 13, 520. [Google Scholar] [CrossRef] [PubMed]
- Scheel, T.K.; Gottwein, J.M.; Mikkelsen, L.S.; Jensen, T.B.; Bukh, J. Recombinant HCV variants with NS5A from genotypes 1-7 have different sensitivities to an NS5A inhibitor but not interferon-α. Gastroenterology 2011, 140, 1032–1042. [Google Scholar] [CrossRef] [PubMed]
- Nakamoto, S.; Kanda, T.; Wu, S.; Shirasawa, H.; Yokosuka, O. Hepatitis C virus NS5A inhibitors and drug resistance mutations. World J Gastroenterol 2014, 20, 2902–2912. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Williford, S.E.; McGivern, D.R.; Burch, C.L.; Hu, F.; Benzine, T.; Ingravallo, P.; Asante-Appiah, E.; Howe, A.Y.M.; Swanstrom, R.; et al. Evolutionary pathways to NS5A inhibitor resistance in genotype 1 hepatitis C virus. Antiviral Res 2018, 158, 45–51. [Google Scholar] [CrossRef]
- Wyles, D.; Mangia, A.; Cheng, W.; Shafran, S.; Schwabe, C.; Ouyang, W.; Hedskog, C.; McNally, J.; Brainard, D.M.; Doehle, B.P.; et al. Long-term persistence of HCV NS5A resistance-associated substitutions after treatment with the HCV NS5A inhibitor, ledipasvir, without sofosbuvir. Antivir Ther 2018, 23, 229–238. [Google Scholar] [CrossRef]
- Kyuregyan, K.K.; Kichatova, V.S.; Karlsen, A.A.; Isaeva, O.V.; Solonin, S.A.; Petkov, S.; Nielsen, M.; Isaguliants, M.G.; Mikhailov, M.I. Factors Influencing the Prevalence of Resistance-Associated Substitutions in NS5A Protein in Treatment-Naive Patients with Chronic Hepatitis C. Biomedicines 2020, 8, 80. [Google Scholar] [CrossRef] [PubMed]
- Nitta, S.; Asahina, Y.; Matsuda, M.; Yamada, N.; Sugiyama, R.; Masaki, T.; Suzuki, R.; Kato, N.; Watanabe, M.; Wakita, T.; et al. Effects of Resistance-Associated NS5A Mutations in Hepatitis C Virus on Viral Production and Susceptibility to Antiviral Reagents. Sci Rep 2016, 6, 34652. [Google Scholar] [CrossRef] [PubMed]
