Submitted:
22 June 2024
Posted:
24 June 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. How Anhydrobiotes Acquire Desiccation Tolerance
2.1. No Pain, No Gain—Low Stress Produces More Tolerance
2.2. Morphological and Physiological Changes in Response to Desiccation Stress
2.2.1. Morphological Changes
2.2.2. Physiological Changes
| GENE ID | GENE NAME | POSSIBLE ROLE IN (DESICCICATION) RESISTANCE | REFERENCES |
|---|---|---|---|
| YJL184W | GON7 | Involved in cell wall mannoprotein biosynthesis and osmotic stress response. | [55] |
| YMR175W | SIP18 | Involved in osmotic and desiccation stress response. | [56,57] |
| YMR260C | TIF11 | Localizes to cytoplasmic stress granule; desiccation resistance increases in mutant. | [8,58] |
| YFL014W | HSP12 | Plant LEA-like protein, involved in plasma membrane organization and response to multiple stresses, including desiccation stress. | [44] |
| YDL213C | NOP6 | Required for desiccation-rehydration process. | [53] |
| YGR008C | STF2 | Involved in cellular response to desiccation, oxidation, and DNA replication stress. | [53] |
| YBR016W | CPP1 | Involved in the adaptive response to hyperosmotic stress. Detailed biological function unknown. | [59] |
| YPL223C | GRE1 | Paralog to SIP18. Involved in response to multiple stresses, including osmotic, oxidative, heat shock and desiccation. | [60] |
| YFL010C | WWM1 | Biological function unknown. Interacts with the caspase-related protease Mca1p. | [61] |
| YJL144W | ROQ1 | Regulator of the Ubr1p E3 ubiquitin ligase; Involved in osmotic, DNA replication and desiccation stress. | [61,62] |
| YNL162W | RPL42A | Subunit of the large ribosomal 60S subunit. Function in desiccation stress unknown. | [53,63] |
| YNL190W | Cell wall protein; essential for desiccation stress response. | [53] |
3. How do Anhydrobiotes Survive in Desiccated State
3.1. Minimal Metabolism but Not Ametabolism
4. Desiccated Yeast as a Model to Study Prion and Other Neurodegenerative Disorders
Funding
Conflicts of Interest
References
- Dargaville, B.L.; Hutmacher, D.W. Water as the often neglected medium at the interface between materials and biology. Nat Commun 2022, 13, 4222. [Google Scholar] [CrossRef] [PubMed]
- Cowan, N.B.; Agol, E.; Meadows, V.S.; Robinson, T.; Livengood, T.A.; Deming, D.; Lisse, C.M.; A’Hearn, M.F.; Wellnitz, D.D.; Seager, S.; et al. ALIEN MAPS OF AN OCEAN-BEARING WORLD. The Astrophysical Journal 2009, 700, 915. [Google Scholar] [CrossRef]
- Wharton, D.A. Anhydrobiosis. Curr Biol 2015, 25, R1114–R1116. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, B.; Blaxter, M. Tardigrades. Curr Biol 2002, 12, R475. [Google Scholar] [CrossRef] [PubMed]
