Submitted:
27 June 2024
Posted:
29 June 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Methods
2.1. Cohort Description
2.2. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
2.3. Statistical Analysis
3. Results
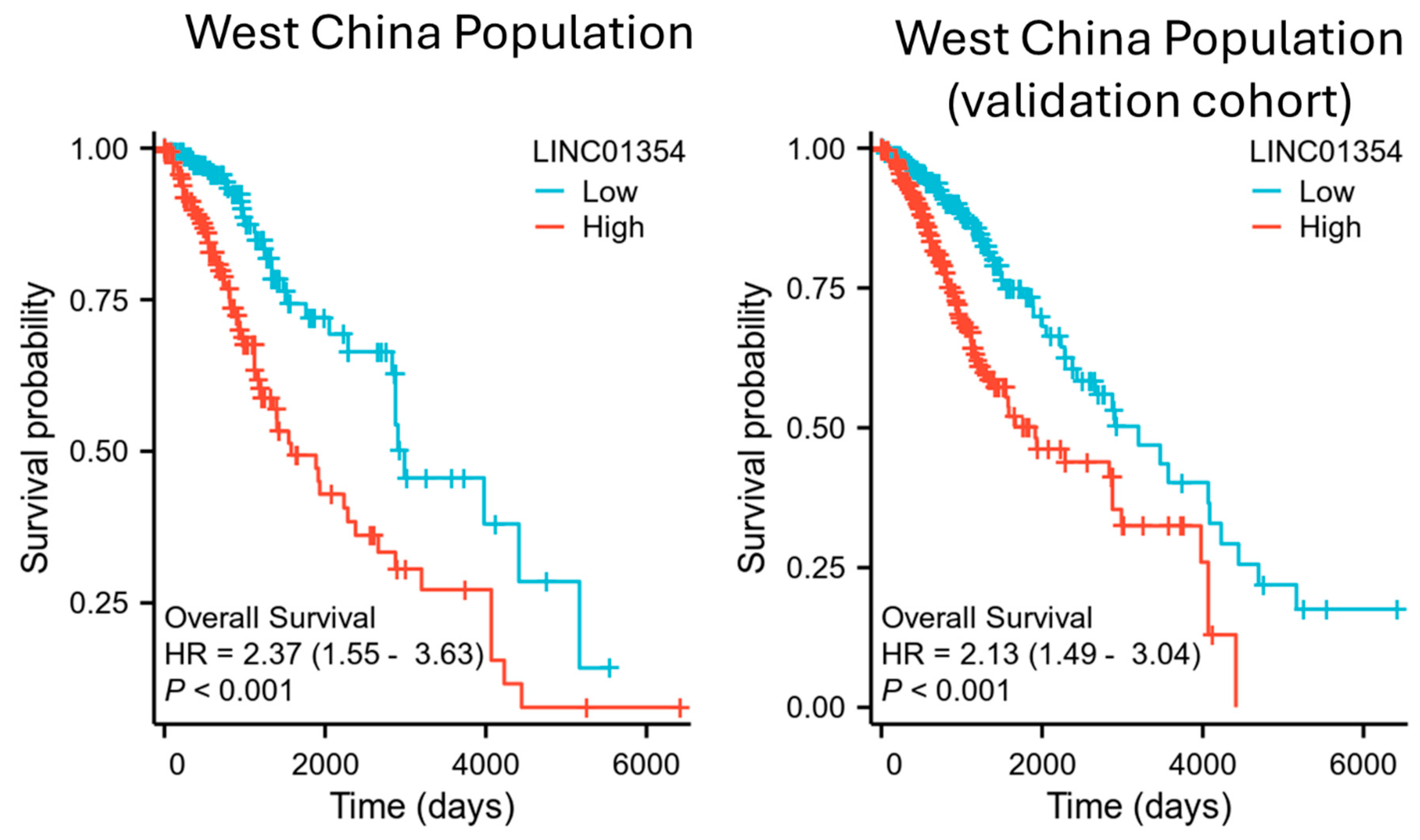
3.1. Survival Association of LINC01354 and Low-Grade Glioma Survival in West China Population
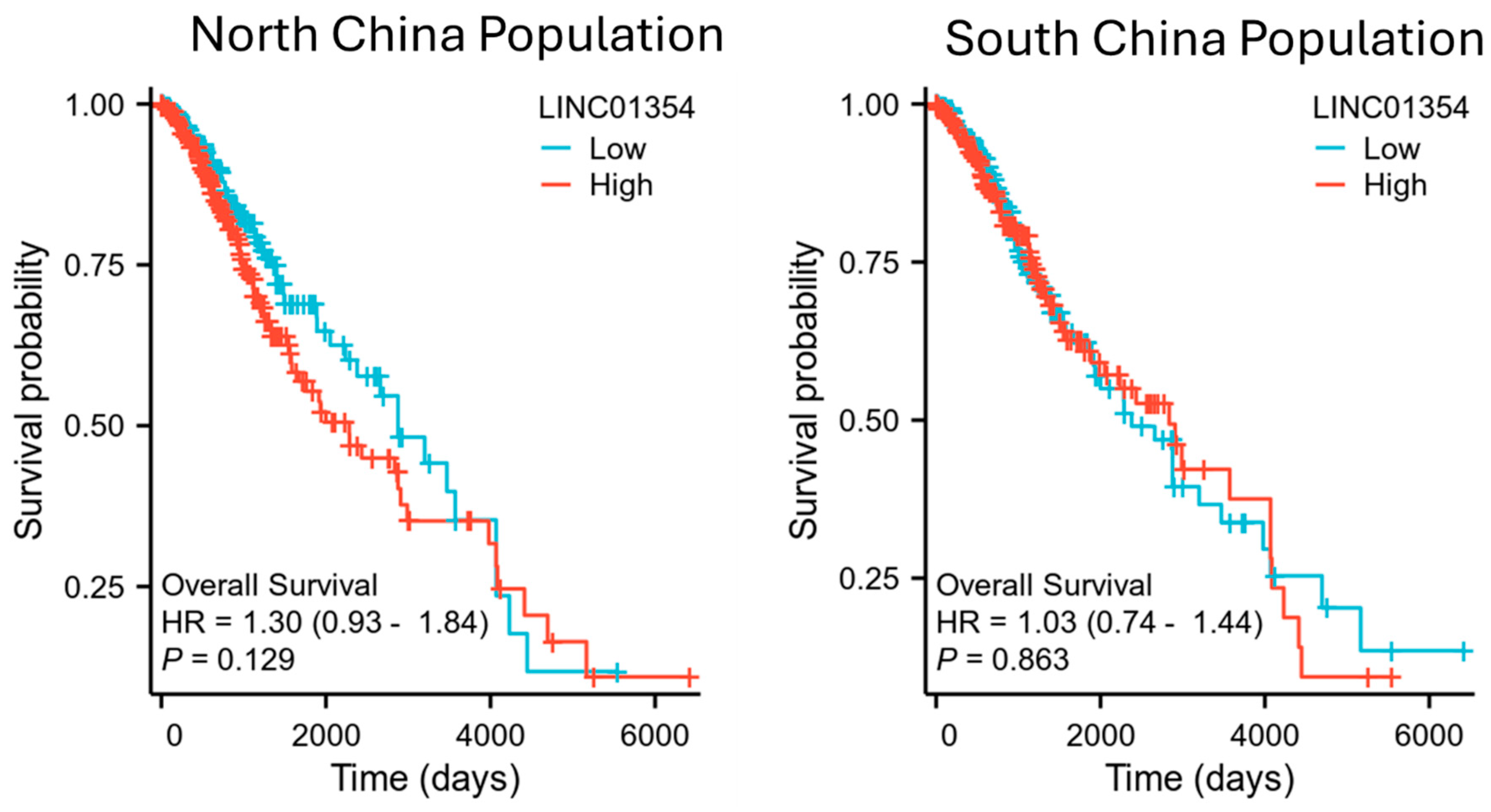
3.2. Survival Association of LINC01354 and Low-Grade Glioma Survival in North and South China Population
3.3. Comparison of the Clinical Information of the Three Cohorts
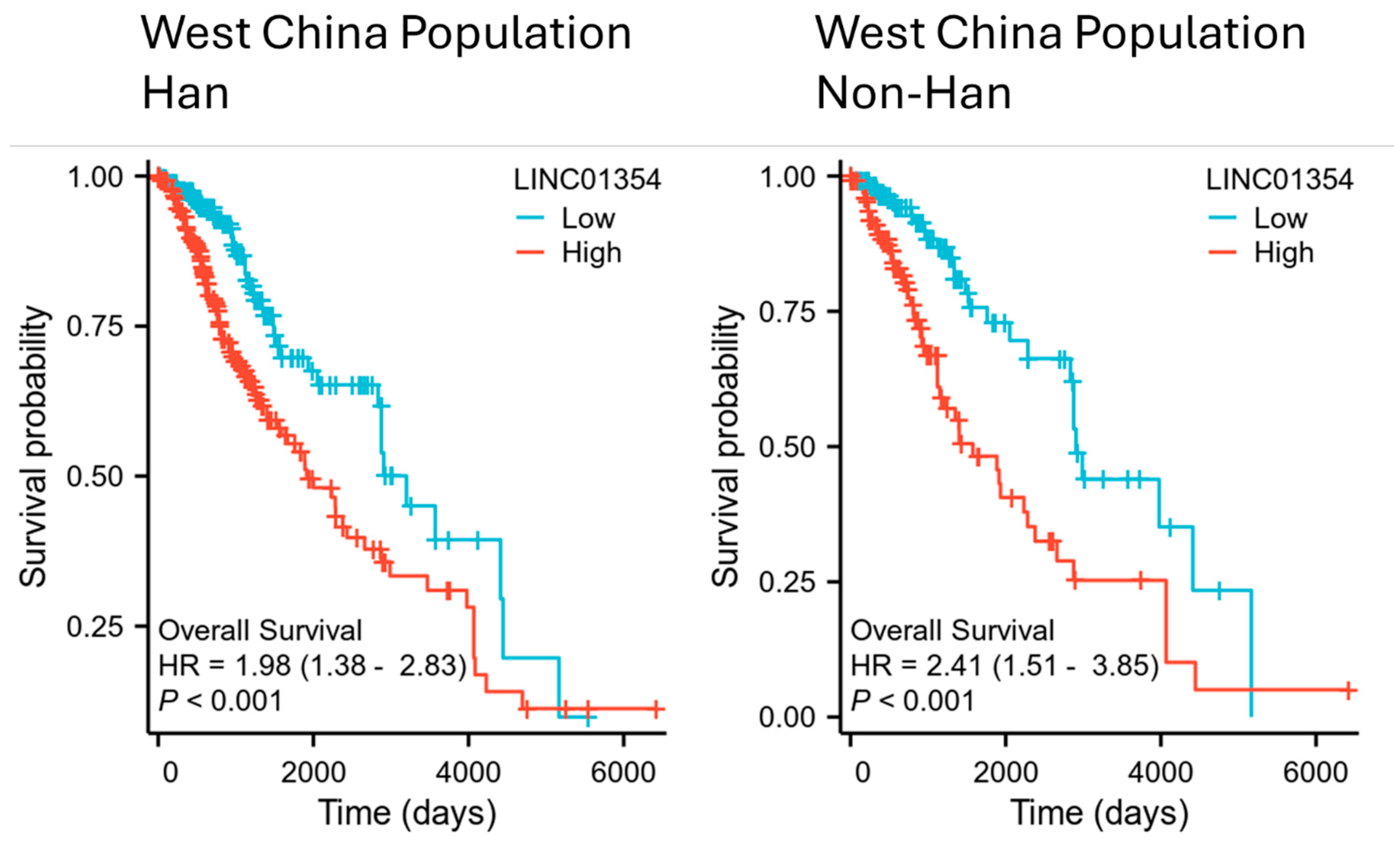
3.4. Comparison of Survival Association of LINC01354 and Low-Grade Glioma Survival in West China Population of Han and Non-Han Patients
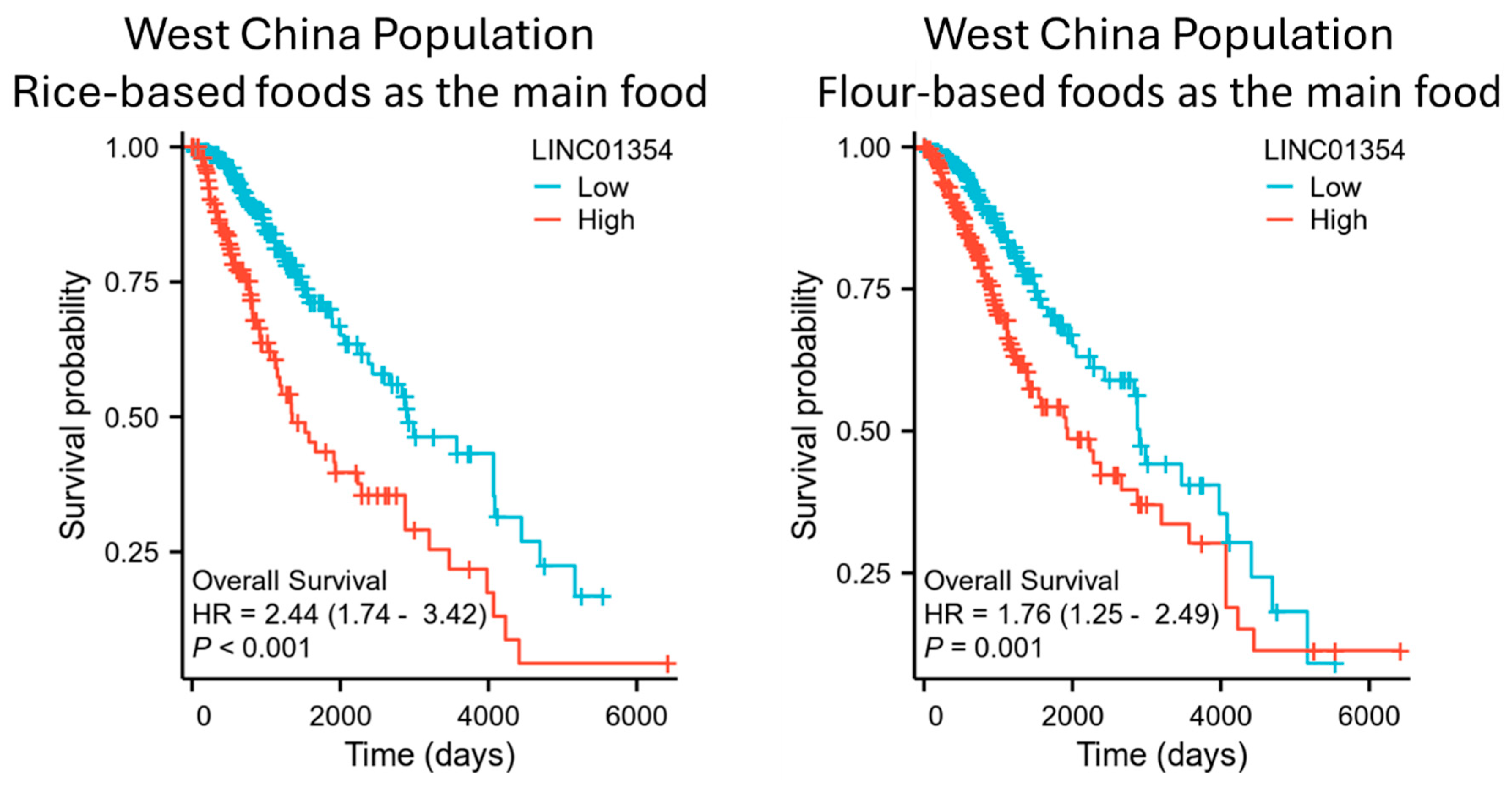
3.5. Comparison of Survival Association of LINC01354 with Low-Grade Glioma in West China Population Based on Main Dietary Staple: Rice-Based vs. Flour-Based Foods
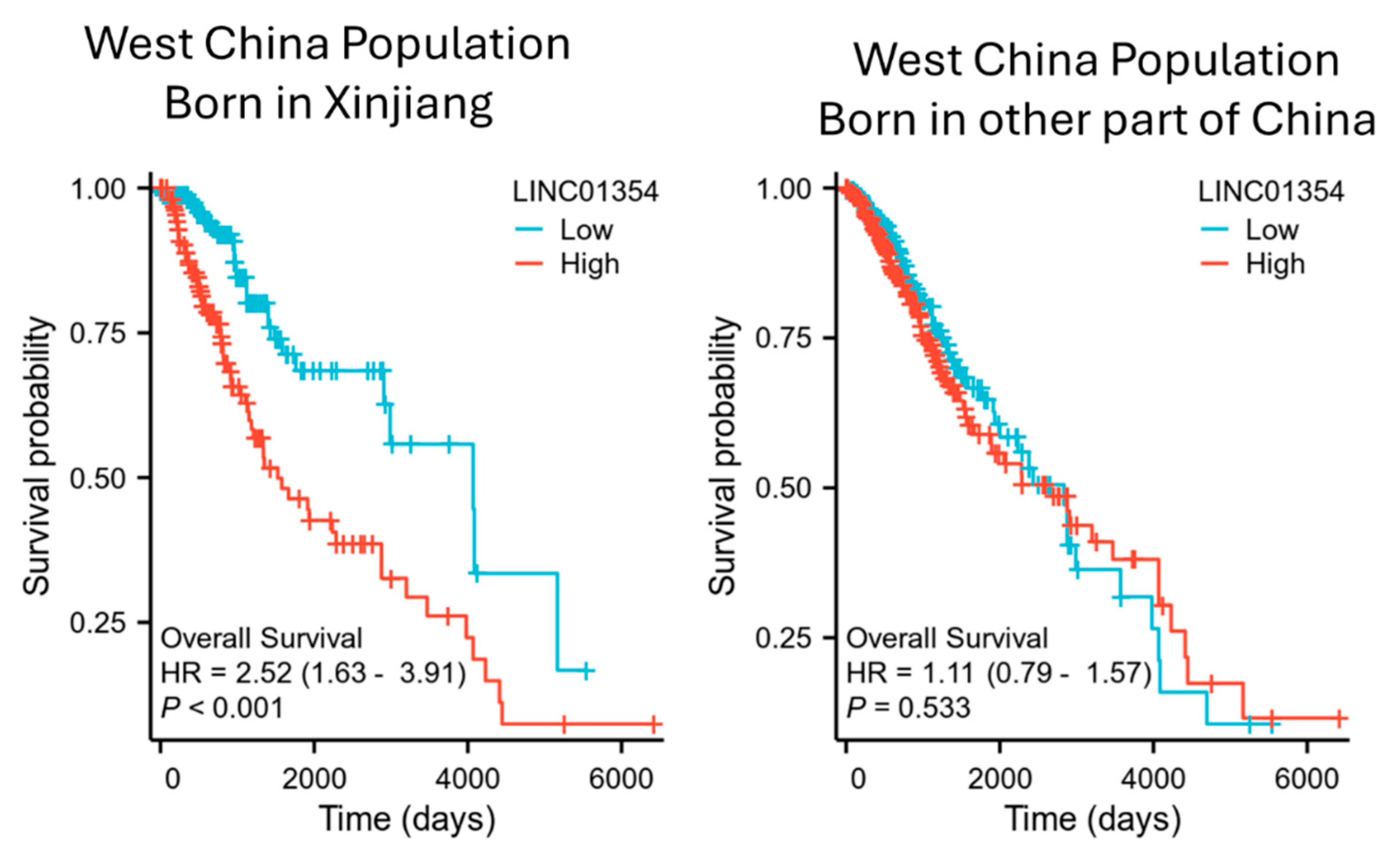
3.6. Comparison of Survival Association of LINC01354 with Low-Grade Glioma in West China Population Based on Birth Locations
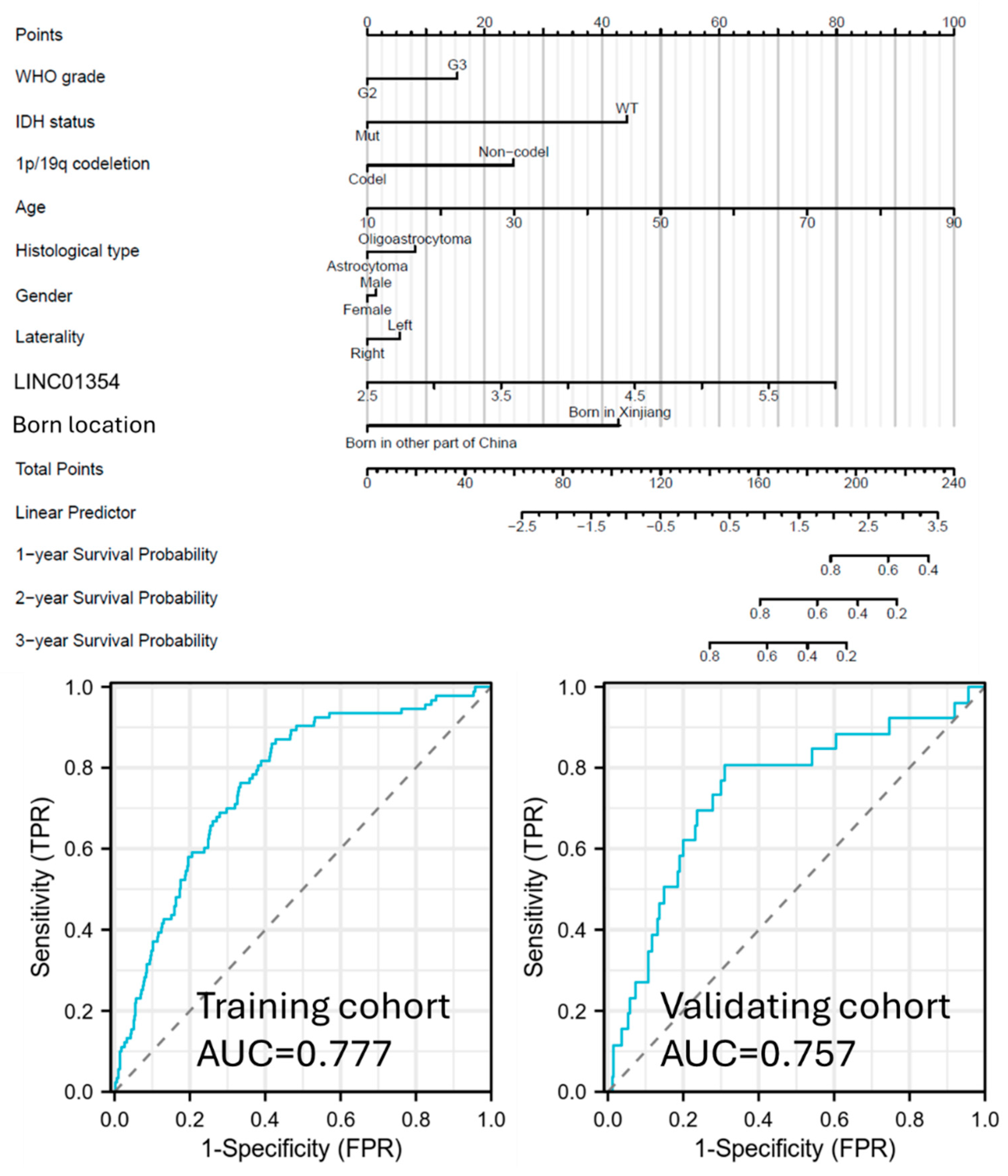
3.7. Clinical Application of LINC01354 as a Prognostic Biomarker for Low-Grade Glioma for West China Population
4. Discussion
Funding
Ethical Declaration
Patient Consent for Publication
Competing Interest
Acknowledgment
Data Availability
References
- Jenkins, R.B.; Wrensch, M.R.; Johnson, D.; Fridley, B.L.; Decker, P.A.; Xiao, Y.; Kollmeyer, T.M.; Rynearson, A.L.; Fink, S.; Rice, T.; et al. Distinct germ line polymorphisms underlie glioma morphologic heterogeneity. Cancer Genet 2011, 204, 13–18. [Google Scholar] [CrossRef] [PubMed]
- Yasinjan, F.; Xing, Y.; Geng, H.; Guo, R.; Yang, L.; Liu, Z.; Wang, H. Immunotherapy: a promising approach for glioma treatment. Frontiers in immunology 2023, 14, 1255611. [Google Scholar] [CrossRef] [PubMed]