- Nitta, S.; Asahina, Y.; Kato, T.; Tsuchiya, J.; Inoue-Shinomiya, E.; Sato, A.; Tsunoda, T.; Miyoshi, M.; Kawai-Kitahata, F.; Murakawa, M.; et al. Impact of novel NS5A resistance-associated substitutions of hepatitis C virus detected in treatment-experienced patients. Sci Rep 2019, 9, 5722. [Google Scholar] [CrossRef] [PubMed]
- Sarrazin, C.; Zeuzem, S. Resistance to direct antiviral agents in patients with hepatitis C virus infection. Gastroenterology 2010, 138, 447–462. [Google Scholar] [CrossRef] [PubMed]
- Pellicelli, A.M.; Romano, M.; Stroffolini, T.; Mazzoni, E.; Mecenate, F.; Monarca, R.; Picardi, A.; Bonaventura, M.E.; Mastropietro, C.; Vignally, P.; et al. HCV genotype 1a shows a better virological response to antiviral therapy than HCV genotype 1b. BMC Gastroenterology 2012, 12, 162. [Google Scholar] [CrossRef] [PubMed]
- Sarrazin, C.; Dvory-Sobol, H.; Svarovskaia, E.S.; Doehle, B.P.; Pang, P.S.; Chuang, S.M.; Ma, J.; Ding, X.; Afdhal, N.H.; Kowdley, K.V.; et al. Prevalence of Resistance-Associated Substitutions in HCV NS5A, NS5B, or NS3 and Outcomes of Treatment With Ledipasvir and Sofosbuvir. Gastroenterology 2016, 151, 501–512. [Google Scholar] [CrossRef]
- Wang, G.P.; Terrault, N.; Reeves, J.D.; Liu, L.; Li, E.; Zhao, L.; Lim, J.K.; Morelli, G.; Kuo, A.; Levitsky, J.; et al. Prevalence and impact of baseline resistance-associated substitutions on the efficacy of ledipasvir/sofosbuvir or simeprevir/sofosbuvir against GT1 HCV infection. Sci Rep 2018, 8, 3199. [Google Scholar] [CrossRef] [PubMed]
- Mizokami, M.; Dvory-Sobol, H.; Izumi, N.; Nishiguchi, S.; Doehle, B.; Svarovskaia, E.S.; De-Oertel, S.; Knox, S.; Brainard, D.M.; Miller, M.D.; et al. Resistance Analyses of Japanese Hepatitis C-Infected Patients Receiving Sofosbuvir or Ledipasvir/Sofosbuvir Containing Regimens in Phase 3 Studies. J Viral Hepat 2016, 23, 780–788. [Google Scholar] [CrossRef]
