Submitted:
17 June 2024
Posted:
18 June 2024
You are already at the latest version
Abstract
Keywords:
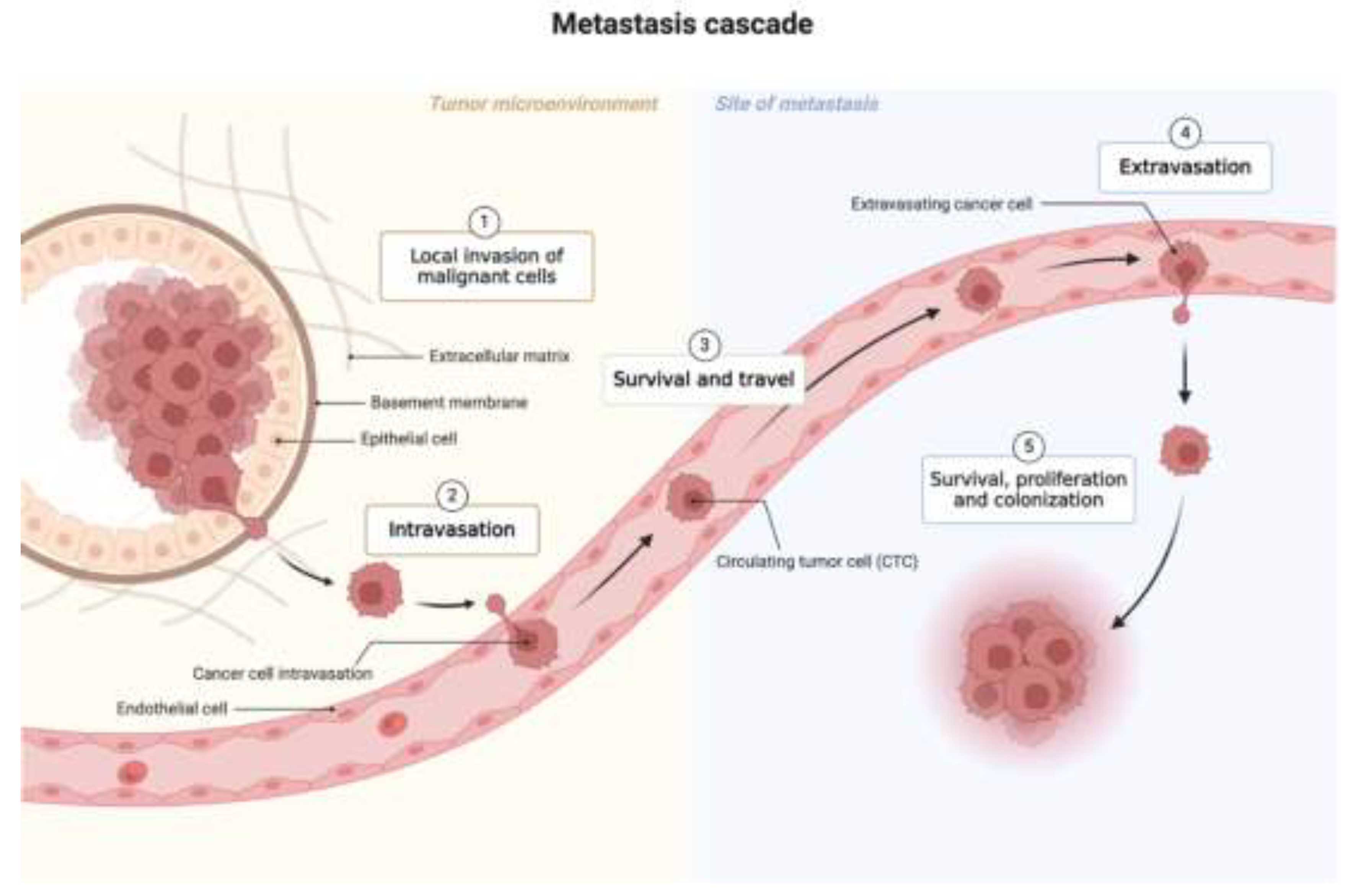
1. Introduction
2. Background
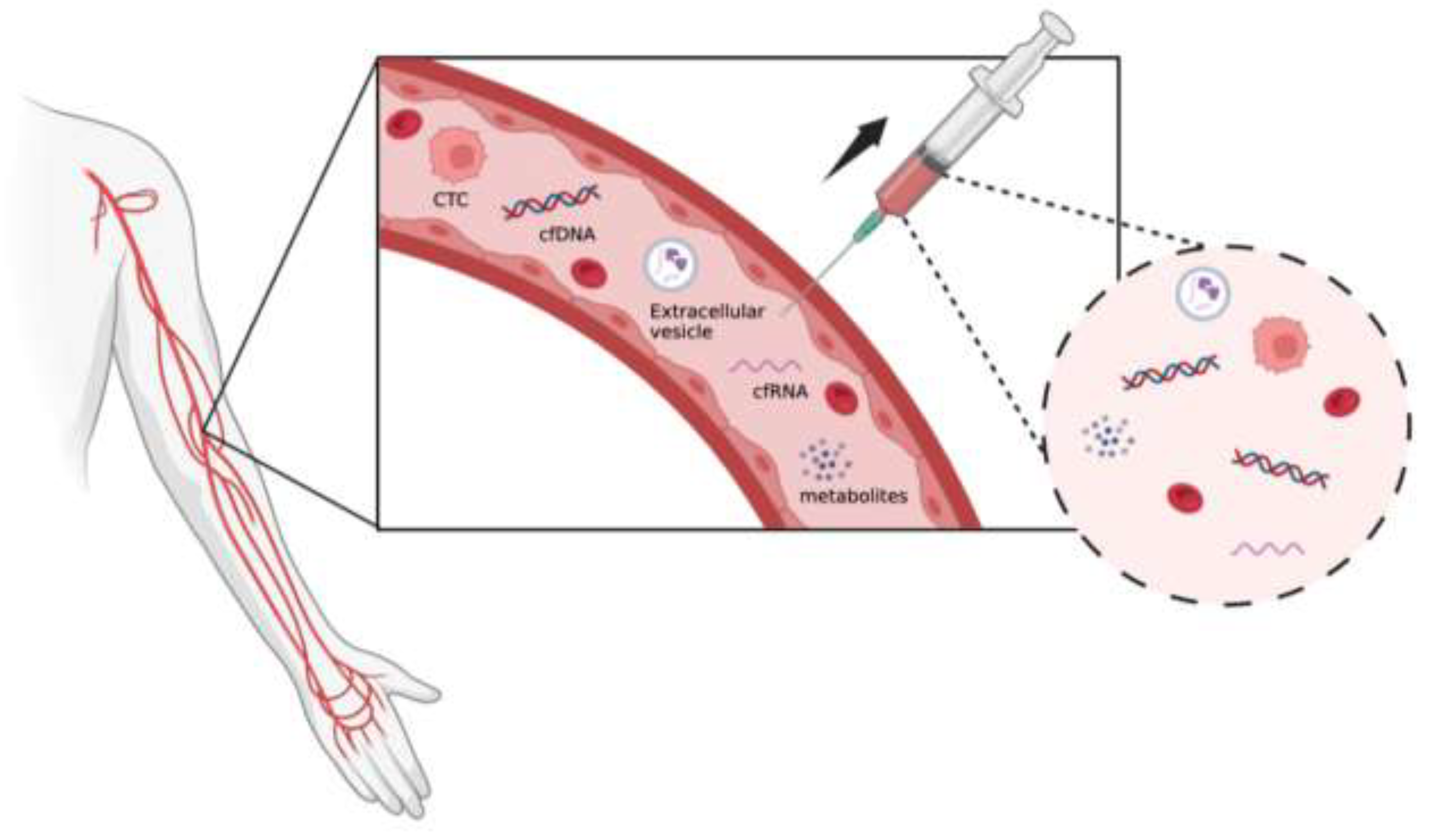
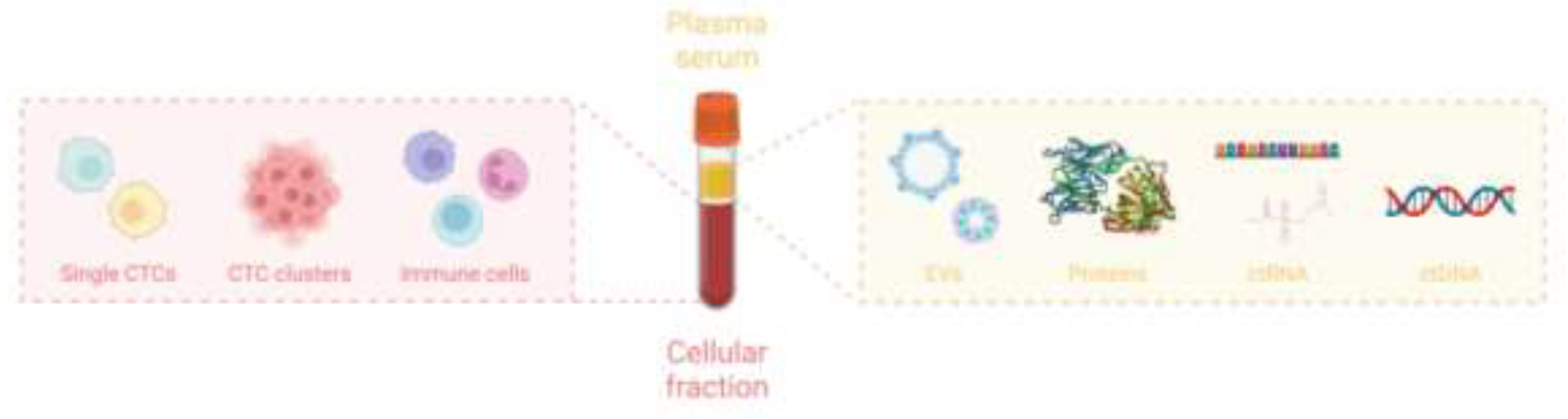
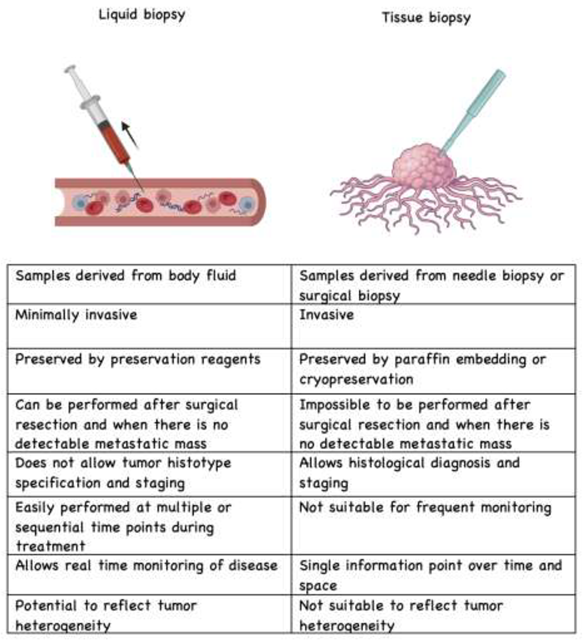
3. Technology to Collect and Detect Liquid Biopsy
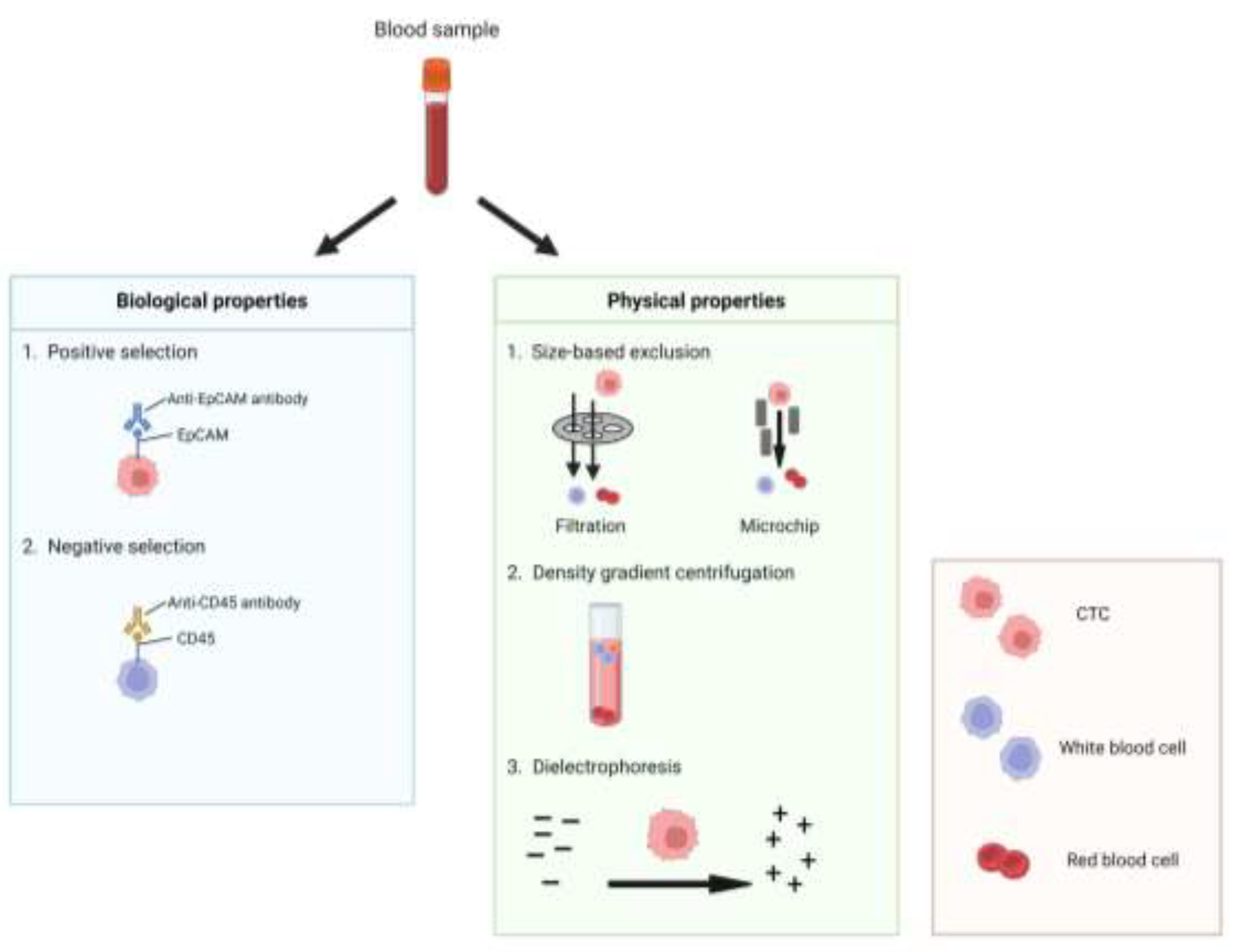
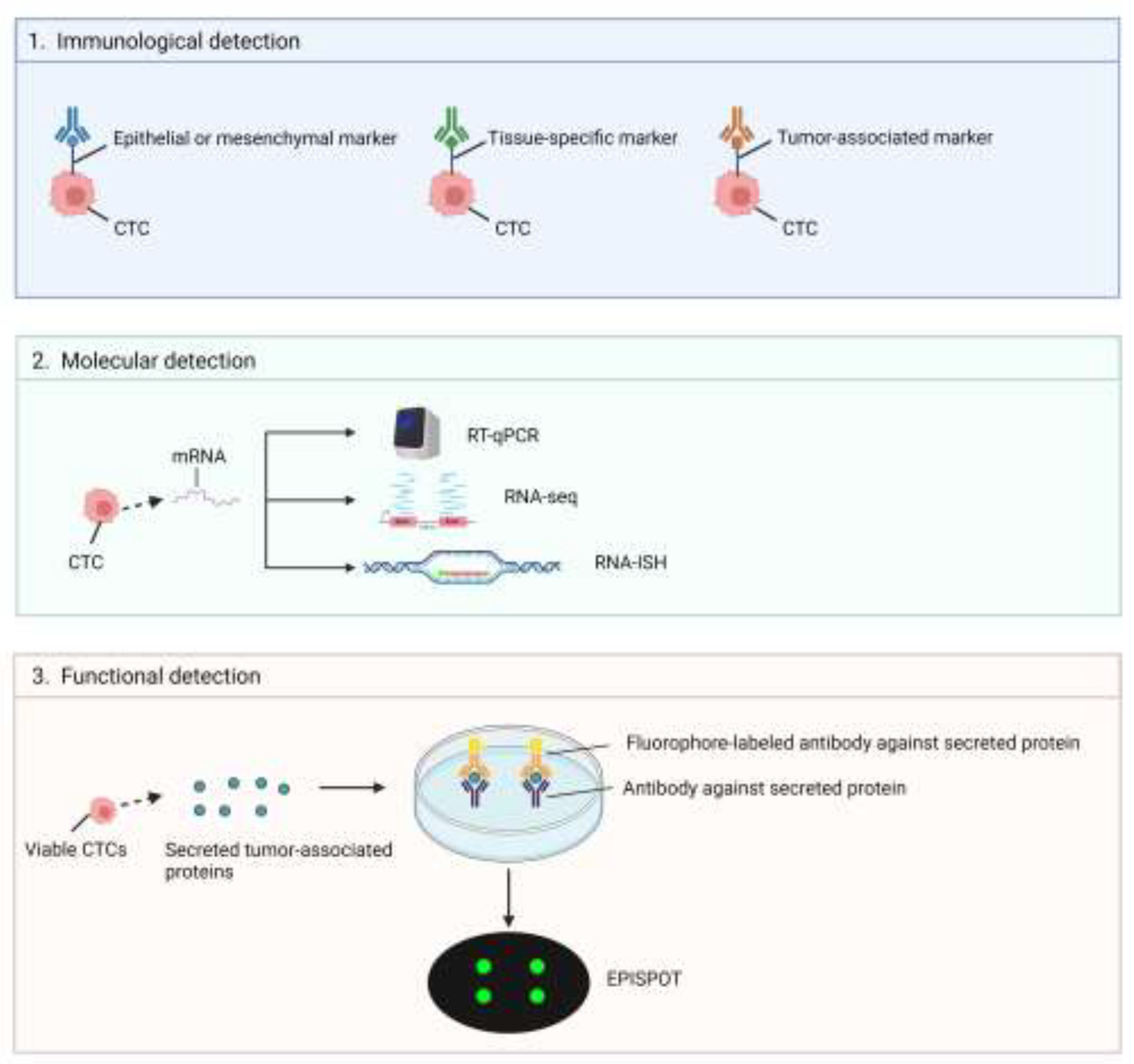
3.1. Circulating Tumor Cells (CTCs)
3.1.1. Capture and Isolation of CTCs
3.1.2. Strategies on CTCs Analysis