- Hakar, M.H.; Wood, M.D. Updates in Pediatric Glioma Pathology. Surg Pathol Clin 2020, 13, 801–816. [Google Scholar] [CrossRef] [PubMed]
- Ghantasala, S.; Gollapalli, K.; Epari, S.; Moiyadi, A.; Srivastava, S. Glioma tumor proteomics: clinically useful protein biomarkers and future perspectives. Expert Rev Proteomics 2020, 17, 221–232. [Google Scholar] [CrossRef] [PubMed]
- Perez, A.; Huse, J.T. The Evolving Classification of Diffuse Gliomas: World Health Organization Updates for 2021. Curr Neurol Neurosci Rep 2021, 21, 67. [Google Scholar] [CrossRef]
- Cooley, L.D.; Lansdon, L.A.; Laurence, K.; Herriges, J.C.; Zhang, L.; Repnikova, E.A.; Joyce, J.; Thakor, P.; Warren, L.; Smith, S.C.; et al. Integrated genetic profiling of archival pediatric high-grade glial tumors and reassessment with 2021 WHO classification of paediatric CNS tumours. Cancer Genet 2023, 274-275, 10–20. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Tang, T. A bioinformatic study of IGFBPs in glioma regarding their diagnostic, prognostic, and therapeutic prediction value. Am J Transl Res 2023, 15, 2140–2155. [Google Scholar] [PubMed]
- Liu, H.; Weng, J. A Comprehensive Bioinformatic Analysis of Cyclin-dependent Kinase 2 (CDK2) in Glioma. Gene 2022, 146325. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Weng, J. A Pan-Cancer Bioinformatic Analysis of RAD51 Regarding the Values for Diagnosis, Prognosis, and Therapeutic Prediction. Frontiers in oncology 2022, 12. [Google Scholar] [CrossRef]
- Liu, H.; Tang, T. Pan-cancer genetic analysis of cuproptosis and copper metabolism-related gene set. Frontiers in oncology 2022, 12, 952290. [Google Scholar] [CrossRef]
- Liu, H.; Tang, T. Pan-cancer genetic analysis of disulfidptosis-related gene set. bioRxiv 2002. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Dilger, J.P.; Lin, J. A pan-cancer-bioinformatic-based literature review of TRPM7 in cancers. Pharmacology & Therapeutics 2022, 108302. [Google Scholar] [CrossRef]