- Mawatari, S.; Oda, K.; Tabu, K.; Ijuin, S.; Kumagai, K.; Fujisaki, K.; Hashiguchi, M.; Inada, Y.; Uto, H.; Hiramine, Y.; et al. The co-existence of NS5A and NS5B resistance-associated substitutions is associated with virologic failure in Hepatitis C Virus genotype 1 patients treated with sofosbuvir and ledipasvir. PLoS One 2018, 13, e0198642. [Google Scholar] [CrossRef] [PubMed]
- Hsu, S.J.; Enkhzaya, S.; Lin, Y.Y.; Tseng, T.C.; Khosbayar, T.; Tsai, C.H.; Wang, T.S.; Enkhtuya, D.; Ivshinkhorol, D.; Naranzul, N.; et al. Resistance-associated substitution and ledipasvir/sofosbuvir therapy in Mongolian chronic hepatitis C patients. J Formos Med Assoc 2020, 119, 712–719. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Cehic, D.; Rando, A.; Rodriguez-Frias, F.; Gregori, J.; Costa, J.G.; Carrión, J.A.; Macenlle, R.; Pamplona, J.; Castro-Iglesias, A.; Cañizares, A.; et al. Resistance-associated substitutions after sofosbuvir/velpatasvir/voxilaprevir triple therapy failure. J Viral Hepat 2021, 28, 1319–1324. [Google Scholar] [CrossRef] [PubMed]
- Dietz, J.; Susser, S.; Vermehren, J.; Peiffer, K.H.; Grammatikos, G.; Berger, A.; Ferenci, P.; Buti, M.; Müllhaupt, B.; Hunyady, B.; et al. Patterns of Resistance-Associated Substitutions in Patients With Chronic HCV Infection Following Treatment With Direct-Acting Antivirals. Gastroenterology 2018, 154, 976–988. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Mao, X.; Yu, K.; Suo, C.; Jin, L.; Zhang, T.; Chen, X. Prevalence of HCV resistance-associated substitutions among treatment-failure patients receiving direct-acting antiviral agents. J Viral Hepat 2020, 27, 585–592. [Google Scholar] [CrossRef] [PubMed]
- Dietz, J.; Müllhaupt, B.; Buggisch, P.; Graf, C.; Peiffer, K.H.; Matschenz, K.; Schattenberg, J.M.; Antoni, C.; Mauss, S.; Niederau, C.; et al. Long-term persistence of HCV resistance-associated substitutions after DAA treatment failure. J Hepatol 2023, 78, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Sagnelli, E.; Starace, M.; Minichini, C.; Pisaturo, M.; Macera, M.; Sagnelli, C.; Coppola, N. Resistance detection and re-treatment options in hepatitis C virus-related chronic liver diseases after DAA-treatment failure. Infection 2018, 46, 761–783. [Google Scholar] [CrossRef] [PubMed]