- Smolikova, G.; Leonova, T.; Vashurina, N.; Frolov, A.; Medvedev, S. Desiccation Tolerance as the Basis of Long-Term Seed Viability. Int J Mol Sci 2020, 22, 101. [Google Scholar] [CrossRef]
- Hesgrove, C.; Boothby, T.C. The biology of tardigrade disordered proteins in extreme stress tolerance. Cell Communication and Signaling 2020, 18, 178. [Google Scholar] [CrossRef]
- Rapoport, A.; Turchetti, B.; Buzzini, P. Application of anhydrobiosis and dehydration of yeasts for non-conventional biotechnological goals. World J Microbiol Biotechnol 2016, 32, 104. [Google Scholar] [CrossRef] [PubMed]
- Welch, A.Z.; Gibney, P.A.; Botstein, D.; Koshland, D.E. TOR and RAS pathways regulate desiccation tolerance in Saccharomyces cerevisiae. Mol Biol Cell 2013, 24, 115–128. [Google Scholar] [CrossRef] [PubMed]
- Gasch, A.P.; Spellman, P.T.; Kao, C.M.; Carmel-Harel, O.; Eisen, M.B.; Storz, G.; Botstein, D.; Brown, P.O. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 2000, 11, 4241–4257. [Google Scholar] [CrossRef] [PubMed]
- Boothby, T.C.; Tapia, H.; Brozena, A.H.; Piszkiewicz, S.; Smith, A.E.; Giovannini, I.; Rebecchi, L.; Pielak, G.J.; Koshland, D.; Goldstein, B. Tardigrades Use Intrinsically Disordered Proteins to Survive Desiccation. Mol Cell 2017, 65, 975–984.e975. [Google Scholar] [CrossRef]
- Kim, S.X.; Çamdere, G.; Hu, X.; Koshland, D.; Tapia, H. Synergy between the small intrinsically disordered protein Hsp12 and trehalose sustain viability after severe desiccation. Elife 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Ren, Q.; Brenner, R.; Boothby, T.C.; Zhang, Z. Membrane and lipid metabolism plays an important role in desiccation resistance in the yeast Saccharomyces cerevisiae. BMC Microbiol 2020, 20, 338. [Google Scholar] [CrossRef] [PubMed]
- Tebele, S.M.; Marks, R.A.; Farrant, J.M. Two Decades of Desiccation Biology: A Systematic Review of the Best Studied Angiosperm Resurrection Plants. Plants 2021, 10, 2784. [Google Scholar] [CrossRef] [PubMed]
- Liebman, S.W.; Chernoff, Y.O. Prions in yeast. Genetics 2012, 191, 1041–1072. [Google Scholar] [CrossRef] [PubMed]
- Collinson, L.P.; Dawes, I.W. Inducibility of the response of yeast cells to peroxide stress. J Gen Microbiol 1992, 138, 329–335. [Google Scholar] [CrossRef]
- Hoekstra, F.A.; Golovina, E.A.; Buitink, J. Mechanisms of plant desiccation tolerance. Trends Plant Sci 2001, 6, 431–438. [Google Scholar] [CrossRef] [PubMed]