- Liu, H. Pan-cancer profiles of the cuproptosis gene set. American journal of cancer research 2022, 12, 4074–4081. [Google Scholar] [PubMed]
- Lin, W.; Zhou, Q.; Wang, C.Q.; Zhu, L.; Bi, C.; Zhang, S.; Wang, X.; Jin, H. LncRNAs regulate metabolism in cancer. International journal of biological sciences 2020, 16, 1194–1206. [Google Scholar] [CrossRef] [PubMed]
- Ghorbani, A.; Hosseinie, F.; Khorshid Sokhangouy, S.; Islampanah, M.; Khojasteh-Leylakoohi, F.; Maftooh, M.; Nassiri, M.; Hassanian, S.M.; Ghayour-Mobarhan, M.; Ferns, G.A.; et al. The prognostic, diagnostic, and therapeutic impact of Long noncoding RNAs in gastric cancer. Cancer Genet 2024, 282-283, 14–26. [Google Scholar] [CrossRef] [PubMed]
- Coonrod, E.; Othoum, G.; Nickless, A.; Zhang, J.; Inkman, M.; Zhao, S.; Dang, H.; Webster, J.; Rozycki, E.; Fontes, S. 67. Long noncoding RNAs encoding peptides in cancer. Cancer Genetics 2022, 268, 22. [Google Scholar] [CrossRef]
- Kciuk, M.; Yahya, E.B.; Mohamed, M.M.I.; Abdulsamad, M.A.; Allaq, A.A.; Gielecińska, A.; Kontek, R. Insights into the Role of LncRNAs and miRNAs in Glioma Progression and Their Potential as Novel Therapeutic Targets. Cancers 2023, 15. [Google Scholar] [CrossRef] [PubMed]
- Peng, W.X.; Koirala, P.; Mo, Y.Y. LncRNA-mediated regulation of cell signaling in cancer. Oncogene 2017, 36, 5661–5667. [Google Scholar] [CrossRef]
- Fonseca Á, Y.G.; González-Giraldo, Y.; Santos, J.G.; Aristizábal-Pachón, A.F. The hsa-miR-516a-5p and hsa-miR-516b-5p microRNAs reduce the migration and invasion on T98G glioblastoma cell line. Cancer Genet 2023, 270-271, 12–21. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, Y.; Wu, J.; Liu, X.; Li, L.; Zhang, J. LncRNA FOXD2-AS1 promotes the growth, invasion and migration of OSCC cells by regulating the MiR-185-5p/PLOD1/Akt/mTOR pathway. Cancer Genet 2024, 284-285, 48–57. [Google Scholar] [CrossRef]