- Jeong, Y.; Jin, B.; Lee, H.W.; Park, H.J.; Park, J.Y.; Kim, D.Y.; Han, K.H.; Ahn, S.H.; Kim, S. Evolution and persistence of resistance-associated substitutions of hepatitis C virus after direct-acting antiviral treatment failures. J Viral Hepat 2018, 25, 1251–1259. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, M.J.; Lipsey, D.; Ganova-Raeva, L.; Punkova, L.T.; Agyemang, L.; Sue, A.; Ramachandran, S.; Khudyakov, Y.; Litwin, A.H. A Phylogenetic Analysis of Hepatitis C Virus Transmission, Relapse, and Reinfection Among People Who Inject Drugs Receiving Opioid Agonist Therapy. J Infect Dis 2020, 222, 488–498. [Google Scholar] [CrossRef] [PubMed]
- Esposito, I.; Marciano, S.; Haddad, L.; Galdame, O.; Franco, A.; Gadano, A.; Flichman, D.; Trinks, J. Prevalence and Factors Related to Natural Resistance-Associated Substitutions to Direct-Acting Antivirals in Patients with Genotype 1 Hepatitis C Virus Infection. Viruses 2018, 11, 3. [Google Scholar] [CrossRef] [PubMed]
- Di Maio, V.C.; Cento, V.; Lenci, I.; Aragri, M.; Rossi, P.; Barbaliscia, S.; Melis, M.; Verucchi, G.; Magni, C.F.; Teti, E.; et al. Multiclass HCV resistance to direct-acting antiviral failure in real-life patients advocates for tailored second-line therapies. Liver Int 2017, 37, 514–528. [Google Scholar] [CrossRef] [PubMed]
- Di Maio, V.C.; Barbaliscia, S.; Teti, E.; Fiorentino, G.; Milana, M.; Paolucci, S.; Pollicino, T.; Morsica, G.; Starace, M.; Bruzzone, B.; et al. Resistance analysis and treatment outcomes in hepatitis C virus genotype 3-infected patients within the Italian network VIRONET-C. Liver Int 2021, 41, 1802–1814. [Google Scholar] [CrossRef] [PubMed]