- Ingram, J.; Bartels, D. THE MOLECULAR BASIS OF DEHYDRATION TOLERANCE IN PLANTS. Annu Rev Plant Physiol Plant Mol Biol 1996, 47, 377–403. [Google Scholar] [CrossRef] [PubMed]
- Crowe, J.H.; Madin, K.A.C. Anhydrobiosis in nematodes: Evaporative water loss and survival. Journal of Experimental Zoology 1755, 193, 11. [Google Scholar] [CrossRef]
- Erkut, C.; Penkov, S.; Khesbak, H.; Vorkel, D.; Verbavatz, J.-M.; Fahmy, K.; Kurzchalia, T.V. Trehalose Renders the Dauer Larva of Caenorhabditis elegans Resistant to Extreme Desiccation. Current Biology 2011, 21, 1331–1336. [Google Scholar] [CrossRef]
- Boothby, T.C.; Tenlen, J.R.; Smith, F.W.; Wang, J.R.; Patanella, K.A.; Nishimura, E.O.; Tintori, S.C.; Li, Q.; Jones, C.D.; Yandell, M.; et al. Evidence for extensive horizontal gene transfer from the draft genome of a tardigrade. Proc Natl Acad Sci U S A 2015, 112, 15976–15981. [Google Scholar] [CrossRef]
- Nguyen, K.; Kc, S.; Gonzalez, T.; Tapia, H.; Boothby, T.C. Trehalose and tardigrade CAHS proteins work synergistically to promote desiccation tolerance. Commun Biol 2022, 5, 1046. [Google Scholar] [CrossRef]
- Calahan, D.; Dunham, M.; DeSevo, C.; Koshland, D.E. Genetic analysis of desiccation tolerance in Sachharomyces cerevisiae. Genetics 2011, 189, 507–519. [Google Scholar] [CrossRef]
- Huang, M.; Hull, C.M. Sporulation: how to survive on planet Earth (and beyond). Curr Genet 2017, 63, 831–838. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, G.R. Chromosome-condensed G1 phase yeast cells are tolerant to desiccation stress. Microb Cell 2022, 9, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Bagniewska-Zadworna, A. The root microtubule cytoskeleton and cell cycle analysis through desiccation of Brassica napus seedlings. Protoplasma 2008, 233, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Kakumanu, A.; Ambavaram, M.M.; Klumas, C.; Krishnan, A.; Batlang, U.; Myers, E.; Grene, R.; Pereira, A. Effects of drought on gene expression in maize reproductive and leaf meristem tissue revealed by RNA-Seq. Plant Physiol 2012, 160, 846–867. [Google Scholar] [CrossRef] [PubMed]
- Rippin, M.; Borchhardt, N.; Karsten, U.; Becker, B. Cold Acclimation Improves the Desiccation Stress Resilience of Polar Strains of Klebsormidium (Streptophyta). Front Microbiol 2019, 10, 1730. [Google Scholar] [CrossRef]
- Slade, D.; Radman, M. Oxidative stress resistance in Deinococcus radiodurans. Microbiol Mol Biol Rev 2011, 75, 133–191. [Google Scholar] [CrossRef]
- Lew, D.J.W.T.; Pringle, J.R. Cell cycle control in Saccharomyces cerevisiae. In The Molecular and Cellular Biology of the Yeast Saccharomyces: Cell Cycle and Cell Biology; Pringle, J.R.B.J.R., Jones, E.W., Eds.; Cold Spring Harbor Laboratory Press, 1997; pp. 607–695. [Google Scholar]