- Chen, Y.; Hu, D.; Wang, F.; Huang, C.; Xie, H.; Jin, L. A systematic framework for identifying prognostic necroptosis-related lncRNAs and verification of lncRNA CRNDE/miR-23b-3p/IDH1 regulatory axis in glioma. Aging 2023, 15, 12296–12313. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Tang, T. MAPK signaling pathway-based glioma subtypes, machine-learning risk model, and key hub proteins identification. Scientific Reports 2023, 13, 19055. [Google Scholar] [CrossRef]
- Liu, H. Expression and potential immune involvement of cuproptosis in kidney renal clear cell carcinoma. Cancer Genetics 2023, 274-275, 21–25. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Li, Y. Potential roles of Cornichon Family AMPA Receptor Auxiliary Protein 4 (CNIH4) in head and neck squamous cell carcinoma. Cancer biomarkers : section A of Disease markers 2022. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, H. Clinical powers of Aminoacyl tRNA Synthetase Complex Interacting Multifunctional Protein 1 (AIMP1) for head-neck squamous cell carcinoma. Cancer biomarkers : section A of Disease markers 2022. [Google Scholar] [CrossRef]
- Chen, G.; Wang, C.; Wang, J.; Yin, S.; Gao, H.; Xiang, L.U.; Liu, H.; Xiong, Y.; Wang, P.; Zhu, X.; et al. Antiosteoporotic effect of icariin in ovariectomized rats is mediated via the Wnt/beta-catenin pathway. Experimental and therapeutic medicine 2016, 12, 279–287. [Google Scholar] [CrossRef]
- Liu, H.; Xiong, Y.; Gao, H.; Yin, S.; Wang, J.; Chen, G.; Wang, C.; Xiang, L.; Wang, P.; Fang, J. Icariin improves osteoporosis, inhibits the expression of PPAR gamma, C/EBP gamma, FABP4 mRNA, N1ICD, and jagged1 proteins and increases Notch2 mRNA in ovariectomized rats. In Proceedings of the International journal of molecular medicine; 2016; p. S77. [Google Scholar]