- Solitano, V.; Plaz Torres, M.C.; Pugliese, N.; Aghemo, A. Management and Treatment of Hepatitis C: Are There Still Unsolved Problems and Unique Populations? Viruses 2021, 13, 1048. [Google Scholar] [CrossRef] [PubMed]
- Graham, C.S.; Swan, T. A path to eradication of hepatitis C in low- and middle-income countries. Antiviral Res 2015, 119, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Ur-Rehman, T.; Ali, M.; Haque, S.; Rasheed, F.; Lougher, E.; Nawaz, M.S.; Paudyal, V. Improving access to the treatment of hepatitis C in low- and middle-income countries: evaluation of a patient assistance programme. International Journal of Clinical Pharmacy 2021, 43, 958–968. [Google Scholar] [CrossRef]
- Zahid, H.; Aslam, K.; Dahl, E.H.; Abbassi, W.; Adan, S.; Van den Bergh, R.; Balinska, M.A.; Luck, N.H. DAA treatment failures in a low-resource setting with a high burden of hepatitis C infections: a case series. Oxf Med Case Reports 2022, 2022, omac049. [Google Scholar] [CrossRef]
- Manns, M.P.; Maasoumy, B. Breakthroughs in hepatitis C research: from discovery to cure. Nature Reviews Gastroenterology & Hepatology 2022, 19, 533–550. [Google Scholar] [CrossRef]
- Jacobson, I.M. The HCV Treatment Revolution Continues: Resistance Considerations, Pangenotypic Efficacy, and Advances in Challenging Populations. Gastroenterol Hepatol (N Y) 2016, 12, 1–11. [Google Scholar] [PubMed]
- Shah, R.; Ahovegbe, L.; Niebel, M.; Shepherd, J.; Thomson, E.C. Non-epidemic HCV genotypes in low- and middle-income countries and the risk of resistance to current direct-acting antiviral regimens. J Hepatol 2021, 75, 462–473. [Google Scholar] [CrossRef] [PubMed]
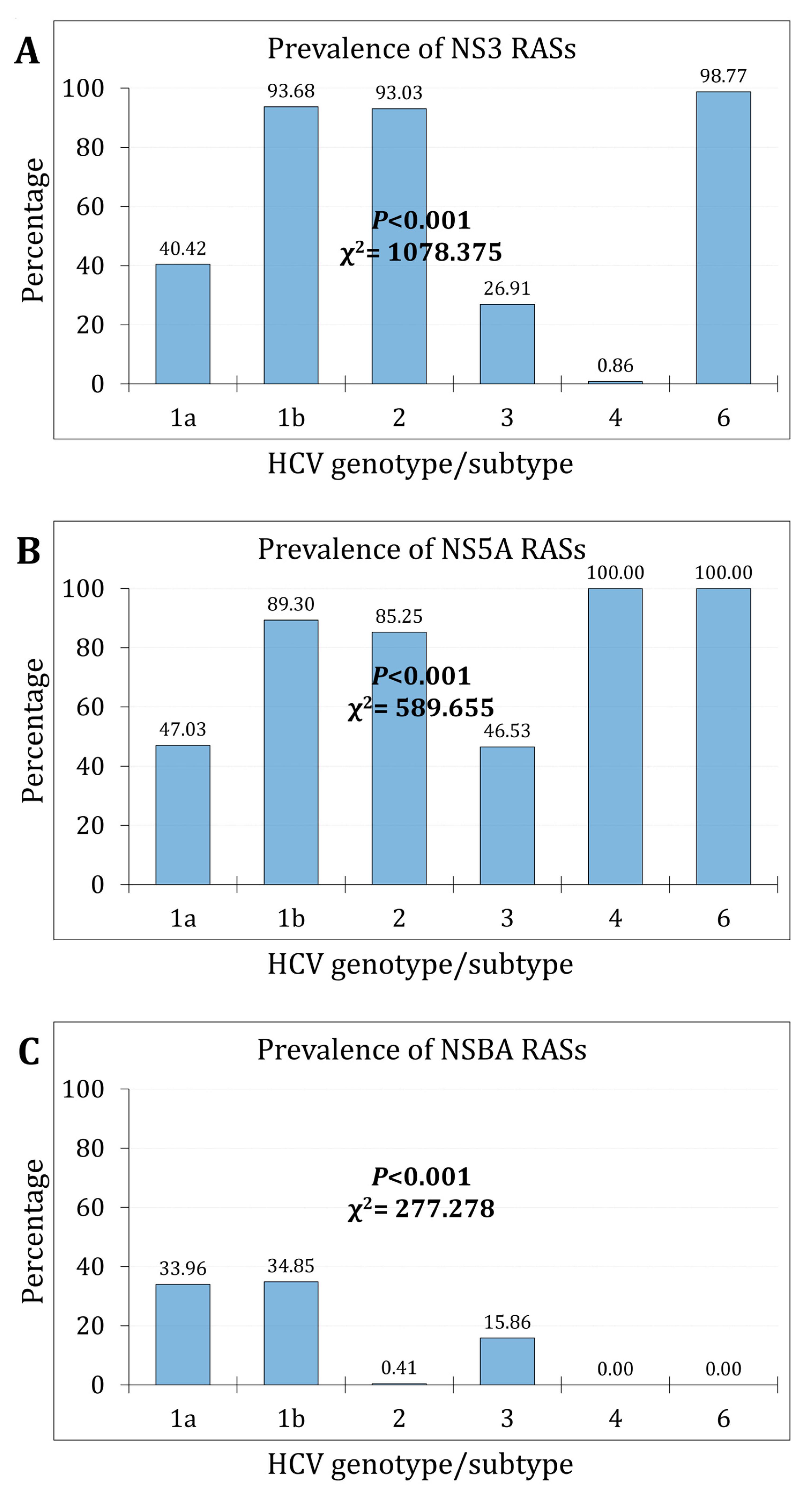
| HCV genotype/subtype: RASs | N (%) | 1a (n=1534) | 1b (n=617) | 2 (n=244) | 3 (n=851) | 4 (n=116) | 6 (n=81) |
|---|---|---|---|---|---|---|---|
| 1a: V36A/C/F/G/L/M 1b: V36A/C/G/L/M 6: V36I |
330 (14.8) | 121 (7.9) | 209 (33.9) | - | - | - | 0 |