- Yang, H.; Ren, Q.; Zhang, Z. Cleavage of Mcd1 by caspase-like protease Esp1 promotes apoptosis in budding yeast. Mol Biol Cell 2008, 19, 2127–2134. [Google Scholar] [CrossRef]
- Rajvanshi, P.K.; Arya, M.; Rajasekharan, R. The stress-regulatory transcription factors Msn2 and Msn4 regulate fatty acid oxidation in budding yeast. J Biol Chem 2017, 292, 18628–18643. [Google Scholar] [CrossRef]
- Werner-Washburne, M.; Braun, E.; Johnston, G.C.; Singer, R.A. Stationary phase in the yeast Saccharomyces cerevisiae. Microbiol Rev 1993, 57, 383–401. [Google Scholar] [CrossRef] [PubMed]
- de Nobel, H.; Ruiz, C.; Martin, H.; Morris, W.; Brul, S.; Molina, M.; Klis, F.M. Cell wall perturbation in yeast results in dual phosphorylation of the Slt2/Mpk1 MAP kinase and in an Slt2-mediated increase in FKS2-lacZ expression, glucanase resistance and thermotolerance. Microbiology (Reading) 2000, 146 Pt 9, 2121–2132. [Google Scholar] [CrossRef]
- Wolinski, H.; Kolb, D.; Hermann, S.; Koning, R.I.; Kohlwein, S.D. A role for seipin in lipid droplet dynamics and inheritance in yeast. J Cell Sci 2011, 124 Pt 22, 3894–3904. [Google Scholar] [CrossRef]
- Kurat, C.F.; Natter, K.; Petschnigg, J.; Wolinski, H.; Scheuringer, K.; Scholz, H.; Zimmermann, R.; Leber, R.; Zechner, R.; Kohlwein, S.D. Obese yeast: triglyceride lipolysis is functionally conserved from mammals to yeast. J Biol Chem 2006, 281, 491–500. [Google Scholar] [CrossRef]
- Beker, M.J.; Rapoport, A.I. Conservation of yeasts by dehydration. In Biotechnology Methods; Springer: Berlin, Heidelberg, 1987; pp. 127–171. [Google Scholar]
- Rapoport, A. Anhydrobiosis and Dehydration of Yeasts. In Biotechnology of Yeasts and Filamentous Fungi; Sibirny, A.A., Ed.; Springer International Publishing, 2017; pp. 87–116. [Google Scholar]
- Rapoport, A.; Golovina, E.A.; Gervais, P.; Dupont, S.; Beney, L. Anhydrobiosis: Inside yeast cells. Biotechnol Adv 2019, 37, 51–67. [Google Scholar] [CrossRef]
- Pereira Ede, J.; Panek, A.D.; Eleutherio, E.C. Protection against oxidation during dehydration of yeast. Cell Stress Chaperones 2003, 8, 120–124. [Google Scholar] [CrossRef] [PubMed]
- Hagymasi, A.T.; Dempsey, J.P.; Srivastava, P.K. Heat-Shock Proteins. Curr Protoc 2022, 2, e592. [Google Scholar] [CrossRef] [PubMed]
- Schlesinger, M.J. Heat shock proteins. J Biol Chem 1990, 265, 12111–12114. [Google Scholar] [CrossRef] [PubMed]