- Wang, C.; Chen, G.; Wang, J.; Liu, H.; Xiong, Y.; Wang, P.; Yang, L.; Zhu, X.; Zhang, R. Effect of Herba Epimedium Extract on Bone Mineral Density and Microstructure in Ovariectomised Rat. Journal of Pharmaceutical and Biomedical Sciences 2016, 6. [Google Scholar]
- Li, X.; Peng, B.; Zhu, X.; Wang, P.; Xiong, Y.; Liu, H.; Sun, K.; Wang, H.; Ou, L.; Wu, Z.; et al. Changes in related circular RNAs following ERbeta knockdown and the relationship to rBMSC osteogenesis. Biochemical and biophysical research communications 2017, 493, 100–107. [Google Scholar] [CrossRef]
- Liu, H.; Xiong, Y.; Zhu, X.; Gao, H.; Yin, S.; Wang, J.; Chen, G.; Wang, C.; Xiang, L.; Wang, P.; et al. Icariin improves osteoporosis, inhibits the expression of PPARgamma, C/EBPalpha, FABP4 mRNA, N1ICD and jagged1 proteins, and increases Notch2 mRNA in ovariectomized rats. Experimental and therapeutic medicine 2017, 13, 1360–1368. [Google Scholar] [CrossRef]
- Wu, Z.; Ou, L.; Wang, C.; Yang, L.; Wang, P.; Liu, H.; Xiong, Y.; Sun, K.; Zhang, R.; Zhu, X. Icaritin induces MC3T3-E1 subclone14 cell differentiation through estrogen receptor-mediated ERK1/2 and p38 signaling activation. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie 2017, 94, 1–9. [Google Scholar] [CrossRef]
- Liu, H.; Xiong, Y.; Wang, H.; Yang, L.; Wang, C.; Liu, X.; Wu, Z.; Li, X.; Ou, L.; Zhang, R.; et al. Effects of water extract from epimedium on neuropeptide signaling in an ovariectomized osteoporosis rat model. Journal of ethnopharmacology 2018, 221, 126–136. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Liu, H.; Xiong, Y.; Yang, L.; Wang, C.; Zhang, R.; Zhu, X. Postmenopausal osteoporosis is associated with the regulation of SP, CGRP, VIP, and NPY. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie 2018, 104, 742–750. [Google Scholar] [CrossRef]