| 1a: Q41R 1b: Q41R 3: Q41K 4: Q41R 6: Q41K/R |
0 | 0 | 0 | - | 0 | 0 | 0 |
| 1a: F43I/L/S/V 1b: F43I/S/V 2: F43V |
0 | 0 | 0 | 0 | - | - | - |
| 1a: T54A/S 1b: T54A/C/G/S |
165 (7.7) | 33 (2.2) | 132 (21.4) | - | - | - | - |
| 1a: V55I 1b: V55A 2: V55A/I |
38 (1.6) | 14 (0.9) | 0 | 24 (9.8) | - | - | - |
| 1a: Y56H 1b: Y56H/L/F 2: Y56H/F 3: Y56H 4: Y56H 6: Y56H |
23 (0.7) | 1 (0.1) | 9 (1.5) | 13 (5.3) | 0 | 0 | 0 |
| 1a: Q80K/L/R 1b: Q80H/K/L/R 3: Q80K/R 4: Q80R 6: L80K/Q |
656 (20.5) | 377 (24.6) | 200 (32.4) | - | 0 | 0 | 79 (97.5) |
| 1a: S122G/N/R 1b: S122A/D/G/I/N/R/T 6: S122T |
216 (9.7) | 162 (10.6) | 50 (8.1) | - | - | - | 4 (4.9) |
| 1a: R155G/I/K/M/Q/S/T/V/W 1b: R155C/G/I/K/L/Q/M/S/T/W 3: R155K 4: R155C/K |
11 (0.4) | 8 (0.5) | 2 (0.3) | - | 1 (0.1) | 0 | - |
| 1a: A156G/P/S/T/V 1b: A156G/P/S/T/V 2: A156L/M/T/V 3: A156G/P/T/V 4: A156G/H/K/L/S/T/V 6: A156T/V |
0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1a: V158I 1b: V158I |
2 (0.1) | 1 (0.1) | 1 (0.2) | - | - | - | - |
| 3: A166S/T/Y | 228 (26.8) | - | - | - | 228 (26.8) | - | - |
| 1a: D168A/C/E/F/G/H/I/K/L/N/Q/R/T/V/Y 1b: D168A/C/E/F/G/H/I/K/L/N/Q/T/V/Y 2: D168A/E/F/G/H/N/S/T/V/Y 3: Q168H/K/L/R 4: D168A/E/G/H/T/V 6: D168A/E/G/H/V/Y |
421 (12.2) | 43 (2.8) | 186 (30.1) | 190 (77.9) | 1 (0.1) | 1 (0.9) | 0 |
| 1a: I/V170T 1b: I/V170A/L/T 6: I170V |
7 (0.3) | 0 | 0 | - | - | - | 7 (8.6) |
| M175L | 551 (89.3) | - | 551 (89.3) | - | - | - | - |
| Sequences with any NS3 RASs | 1735 (50.4) | 620 (40.4) | 578 (93.7) | 227 (93.0) | 229 (26.9) | 1 (0.9) | 80 (98.8) |
| HCV genotype/subtype: RASs | N (%) | 1a (n=1533) | 1b (n=617) | 2 (n=244) | 3 (n=851) | 4 (n=116) | 6 (n=81) |
|---|---|---|---|---|---|---|---|
| 1a: K24E/Q/R/T 1b: Q24K 2: T24A/S 3: S24F 6: Q24H |
1051 (31.6) | 502 (32.7) | 344 (55.8) | 205 (84.0) | 0 | - | 0 |
| 1a: K26E | 0 | 0 | - | - | - | - | - |