- De Maio, A. Heat shock proteins: facts, thoughts, and dreams. Shock 1999, 11, 1–12. [Google Scholar] [CrossRef]
- Hu, C.; Yang, J.; Qi, Z.; Wu, H.; Wang, B.; Zou, F.; Mei, H.; Liu, J.; Wang, W.; Liu, Q. Heat shock proteins: Biological functions, pathological roles, and therapeutic opportunities. MedComm (2020) 2022, 3, e161. [Google Scholar] [CrossRef]
- Sales, K.; Brandt, W.; Rumbak, E.; Lindsey, G. The LEA-like protein HSP 12 in Saccharomyces cerevisiae has a plasma membrane location and protects membranes against desiccation and ethanol-induced stress. Biochim Biophys Acta 2000, 1463, 267–278. [Google Scholar] [CrossRef] [PubMed]
- Tapia, H.; Young, L.; Fox, D.; Bertozzi, C.R.; Koshland, D. Increasing intracellular trehalose is sufficient to confer desiccation tolerance to Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 2015, 112, 6122–6127. [Google Scholar] [CrossRef] [PubMed]
- Guzhova, I.; Krallish, I.; Khroustalyova, G.; Margulis, B.; Rapoport, A. Dehydration of yeast: Changes in the intracellular content of Hsp70 family proteins. Process Biochemistry 2008, 43, 1138–1141. [Google Scholar] [CrossRef]
- Iryani, M.T.M.; Sorgeloos, P.; Danish-Daniel, M.; Tan, M.P.; Wong, L.L.; Mok, W.J.; Satyantini, W.H.; Mahasri, G.; Sung, Y.Y. Cyst viability and stress tolerance upon heat shock protein 70 knockdown in the brine shrimp Artemia franciscana. Cell Stress Chaperones 2020, 25, 1099–1103. [Google Scholar] [CrossRef]
- Trivedi, R.; Nagarajaram, H.A. Intrinsically Disordered Proteins: An Overview. Int J Mol Sci 2022, 23, 14050. [Google Scholar] [CrossRef]
- Chakrabortee, S.; Tripathi, R.; Watson, M.; Schierle, G.S.; Kurniawan, D.P.; Kaminski, C.F.; Wise, M.J.; Tunnacliffe, A. Intrinsically disordered proteins as molecular shields. Mol Biosyst 2012, 8, 210–219. [Google Scholar] [CrossRef]
- Hincha, D.K.; Thalhammer, A. LEA proteins: IDPs with versatile functions in cellular dehydration tolerance. Biochem Soc Trans 2012, 40, 1000–1003. [Google Scholar] [CrossRef]
- Garay-Arroyo, A.; Colmenero-Flores, J.M.; Garciarrubio, A.; Covarrubias, A.A. Highly hydrophilic proteins in prokaryotes and eukaryotes are common during conditions of water deficit. J Biol Chem 2000, 275, 5668–5674. [Google Scholar] [CrossRef]
- Rodríguez-Porrata, B.; Carmona-Gutierrez, D.; Reisenbichler, A.; Bauer, M.; Lopez, G.; Escoté, X.; Mas, A.; Madeo, F.; Cordero-Otero, R. Sip18 hydrophilin prevents yeast cell death during desiccation stress. J Appl Microbiol 2012, 112, 512–525. [Google Scholar] [CrossRef]