- Hazra, R.; Utama, R.; Naik, P.; Dobin, A.; Spector, D.L. Identification of glioblastoma stem cell-associated lncRNAs using single-cell RNA sequencing datasets. Stem Cell Reports 2023, 18, 2056–2070. [Google Scholar] [CrossRef] [PubMed]
- Saw, P.E.; Xu, X.; Chen, J.; Song, E.W. Non-coding RNAs: the new central dogma of cancer biology. Sci China Life Sci 2021, 64, 22–50. [Google Scholar] [CrossRef] [PubMed]
- Zi, H.; Tuo, Z.; He, Q.; Meng, J.; Hu, Y.; Li, Y.; Yang, K. Comprehensive Bioinformatics Analysis of Gasdermin Family of Glioma. Comput Intell Neurosci 2022, 2022, 9046507. [Google Scholar] [CrossRef] [PubMed]
- Su, F.; Duan, J.; Zhu, J.; Fu, H.; Zheng, X.; Ge, C. Long non-coding RNA nuclear paraspeckle assembly transcript 1 regulates ionizing radiation-induced pyroptosis via microRNA-448/gasdermin E in colorectal cancer cells. Int J Oncol 2021, 59. [Google Scholar] [CrossRef]
- Li, C.; Song, H.; Chen, C.; Chen, S.; Zhang, Q.; Liu, D.; Li, J.; Dong, H.; Wu, Y.; Liu, Y. LncRNA PVT1 Knockdown Ameliorates Myocardial Ischemia Reperfusion Damage via Suppressing Gasdermin D-Mediated Pyroptosis in Cardiomyocytes. Front Cardiovasc Med 2021, 8, 747802. [Google Scholar] [CrossRef]
- Zhang, Y.; Song, Z.; Li, X.; Xu, S.; Zhou, S.; Jin, X.; Zhang, H. Long noncoding RNA KCNQ1OT1 induces pyroptosis in diabetic corneal endothelial keratopathy. American journal of physiology. Cell physiology 2020, 318, C346–c359. [Google Scholar] [CrossRef] [PubMed]
- Wu, A.; Sun, W.; Mou, F. lncRNA-MALAT1 promotes high glucose-induced H9C2 cardiomyocyte pyroptosis by downregulating miR-141-3p expression. Mol Med Rep 2021, 23. [Google Scholar] [CrossRef]
- Ren, N.; Jiang, T.; Wang, C.; Xie, S.; Xing, Y.; Piao, D.; Zhang, T.; Zhu, Y. LncRNA ADAMTS9-AS2 inhibits gastric cancer (GC) development and sensitizes chemoresistant GC cells to cisplatin by regulating miR-223-3p/NLRP3 axis. Aging 2020, 12, 11025–11041. [Google Scholar] [CrossRef]