| 1a: M28A/G/S/T/V 1b: L28A/M/T 2: L/F28C/S 3: M28T/K 4: L28M/S/T/V 6: F/L28A/I/L/M/T/V |
757 (22.7) | 67 (4.4) | 522 (84.6) | 0 | 0 | 84 (72.4) | 81 (100.0) |
| 1a: P29R 1b: P29S, del29 2: P29S |
0 | 0 | 0 | 0 | - | - | - |
| 1a: Q30C/D/E/G/H/K/L/N/R/T/Y, del30 1b: R30G/H/P/Q/S 2: L30H/S 3: A30D/E/K/S 4: L30F/G/H/R/S 6: R30E/H/N/S |
1105 (33.1) | 491 (32.0) | 351 (56.9) | 3 (1.2) | 230 (27.0) | 27 (23.3) | 3 (3.7) |
| 1a: L31I/F/M/P/V 1b: L31F/I/M/V/W 2: L31I/M/V 3: L31F/I/M/P/V 4: M/L31I/V 6: L31I/M/V |
251 (7.5) | 6 (0.4) | 21 (3.4) | 15 (6.1) | 190 (22.3) | 0 | 19 (23.5) |
| 1a: P32L/S, del32 1b: P32F/L/S, del32 6: P32A/L/Q/R/S |
0 | 0 | 0 | - | - | - | 0 |
| 1a: S38F | 0 | 0 | - | - | - | - | - |
| 1a: H58C/D/L/P/R 1b: P58A/D/L/S/R/T 4: T58A/P/S 6: T58A/G/H/N/S |
750 (32.0) | 637 (41.6) | 3 (0.5) | - | - | 76 (65.5) | 34 (42.0) |
| 1b: Q/E62D 3: S62L |
23 (1.6) | - | 6 (1.0) | - | 17 (2.0) | - | - |
| 1a: A92K/T 1b: A92E/K/T/V 2: C92R/S/T/W 3: E92K 6: E92T |
200 (6.0) | 9 (0.6) | 189 (30.6) | 2 (0.8) | 0 | - | 0 |
| 1a: Y93C/F/H/L/N/R/S/T/W 1b: Y93C/H/N/R/S/T 2: Y93F/N/H 3: Y93H/N/S 4: Y93C/H/N/S/R/W 6: T93A/H/N/S |
166 (5.0) | 41 (2.7) | 17 (2.8) | 48 (19.7) | 52 (6.1) | 4 (3.4) | 4 (4.9) |
| Sequences with any NS5A RASs | 2073 (62.0) | 721 (47.0) | 551 (89.3) | 208 (85.2) | 396 (46.5) | 116 (100.0) | 81 (100.0) |
| HCV genotype/subtype: RASs | N (%) | 1a (n=1534) | 1b (n=617) | 2 (n=244) | 3 (n=851) | 4 (n=116) | 6 (n=81) |
|---|---|---|---|---|---|---|---|
| NIs | |||||||
| 3: A150V | 100 (11.8) | - | - | - | 100 (11.8) | - | - |
| 1a: L159F 1b: L159F 2: L159F 3: L159F |
45 (1.4) | 40 (2.6) | 3 (0.5) | 0 | 2 (0.2) | - | - |
| 3: K206E | 43 (5.1) | - | - | - | 43 (5.1) | - | - |
| 1a: S282G/R/T 1b: S282G/R/T 2: S282G/R/T 3: S282G/R/T 4: S282C/G/R/T 6: S282G/R/T |
1/3443 (0.03) | 0 | 0 | 1 (0.4) | 0 | 0 | 0 |
| 1a: C316H/R 1b: C316F/H/N |
9 (0.4) | 5 (0.3) | 4 (0.6) | - | - | - | - |
| 1a: L320I/F/V | 0 | 0 | - | - | - | - | - |