- López-Martínez, G.; Rodríguez-Porrata, B.; Margalef-Català, M.; Cordero-Otero, R. The STF2p hydrophilin from Saccharomyces cerevisiae is required for dehydration stress tolerance. PLoS One 2012, 7, e33324. [Google Scholar] [CrossRef]
- Dauss, E.; Papoušková, K.; Sychrová, H.; Rapoport, A. Anhydrobiosis in yeast: role of cortical endoplasmic reticulum protein Ist2 in Saccharomyces cerevisiae cells during dehydration and subsequent rehydration. Antonie van Leeuwenhoek 2021, 114, 1069–1077. [Google Scholar] [CrossRef] [PubMed]
- Ando, A.; Tanaka, F.; Murata, Y.; Takagi, H.; Shima, J. Identification and classification of genes required for tolerance to high-sucrose stress revealed by genome-wide screening of Saccharomyces cerevisiae. FEMS Yeast Res 2006, 6, 249–267. [Google Scholar] [CrossRef] [PubMed]
- Miralles, V.J.; Serrano, R. A genomic locus in Saccharomyces cerevisiae with four genes up-regulated by osmotic stress. Mol Microbiol 1995, 17, 653–662. [Google Scholar] [CrossRef]
- Dang, N.X.; Hincha, D.K. Identification of two hydrophilins that contribute to the desiccation and freezing tolerance of yeast (Saccharomyces cerevisiae) cells. Cryobiology 2011, 62, 188–193. [Google Scholar] [CrossRef] [PubMed]
- Jain, S.; Wheeler, J.R.; Walters, R.W.; Agrawal, A.; Barsic, A.; Parker, R. ATPase-Modulated Stress Granules Contain a Diverse Proteome and Substructure. Cell 2016, 164, 487–498. [Google Scholar] [CrossRef]
- Venancio, T.M.; Aravind, L. CYSTM, a novel cysteine-rich transmembrane module with a role in stress tolerance across eukaryotes. Bioinformatics 2010, 26, 149–152. [Google Scholar] [CrossRef] [PubMed]
- Garay-Arroyo, A.; Covarrubias, A.A. Three genes whose expression is induced by stress in Saccharomyces cerevisiae. Yeast 1999, 15, 879–892. [Google Scholar] [CrossRef]
- Szoradi, T.; Schaeff, K.; Garcia-Rivera, E.M.; Itzhak, D.N.; Schmidt, R.M.; Bircham, P.W.; Leiss, K.; Diaz-Miyar, J.; Chen, V.K.; Muzzey, D.; et al. SHRED Is a Regulatory Cascade that Reprograms Ubr1 Substrate Specificity for Enhanced Protein Quality Control during Stress. Mol Cell 2018, 70, 1025–1037.e1025. [Google Scholar] [CrossRef]
- Tkach, J.M.; Yimit, A.; Lee, A.Y.; Riffle, M.; Costanzo, M.; Jaschob, D.; Hendry, J.A.; Ou, J.; Moffat, J.; Boone, C.; et al. Dissecting DNA damage response pathways by analysing protein localization and abundance changes during DNA replication stress. Nat Cell Biol 2012, 14, 966–976. [Google Scholar] [CrossRef]
- Planta, R.J.; Mager, W.H. The list of cytoplasmic ribosomal proteins of Saccharomyces cerevisiae. Yeast 1998, 14, 471–477. [Google Scholar] [CrossRef]
- Jain, N.K.; Roy, I. Effect of trehalose on protein structure. Protein Sci 2009, 18, 24–36. [Google Scholar] [CrossRef] [PubMed]