3.2. Circulating Nucleic Acids
3.3. Circulating Tumor DNA (ctDNA)
3.4. Circulating Tumor RNA (ctRNA)
3.4.1. Isolation of Circulating Cell-Free Nucleic Acids
3.4.2. Circulating Nucleic Acids Detection and Analysis
3.5. Extracellular Vesicles (EV)
3.5.1. EV Isolation and Characterization Technologies
3.6. Metabolites
4. Applications in Cancer Management
4.1. Early Detection and Diagnosis
4.2. Prognostication and Predictive Biomarkers
4.3. Treatment Selection and Personalized Medicine
4.4. Lung Cancer
4.5. Breast Cancer
4.6. Prostate Cancer
5. Key Clinical Trials Evaluating Liquid Biopsy for Cancer
6. Liquid Biopsy Regulatory Considerations and Challenges.
8. Conclusions
References
- Aceto, N.; Bardia, A.; Miyamoto, D.T.; Donaldson, M.C.; Wittner, B.S.; Spencer, J.A.; Yu, M.; Pely, A.; Engstrom, A.; Zhu, H.; et al. Circulating Tumor Cell Clusters Are Oligoclonal Precursors of Breast Cancer Metastasis. Cell 2014, 158, 1110–1122. [Google Scholar] [CrossRef] [PubMed]
- Adhit, K.K.; Wanjari, A.; Menon, S.; K, S. Liquid Biopsy: An Evolving Paradigm for Non-invasive Disease Diagnosis and Monitoring in Medicine. Cureus 2023, 15, e50176. [Google Scholar] [CrossRef] [PubMed]
- Agashe, R.; Kurzrock, R. Circulating Tumor Cells: From the Laboratory to the Cancer Clinic. Cancers 2020, 12, 2361. [Google Scholar] [CrossRef] [PubMed]
- ALIX-PANABIèRES, C. 2012. EPISPOT Assay: Detection of Viable DTCs/CTCs in Solid Tumor Patients. In: IGNATIADIS, M., SOTIRIOU, C. & PANTEL, K. (eds.) Minimal Residual Disease and Circulating Tumor Cells in Breast Cancer. Berlin, Heidelberg: Springer Berlin Heidelberg.