| 1a: V321A 1b: V321I 3: V321A 4: V321A |
0 | 0 | 0 | - | 0 | 0 | - |
| NNI | |||||||
| 1a: L314H | 0 | 0 | - | - | - | - | - |
| 1a: C316Y 1b: C316H/N/Y/W |
5 (0.2) | 0 | 5 (0.8) | - | - | - | - |
| 1b: S368T | 0 | - | 0 | - | - | - | - |
| 1a: A395G | 0 | 0 | - | - | - | - | - |
| 1b: N411S | 0 | - | 0 | - | - | - | - |
| 1a: M414I/T/V 1b: M414I/T/V |
8 (0.4) | 7 (0.5) | 1 (0.2) | - | - | - | - |
| C445F/Y | 203 (32.9) | - | 203 (32.9) | - | - | - | - |
| E446K/Q | 418 (27.2) | 418 (27.2) | - | - | - | - | - |
| 1a: Y448C/H 1b: Y448C/H |
6 (0.3) | 4 (0.3) | 2 (0.3) | - | - | - | - |
| 1a: A553T/V 1b: A553V |
280 (13.0) | 85 (5.5) | 195 (31.6) | - | - | - | - |
| 1a: G554S 1b: G554S |
4 (0.2) | 3 (0.2) | 1 (0.2) | - | - | - | - |
| 1a: Y555H | 0 | 0 | - | - | - | - | - |
| 1a: S556G/R 1b: S556G/R |
343 (15.9) | 142 (9.3) | 201 (32.6) | - | - | - | - |
| 1a: G557R | 0 | 0 | - | - | - | - | - |
| 1a: G558R 1b: G558R |
0 | 0 | 0 | - | - | - | - |
| 1a: D559G/N 1b: D559G/N |
2 (0.09) | 2 (0.1) | 0 | - | - | - | - |
| 1a: Y561H/N | 1 (0.07) | 1 (0.07) | - | - | - | - | - |
| 1a: S565F | 0 | 0 | - | - | - | - | - |
| Sequences with any NS5B RASs | 872 (25.3) | 521 (34.0) | 215 (34.8) | 1 (0.4) | 135 (15.9) | 0 | 0 |
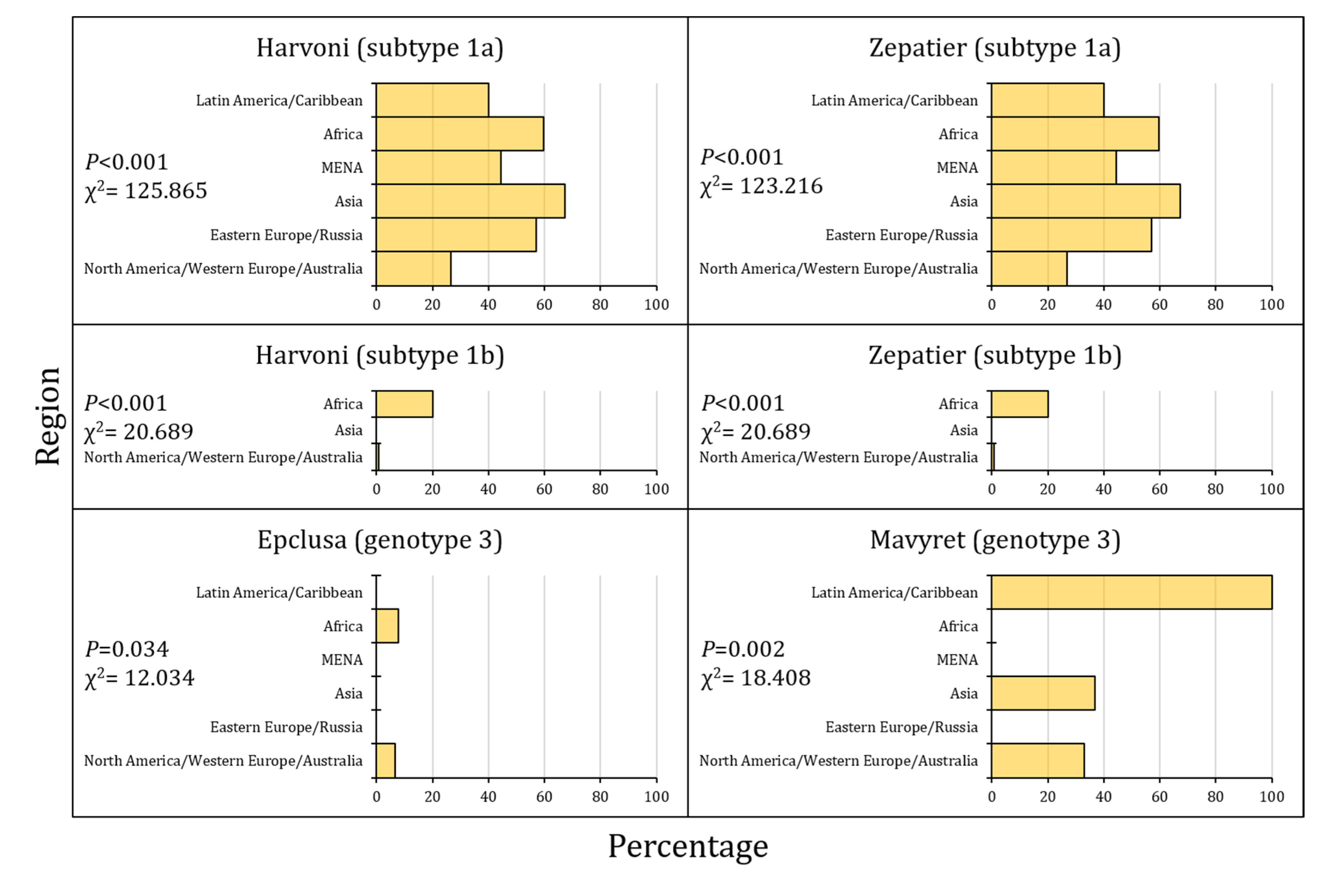
| aa Position in NS5A | 28 | 30 | 31 | 93 |
|---|---|---|---|---|
| Regimen | ||||
| Harvoni (subtype 1a) | - | Q30H/R | L31M/V | Y93C/H/N |
| N (%) | 426 (27.8) | 83 (5.4) | 29 (1.9) | |
| Harvoni (subtype 1b) | - | - | L31V | Y93H |
| N (%) | 0 | 17 (2.8) | ||
| Zepatier (subtype 1a) | M28A/T | Q30H/R | L31M/V | Y93C/H/N |
| N (%) | 6 (0.4) | 426 (27.8) | 83 (5.4) | 29 (1.9) |
| Zepatier (subtype 1b) | - | - | - | Y93H |
| N (%) | 17 (2.8) | |||
| Epclusa (genotype 3) | - | - | - | Y93H |
| N (%) | 35 (4.1) | |||
| Mavyret (genotype 3) | - | A30K | - | - |
| N (%) | 211 (24.8) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