- Malferrari, M.; Nalepa, A.; Venturoli, G.; Francia, F.; Lubitz, W.; Möbius, K.; Savitsky, A. Structural and dynamical characteristics of trehalose and sucrose matrices at different hydration levels as probed by FTIR and high-field EPR. Physical Chemistry Chemical Physics 2014, 16, 9831–9848. [Google Scholar] [CrossRef] [PubMed]
- Crowe, J.H. Trehalose as a “chemical chaperone”: fact and fantasy. Adv Exp Med Biol 2007, 594, 143–158. [Google Scholar] [CrossRef]
- Dickinson, J.R.; Schweizer, M. Metabolism and Molecular Physiology of Saccharomyces Cerevisiae; CRC Press, 2004. [Google Scholar] [CrossRef]
- François, J.; Parrou, J.L. Reserve carbohydrates metabolism in the yeast Saccharomyces cerevisiae. FEMS Microbiol Rev 2001, 25, 125–145. [Google Scholar] [CrossRef] [PubMed]
- Erkut, C.; Gade, V.R.; Laxman, S.; Kurzchalia, T.V. The glyoxylate shunt is essential for desiccation tolerance in C. elegans and budding yeast. Elife 2016, 5. [Google Scholar] [CrossRef]
- Ahn, S.; Jung, J.; Jang, I.A.; Madsen, E.L.; Park, W. Role of Glyoxylate Shunt in Oxidative Stress Response. J Biol Chem 2016, 291, 11928–11938. [Google Scholar] [CrossRef] [PubMed]
- de Nadal, E.; Posas, F. The HOG pathway and the regulation of osmoadaptive responses in yeast. FEMS Yeast Res 2022, 22. [Google Scholar] [CrossRef] [PubMed]
- Semkiv, M.; Ternavska, O.T.; Dmytruk, K.V.; Sybirny, A.A. Effect of Trehalose and Glycerol on the Resistance of Recombinant Saccharomyces cerevisiae Strains to Desiccation, Freeze-Thaw and Osmotic Stresses. Sci. Innov. 2018, 14, 22. [Google Scholar] [CrossRef]
- Takagi, H. Proline as a stress protectant in yeast: physiological functions, metabolic regulations, and biotechnological applications. Appl Microbiol Biotechnol 2008, 81, 211–223. [Google Scholar] [CrossRef] [PubMed]
- Takagi, H.; Sakai, K.; Morida, K.; Nakamori, S. Proline accumulation by mutation or disruption of the proline oxidase gene improves resistance to freezing and desiccation stresses in Saccharomyces cerevisiae. FEMS Microbiol Lett 2000, 184, 103–108. [Google Scholar] [CrossRef] [PubMed]
- Grzyb, T.; Skłodowska, A. Introduction to Bacterial Anhydrobiosis: A General Perspective and the Mechanisms of Desiccation-Associated Damage. Microorganisms 2022, 10, 432. [Google Scholar] [CrossRef] [PubMed]
- Candotto Carniel, F.; Fernandez-Marín, B.; Arc, E.; Craighero, T.; Laza, J.M.; Incerti, G.; Tretiach, M.; Kranner, I. How dry is dry? Molecular mobility in relation to thallus water content in a lichen. J Exp Bot 2021, 72, 1576–1588. [Google Scholar] [CrossRef]