- Alix-Panabières, C.; Pantel, K. Challenges in circulating tumour cell research. Nat. Rev. Cancer 2014, 14, 623–631. [Google Scholar] [CrossRef]
- Alix-Panabières, C.; Rebillard, X.; Brouillet, J.-P.; Barbotte, E.; Iborra, F.; Segui, B.; Maudelonde, T.; Jolivet-Reynaud, C.; Vendrell, J.-P. Detection of Circulating Prostate-Specific Antigen–Secreting Cells in Prostate Cancer Patients. Clin. Chem. 2005, 51, 1538–1541. [Google Scholar] [CrossRef] [PubMed]
- Alix-Panabières, C.; Riethdorf, S.; Pantel, K. Circulating Tumor Cells and Bone Marrow Micrometastasis. Clin. Cancer Res. 2008, 14, 5013–5021. [Google Scholar] [CrossRef]
- Amintas, S.; Bedel, A.; Moreau-Gaudry, F.; Boutin, J.; Buscail, L.; Merlio, J.-P.; Vendrely, V.; Dabernat, S.; Buscail, E. Circulating Tumor Cell Clusters: United We Stand Divided We Fall. Int. J. Mol. Sci. 2020, 21, 2653. [Google Scholar] [CrossRef] [PubMed]
- ANDERGASSEN, U. , KöLBL, A. C., HUTTER, S., FRIESE, K. & JESCHKE, U. 2013. Detection of Circulating Tumour Cells from Blood of Breast Cancer Patients via RT-qPCR. Cancers, 5, 1212-1220.
- Annala, M.; Vandekerkhove, G.; Khalaf, D.; Taavitsainen, S.; Beja, K.; Warner, E.W.; Sunderland, K.; Kollmannsberger, C.; Eigl, B.J.; Finch, D.; et al. Circulating Tumor DNA Genomics Correlate with Resistance to Abiraterone and Enzalutamide in Prostate Cancer. Cancer Discov. 2018, 8, 444–457. [Google Scholar] [CrossRef] [PubMed]
- Antounians, L.; Tzanetakis, A.; Pellerito, O.; Catania, V.D.; Sulistyo, A.; Montalva, L.; McVey, M.J.; Zani, A. The Regenerative Potential of Amniotic Fluid Stem Cell Extracellular Vesicles: Lessons Learned by Comparing Different Isolation Techniques. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef]
- Armakolas, A.; Kotsari, M.; Koskinas, J. Liquid Biopsies, Novel Approaches and Future Directions. Cancers 2023, 15, 1579. [Google Scholar] [CrossRef]
- AZAD, A. A. , VOLIK, S. V., WYATT, A. W., HAEGERT, A., LE BIHAN, S., BELL, R. H., ANDERSON, S. A., MCCONEGHY, B., SHUKIN, R., BAZOV, J., YOUNGREN, J., PARIS, P., THOMAS, G., SMALL, E. J., WANG, Y., GLEAVE, M. E., COLLINS, C. C. & CHI, K. N. 2015. Androgen Receptor Gene Aberrations in Circulating Cell-Free DNA: Biomarkers of Therapeutic Resistance in Castration-Resistant Prostate Cancer. Clin Cancer Res, 21, 2315-24.
- BAGHERI, A. , KHORSHID, H. R. K., TAVALLAIE, M., MOWLA, S. J., SHERAFATIAN, M., RASHIDI, M., ZARGARI, M., BOROUJENI, M. E. & HOSSEINI, S. M. 2019. A panel of noncoding RNAs in non-small-cell lung cancer. J Cell Biochem, 120, 8280-8290.