- Yang, G.; Yang, C.; She, Y.; Shen, Z.; Gao, P. LINC01354 enhances the proliferation and invasion of lung cancer cells by regulating miR-340-5p/ATF1 signaling pathway. Artificial cells, nanomedicine, and biotechnology 2019, 47, 3737–3744. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhao, W.; Na, F.; Li, M.; Tong, S. LINC01354/microRNA-216b/KRAS Axis Promotes the Occurrence and Metastasis of Endometrial Cancer. Nanoscale Res Lett 2022, 17, 21. [Google Scholar] [CrossRef]
- Li, J.; He, M.; Xu, W.; Huang, S. LINC01354 interacting with hnRNP-D contributes to the proliferation and metastasis in colorectal cancer through activating Wnt/β-catenin signaling pathway. Journal of experimental & clinical cancer research : CR 2019, 38, 161. [Google Scholar] [CrossRef]
- Bayraktar, E.; Bayraktar, R.; Oztatlici, H.; Lopez-Berestein, G.; Amero, P.; Rodriguez-Aguayo, C. Targeting miRNAs and Other Non-Coding RNAs as a Therapeutic Approach: An Update. Noncoding RNA 2023, 9. [Google Scholar] [CrossRef]
- He, G.N.; Bao, N.R.; Wang, S.; Xi, M.; Zhang, T.H.; Chen, F.S. Ketamine Induces Ferroptosis of Liver Cancer Cells by Targeting lncRNA PVT1/miR-214-3p/GPX4. Drug design, development and therapy 2021, 15, 3965–3978. [Google Scholar] [CrossRef]
- Xu, M.; Chen, X.; Lin, K.; Zeng, K.; Liu, X.; Xu, X.; Pan, B.; Xu, T.; Sun, L.; He, B.; et al. lncRNA SNHG6 regulates EZH2 expression by sponging miR-26a/b and miR-214 in colorectal cancer. Journal of hematology & oncology 2019, 12, 3. [Google Scholar] [CrossRef]
- Zhang, H.; Sun, P.; Wang, Y.L.; Yu, X.F.; Tong, J.J. MiR-214 promotes proliferation and inhibits apoptosis of oral cancer cells through MAPK/ERK signaling pathway. European review for medical and pharmacological sciences 2020, 24, 3710–3716. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Liu, P.Y.; Bao, W.; Chen, S.J.; Wu, F.S.; Zhu, P.Y. Hydrogen inhibits endometrial cancer growth via a ROS/NLRP3/caspase-1/GSDMD-mediated pyroptotic pathway. BMC cancer 2020, 20, 28. [Google Scholar] [CrossRef]