- Mellanby, K. Metabolic Water and Desiccation. Nature 1942, 150, 21–21. [Google Scholar] [CrossRef]
- Schmidt-Nielsen, K. Animal Physiology: Adaptation and Environment; Cambridge University Press, 1997. [Google Scholar]
- Klaassen, M. Metabolic constraints on long-distance migration in birds. J Exp Biol 1996, 199 Pt 1, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Takei, Y. Metabolic Water As a Route for Water Acquisition in Vertebrates Inhabiting Dehydrating Environments. Zoolog Sci 2024, 41, 132–139. [Google Scholar] [CrossRef]
- Ross, C.A.; Poirier, M.A. Protein aggregation and neurodegenerative disease. Nat Med 2004, 10 Suppl, S10–S17. [Google Scholar] [CrossRef]
- Soto, C.; Satani, N. The intricate mechanisms of neurodegeneration in prion diseases. Trends Mol Med 2011, 17, 14–24. [Google Scholar] [CrossRef]
- Rossi, M.; Baiardi, S.; Parchi, P. Understanding Prion Strains: Evidence from Studies of the Disease Forms Affecting Humans. Viruses 2019, 11, 309. [Google Scholar] [CrossRef]
- Wickner, R.B. [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science 1994, 264, 566–569. [Google Scholar] [CrossRef] [PubMed]
- Kushnirov, V.V.; Kochneva-Pervukhova, N.V.; Chechenova, M.B.; Frolova, N.S.; Ter-Avanesyan, M.D. Prion properties of the Sup35 protein of yeast Pichia methanolica. Embo J 2000, 19, 324–331. [Google Scholar] [CrossRef]
- Derkatch, I.L.; Chernoff, Y.O.; Kushnirov, V.V.; Inge-Vechtomov, S.G.; Liebman, S.W. Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics 1996, 144, 1375–1386. [Google Scholar] [CrossRef] [PubMed]
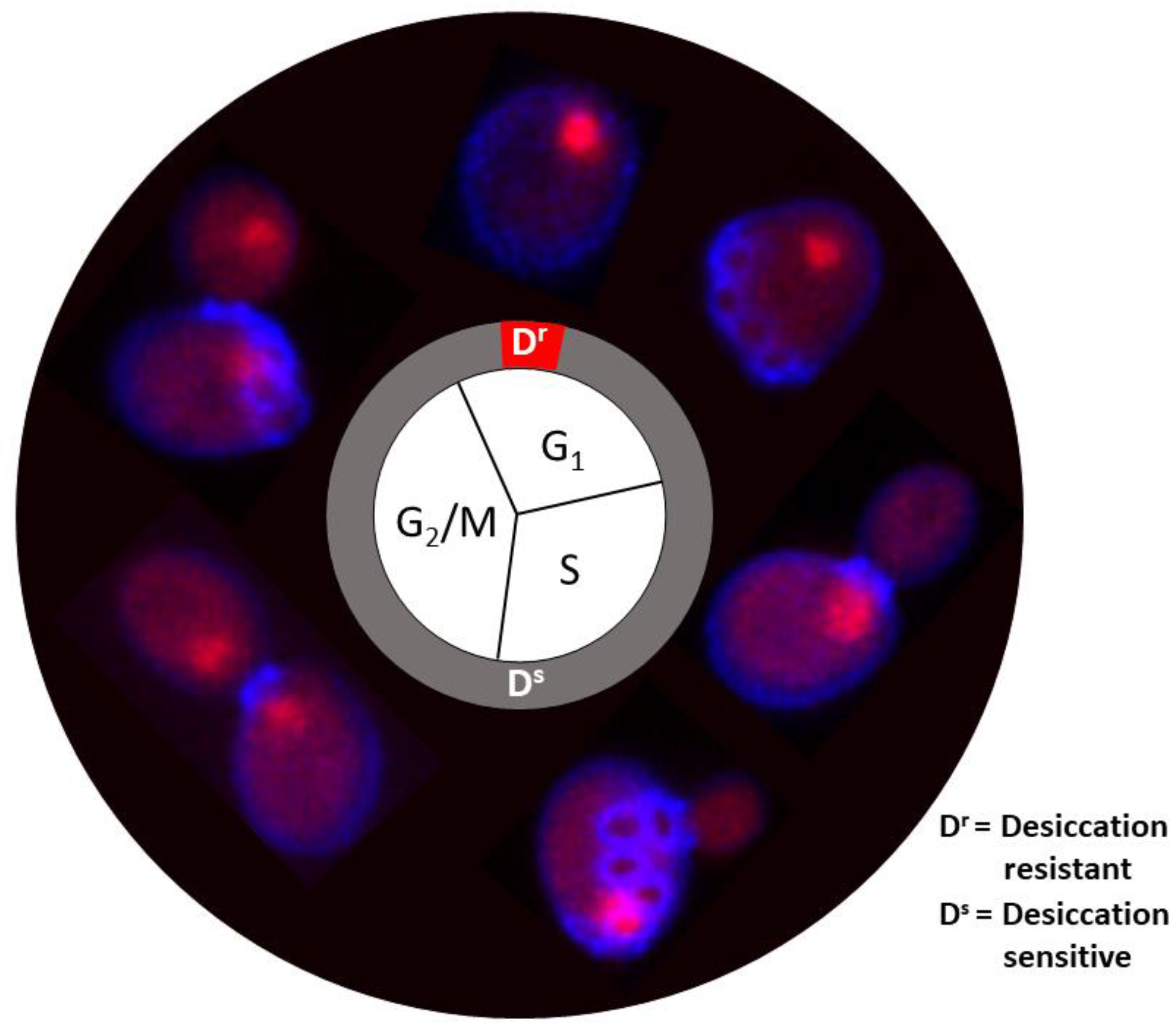
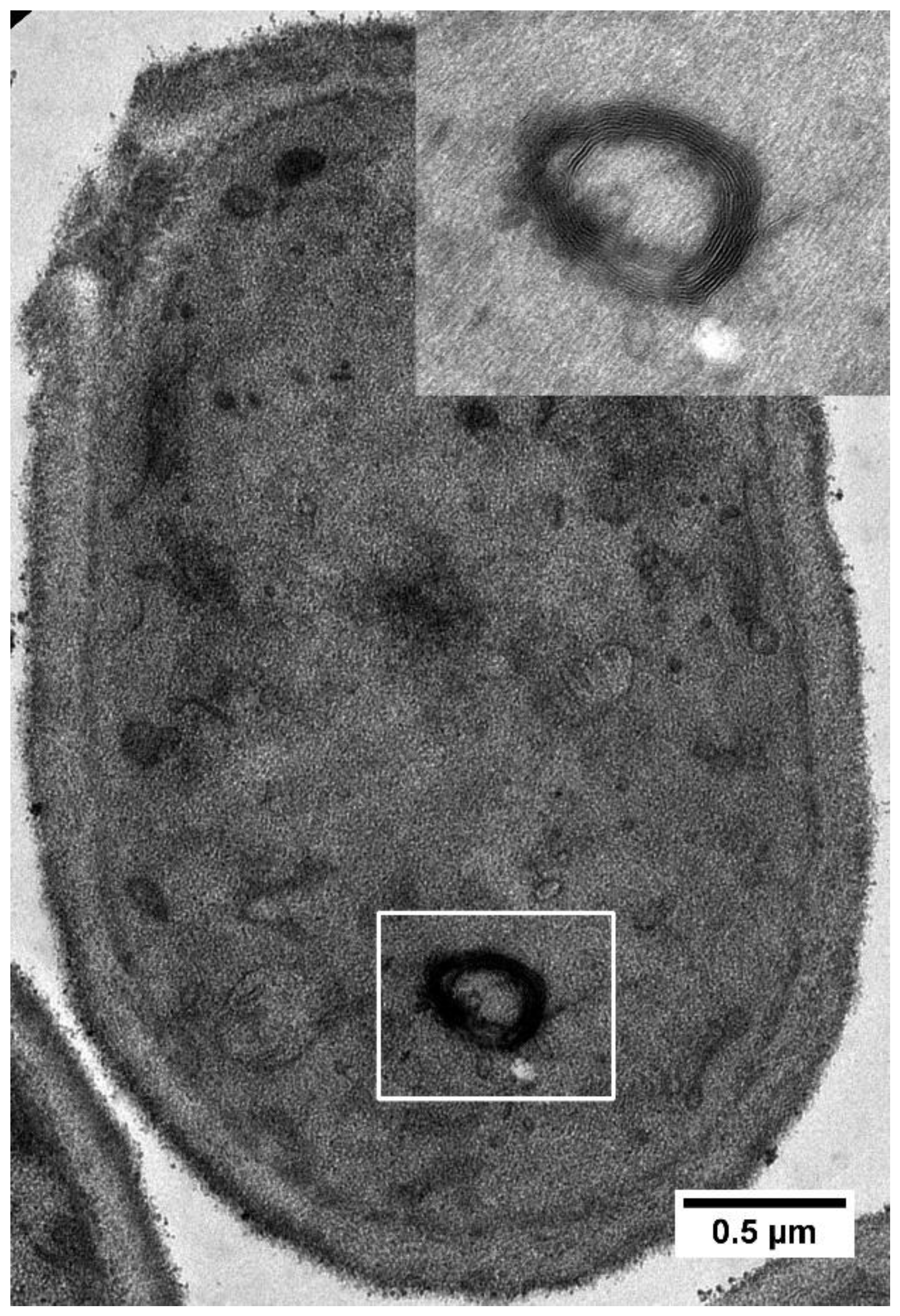
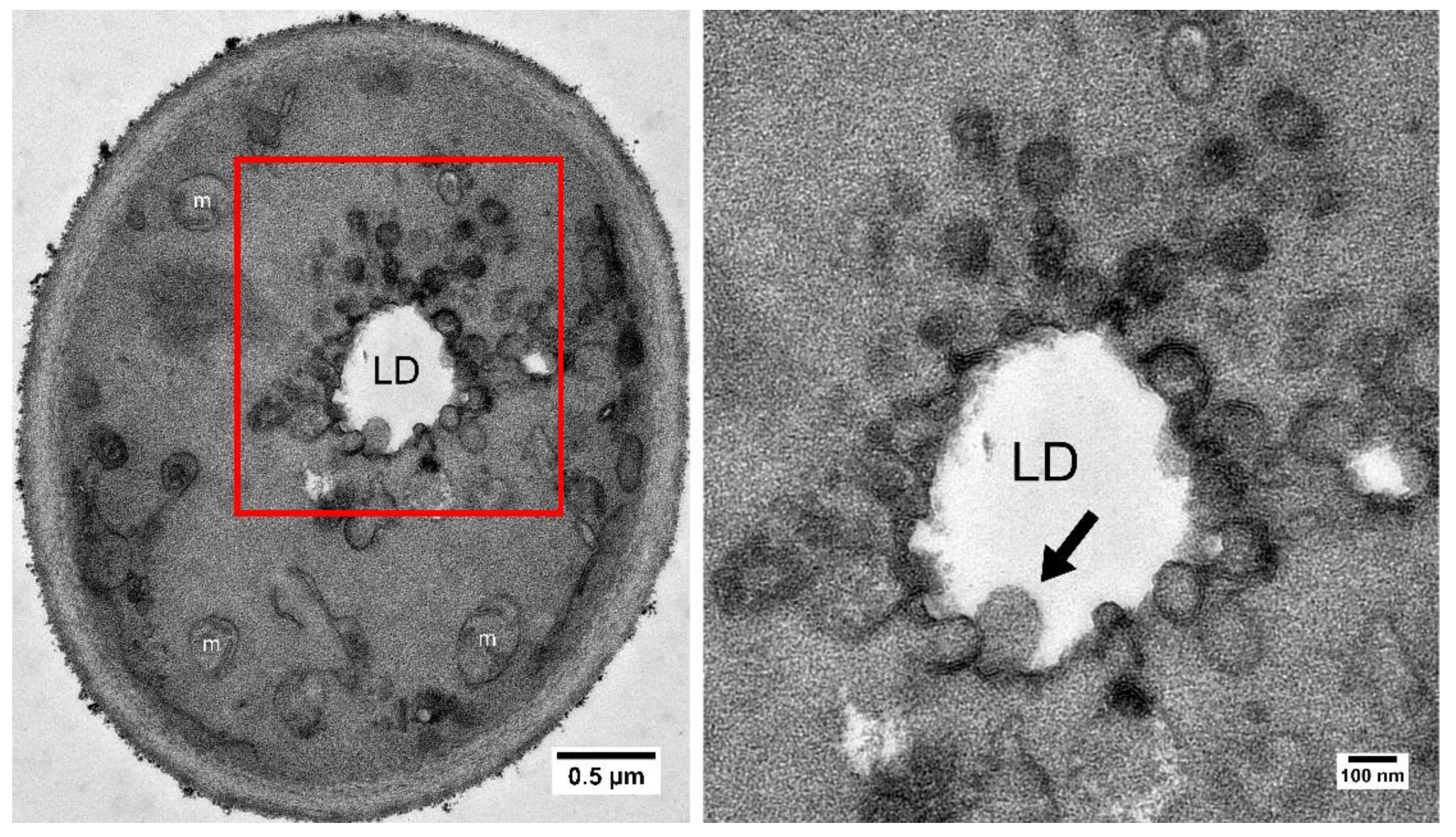
| Gene ID | Gene Name | Description |
|---|---|---|
| YER015W | LPX1 | Peroxisomal matrix-localized lipase; required for normal peroxisomal morphology. |
| YGL205W | POX1 | Fatty-acyl coenzyme A oxidase; involved in the fatty acid beta-oxidation pathway. |
| YIL160C | POT1 | 3-ketoacyl-CoA thiolase with broad chain length specificity; cleaves 3-ketoacyl-CoA into acyl-CoA and acetyl-CoA during beta-oxidation of fatty acids. |
| YKR009C | FOX2 | 3-hydroxyacyl-CoA dehydrogenase and enoyl-CoA hydratase; multifunctional enzyme of the peroxisomal fatty acid beta-oxidation pathway. |
| YLR284C | ECI1 | Peroxisomal delta3, delta2-enoyl-CoA isomerase; essential for the beta-oxidation of unsaturated fatty acids. |
| YOR180C | DCI1 | Peroxisomal protein involved in fatty acid metabolism. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