- Basch, E.; Loblaw, D.A.; Oliver, T.K.; Carducci, M.; Chen, R.C.; Frame, J.N.; Garrels, K.; Hotte, S.; Kattan, M.W.; Raghavan, D.; et al. Systemic Therapy in Men With Metastatic Castration-Resistant Prostate Cancer: American Society of Clinical Oncology and Cancer Care Ontario Clinical Practice Guideline. J. Clin. Oncol. 2014, 32, 3436–3448. [Google Scholar] [CrossRef] [PubMed]
- Belli, R.; Ferraro, E.; Molfino, A.; Carletti, R.; Tambaro, F.; Costelli, P.; Muscaritoli, M. Liquid Biopsy for Cancer Cachexia: Focus on Muscle-Derived microRNAs. Int. J. Mol. Sci. 2021, 22, 9007. [Google Scholar] [CrossRef] [PubMed]
- Benecke, L.; Chiang, D.M.; Ebnoether, E.; Pfaffl, M.W.; Muller, L. Isolation and analysis of tumor-derived extracellular vesicles from head and neck squamous cell carcinoma plasma by galectin-based glycan recognition particles. Int. J. Oncol. 2022, 61, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Beylerli, O.; Gareev, I.; Sufianov, A.; Ilyasova, T.; Guang, Y. Long noncoding RNAs as promising biomarkers in cancer. Non-coding RNA Res. 2022, 7, 66–70. [Google Scholar] [CrossRef] [PubMed]
- James, K.; Bryl-Gorecka, P.; Olde, B.; Gidlof, O.; Torngren, K.; Erlinge, D. Increased expression of miR-224-5p in circulating extracellular vesicles of patients with reduced coronary flow reserve. BMC Cardiovasc. Disord. 2022, 22, 1–10. [Google Scholar] [CrossRef] [PubMed]
- BURR, R. , EDD, J. F., CHIRN, B., MISHRA, A., HABER, D. A., TONER, M. & MAHESWARAN, S. 2022. Negative-Selection Enrichment of Circulating Tumor Cells from Peripheral Blood Using the Microfluidic CTC-iChip. In: VIVANCO, M. D. (ed.) Mammary Stem Cells: Methods and Protocols. New York, NY: Springer US.
- Cabezas-Camarero, S.; Pérez-Segura, P. Liquid Biopsy in Head and Neck Cancer: Current Evidence and Future Perspective on Squamous Cell, Salivary Gland, Paranasal Sinus and Nasopharyngeal Cancers. Cancers 2022, 14, 2858. [Google Scholar] [CrossRef] [PubMed]
- Castro-Giner, F.; Scheidmann, M.C.; Aceto, N. Beyond enumeration: Functional and Computational Analysis of Circulating Tumor Cells to investigate Cancer Metastasis. Front. Med. 2018, 5, 34. [Google Scholar] [CrossRef] [PubMed]
- Cayrefourcq, L.; De Roeck, A.; Garcia, C.; Stoebner, P.-E.; Fichel, F.; Garima, F.; Perriard, F.; Daures, J.-P.; Meunier, L.; Alix-Panabières, C. S100-EPISPOT: A New Tool to Detect Viable Circulating Melanoma Cells. Cells 2019, 8, 755. [Google Scholar] [CrossRef] [PubMed]
- Zeid, F.A.; Charrier, H.; Beseme, O.; Michel, J.-B.; Mulder, P.; Amouyel, P.; Pinet, F.; Turkieh, A. Lim Domain Binding 3 (Ldb3) Identified as a Potential Marker of Cardiac Extracellular Vesicles. Int. J. Mol. Sci. 2022, 23, 7374. [Google Scholar] [CrossRef]
- Cheng, Y.; Zhang, S.; Qin, L.; Zhao, J.; Song, H.; Yuan, Y.; Sun, J.; Tian, F.; Liu, C. Poly(ethylene oxide) Concentration Gradient-Based Microfluidic Isolation of Circulating Tumor Cells. Anal. Chem. 2023, 95, 3468–3475. [Google Scholar] [CrossRef]
- Chu, P.-Y.; Hsieh, C.-H.; Wu, M.-H. The Combination of Immunomagnetic Bead-Based Cell Isolation and Optically Induced Dielectrophoresis (ODEP)-Based Microfluidic Device for the Negative Selection-Based Isolation of Circulating Tumor Cells (CTCs). Front. Bioeng. Biotechnol. 2020, 8, 921. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J.D.; Li, L.; Wang, Y.; Thoburn, C.; Afsari, B.; Danilova, L.; Douville, C.; Javed, A.A.; Wong, F.; Mattox, A.; et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018, 359, 926–930. [Google Scholar] [CrossRef] [PubMed]
- Connal, S.; Cameron, J.M.; Sala, A.; Brennan, P.M.; Palmer, D.S.; Palmer, J.D.; Perlow, H.; Baker, M.J. Liquid biopsies: the future of cancer early detection. J. Transl. Med. 2023, 21, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Conteduca, V.; Wetterskog, D.; Sharabiani, M.T.A.; Grande, E.; Fernandez-Pérez, M.P.; Jayaram, A.; Salvi, S.; Castellano, D.; Romanel, A.; Lolli, C.; et al. Androgen receptor gene status in plasma DNA associates with worse outcome on enzalutamide or abiraterone for castration-resistant prostate cancer: a multi-institution correlative biomarker study. Ann. Oncol. 2017, 28, 1508–1516. [Google Scholar] [CrossRef] [PubMed]
- De Bono, J.S.; Scher, H.I.; Montgomery, R.B.; Parker, C.; Miller, M.C.; Tissing, H.; Doyle, G.V.; Terstappen, L.W.W.M.; Pienta, K.J.; Raghavan, D. Circulating Tumor Cells Predict Survival Benefit from Treatment in Metastatic Castration-Resistant Prostate Cancer. Clin. Cancer Res. 2008, 14, 6302–6309. [Google Scholar] [CrossRef]
- DE MIRANDA, F. S. , BARAUNA, V. G., DOS SANTOS, L., COSTA, G., VASSALLO, P. F. & CAMPOS, L. C. G. 2021. Properties and Application of Cell-Free DNA as a Clinical Biomarker. Int J Mol Sci, 22.