- Jiang, S.; Zhang, H.; Li, X.; Yi, B.; Huang, L.; Hu, Z.; Li, A.; Du, J.; Li, Y.; Zhang, W. Vitamin D/VDR attenuate cisplatin-induced AKI by down-regulating NLRP3/Caspase-1/GSDMD pyroptosis pathway. J Steroid Biochem Mol Biol 2021, 206, 105789. [Google Scholar] [CrossRef]
- Yan, H.; Luo, B.; Wu, X.; Guan, F.; Yu, X.; Zhao, L.; Ke, X.; Wu, J.; Yuan, J. Cisplatin Induces Pyroptosis via Activation of MEG3/NLRP3/caspase-1/GSDMD Pathway in Triple-Negative Breast Cancer. International journal of biological sciences 2021, 17, 2606–2621. [Google Scholar] [CrossRef]
- Wang, C.; Wang, L.; Huang, C.; Liu, Y.; Liu, J.; Kuang, H.; Pang, Q.; Han, H.; Fan, R. Involvement of NLRP3/Caspase-1/GSDMD-Dependent pyroptosis in BPA-Induced apoptosis of human neuroblastoma cells. Biochemical pharmacology 2022, 200, 115042. [Google Scholar] [CrossRef] [PubMed]
| characteristics | West | North | South | p value |
|---|---|---|---|---|
| n | 667 | 432 | 332 | |
| WHO grade, (%) | ||||
| G2 | 48% | 53% | 53% | 0.32 |
| G3 | 52% | 47% | 52% | |
| IDH status, (%) | ||||
| WT | 18% | 20% | 23% | 0.15 |
| Mut | 82% | 80% | 79% | |
| 1p/19q codeletion, (%) | ||||
| Non-codel | 68% | 74% | 79% | 0.12 |
| Codel | 32% | 26% | 27% | |
| Gender, (%) | ||||
| Female | 45% | 49% | 54% | 0.23 |
| Male | 55% | 51% | 50% | |
| Age, median | 41 | 39 | 42 | 0.06 |
| Histological type, (%) | ||||
| Astrocytoma | 59% | 66% | 72% | 0.06 |
| Oligoastrocytoma | 41% | 34% | 35% | |
| Laterality, (%) | ||||
| Left | 50% | 59% | 66% | 0.05 |
| Right | 50% | 41% | 43% | |
| Normalized expression level | ||||
| LINC01354 | 1 | 1.052 | 0.989 | 0.45 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