- DELMONICO, L. , ALVES, G. & BINES, J. 2020. Cell free DNA biology and its involvement in breast carcinogenesis. Adv Clin Chem, 97, 171-223.
- Denève, E.; Riethdorf, S.; Ramos, J.; Nocca, D.; Coffy, A.; Daurès, J.-P.; Maudelonde, T.; Fabre, J.-M.; Pantel, K.; Alix-Panabières, C. Capture of Viable Circulating Tumor Cells in the Liver of Colorectal Cancer Patients. Clin. Chem. 2013, 59, 1384–1392. [Google Scholar] [CrossRef] [PubMed]
- Diehl, F.; Schmidt, K.; Choti, M.A.; Romans, K.; Goodman, S.; Li, M.; Thornton, K.; Agrawal, N.; Sokoll, L.; Szabo, S.A.; et al. Circulating mutant DNA to assess tumor dynamics. Nat. Med. 2008, 14, 985–990. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Zhao, W. The Application of Liquid Biopsy Techniques in High-Risk Population for Hepatocellular Carcinoma. Cancer Manag. Res. 2022, ume 14, 2735–2748. [Google Scholar] [CrossRef]
- Ding, Y.; Li, W.; Wang, K.; Xu, C.; Hao, M.; Ding, L. Perspectives of the Application of Liquid Biopsy in Colorectal Cancer. BioMed Res. Int. 2020, 2020, 1–13. [Google Scholar] [CrossRef]
- Eigeliene, N.; Saarenheimo, J.; Jekunen, A. Potential of Liquid Biopsies for Breast Cancer Screening, Diagnosis, and Response to Treatment. Oncology 2019, 96, 115–124. [Google Scholar] [CrossRef]
- EL MESSAOUDI, S. , ROLET, F., MOULIERE, F. & THIERRY, A. R. 2013. Circulating cell free DNA: Preanalytical considerations. Clinica Chimica Acta, 424, 222-230.
- Elazezy, M.; Joosse, S.A. Techniques of using circulating tumor DNA as a liquid biopsy component in cancer management. Comput. Struct. Biotechnol. J. 2018, 16, 370–378. [Google Scholar] [CrossRef]
- Eslami-S, Z.; Cortés-Hernández, L.E.; Thomas, F.; Pantel, K.; Alix-Panabières, C. Functional analysis of circulating tumour cells: the KEY to understand the biology of the metastatic cascade. Br. J. Cancer 2022, 127, 800–810. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Chen, Y.; Yang, J.; Zhuo, C.; Huang, S.; Zhang, H.; Shi, Y. Clinical Perspectives on Liquid Biopsy in Metastatic Colorectal Cancer. Front. Genet. 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- Garrel, R.; Mazel, M.; Perriard, F.; Vinches, M.; Cayrefourcq, L.; Guigay, J.; Digue, L.; Aubry, K.; Alfonsi, M.; Delord, J.-P.; et al. Circulating Tumor Cells as a Prognostic Factor in Recurrent or Metastatic Head and Neck Squamous Cell Carcinoma: The CIRCUTEC Prospective Study. Clin. Chem. 2019, 65, 1267–1275. [Google Scholar] [CrossRef] [PubMed]
- Gawel, S.H.; Jackson, L.; Jeanblanc, N.; Davis, G.J. Current and future opportunities for liquid biopsy of circulating biomarkers to aid in early cancer detection. J. Cancer Metastasis Treat. 2022, 8, 26. [Google Scholar] [CrossRef]
- Geijsen, A.J.; van Roekel, E.H.; van Duijnhoven, F.J.; Achaintre, D.; Bachleitner-Hofmann, T.; Baierl, A.; Bergmann, M.M.; Boehm, J.; Bours, M.J.; Brenner, H.; et al. Plasma metabolites associated with colorectal cancer stage: Findings from an international consortium. Int. J. Cancer 2019, 146, 3256–3266. [Google Scholar] [CrossRef]
- George, J.; Lim, J.S.; Jang, S.J.; Cun, Y.; Ozretić, L.; Kong, G.; Leenders, F.; Lu, X.; Fernández-Cuesta, L.; Bosco, G.; et al. Comprehensive genomic profiles of small cell lung cancer. Nature 2015, 524, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Goodsaid, F.M. The Labyrinth of Product Development and Regulatory Approvals in Liquid Biopsy Diagnostics. Clin. Transl. Sci. 2019, 12, 431–439. [Google Scholar] [CrossRef]
- Grölz, D.; Hauch, S.; Schlumpberger, M.; Guenther, K.; Voss, T.; Sprenger-Haussels, M.; Oelmüller, U. Liquid Biopsy Preservation Solutions for Standardized Pre-Analytical Workflows—Venous Whole Blood and Plasma. Curr. Pathobiol. Rep. 2018, 6, 275–286. [Google Scholar] [CrossRef]
- Guo, W.; Li, Y.; Pang, W.; Shen, H. Exosomes: A Potential Therapeutic Tool Targeting Communications between Tumor Cells and Macrophages. Mol. Ther. 2020, 28, 1953–1964. [Google Scholar] [CrossRef]
- Habli, Z.; AlChamaa, W.; Saab, R.; Kadara, H.; Khraiche, M.L. Circulating Tumor Cell Detection Technologies and Clinical Utility: Challenges and Opportunities. Cancers 2020, 12, 1930. [Google Scholar] [CrossRef]
- Hassan, S.; Blick, T.; Williams, E.D.; Thompson, E.W. Applications of RNA characterisation in circulating tumour cells. Front. Biosci. 2020, 25, 874–892. [Google Scholar] [CrossRef]
- Havers, M.; Broman, A.; Lenshof, A.; Laurell, T. Advancement and obstacles in microfluidics-based isolation of extracellular vesicles. Anal. Bioanal. Chem. 2022, 415, 1265–1285. [Google Scholar] [CrossRef]
- HE, J. , XI, N., HAN, Z., LUO, W., SHEN, J., WANG, S., LI, J., GUO, Z. & CHENG, H. 2022. The Role of Liquid Biopsy Analytes in Diagnosis, Treatment and Prognosis of Colorectal Cancer. Front Endocrinol (Lausanne), 13, 875442.
- Heller, G.; Fizazi, K.; McCormack, R.; Molina, A.; MacLean, D.; Webb, I.J.; Saad, F.; de Bono, J.S.; Scher, H.I. The Added Value of Circulating Tumor Cell Enumeration to Standard Markers in Assessing Prognosis in a Metastatic Castration-Resistant Prostate Cancer Population. Clin. Cancer Res. 2017, 23, 1967–1973. [Google Scholar] [CrossRef] [PubMed]
- Heller, G.; McCormack, R.; Kheoh, T.; Molina, A.; Smith, M.R.; Dreicer, R.; Saad, F.; de Wit, R.; Aftab, D.T.; Hirmand, M.; et al. Circulating Tumor Cell Number as a Response Measure of Prolonged Survival for Metastatic Castration-Resistant Prostate Cancer: A Comparison With Prostate-Specific Antigen Across Five Randomized Phase III Clinical Trials. J. Clin. Oncol. 2018, 36, 572–580. [Google Scholar] [CrossRef] [PubMed]
- HIRAHATA, T. , UL QURAISH, R., QURAISH, A. U., UL QURAISH, S., NAZ, M. & RAZZAQ, M. A. 2022. Liquid Biopsy: A Distinctive Approach to the Diagnosis and Prognosis of Cancer. Cancer Inform, 21, 11769351221076062.
- Huang, Y.; Li, X.; Hou, J.; Luo, Z.; Yang, G.; Zhou, S. Conductive Nanofibers-Enhanced Microfluidic Device for the Efficient Capture and Electrical Stimulation-Triggered Rapid Release of Circulating Tumor Cells. Biosensors 2023, 13, 497. [Google Scholar] [CrossRef]
- ILIE, M. & HOFMAN, P. 2016. Pros: Can tissue biopsy be replaced by liquid biopsy? Transl Lung Cancer Res, 5, 420-3.
- Jin, N.; Kan, C.-M.; Pei, X.M.; Cheung, W.L.; Ng, S.S.M.; Wong, H.T.; Cheng, H.Y.-L.; Leung, W.W.; Ni Wong, Y.; Tsang, H.F.; et al. Cell-free circulating tumor RNAs in plasma as the potential prognostic biomarkers in colorectal cancer. Front. Oncol. 2023, 13. [Google Scholar] [CrossRef] [PubMed]
- Jin, N.; Kan, C.-M.; Pei, X.M.; Cheung, W.L.; Ng, S.S.M.; Wong, H.T.; Cheng, H.Y.-L.; Leung, W.W.; Ni Wong, Y.; Tsang, H.F.; et al. Cell-free circulating tumor RNAs in plasma as the potential prognostic biomarkers in colorectal cancer. Front. Oncol. 2023, 13. [Google Scholar] [CrossRef]
- Jorgez, C.J.; Dang, D.D.; Simpson, J.L.; Lewis, D.E.; Bischoff, F.Z. Quantity versus quality: Optimal methods for cell-free DNA isolation from plasma of pregnant women. Anesthesia Analg. 2006, 8, 615–619. [Google Scholar] [CrossRef]
- Kan, C.-M.; Pei, X.M.; Yeung, M.H.Y.; Jin, N.; Ng, S.S.M.; Tsang, H.F.; Cho, W.C.S.; Yim, A.K.-Y.; Yu, A.C.-S.; Wong, S.C.C. Exploring the Role of Circulating Cell-Free RNA in the Development of Colorectal Cancer. Int. J. Mol. Sci. 2023, 24, 11026. [Google Scholar] [CrossRef]
- Kan, C.-M.; Tsang, H.F.; Pei, X.M.; Ng, S.S.M.; Yim, A.K.-Y.; Yu, A.C.-S.; Wong, S.C.C. Enhancing Clinical Utility: Utilization of International Standards and Guidelines for Metagenomic Sequencing in Infectious Disease Diagnosis. Int. J. Mol. Sci. 2024, 25, 3333. [Google Scholar] [CrossRef]
- Khachfe, H.H. Use of liquid biopsies in gastrointestinal cancers. World J. Gastrointest. Oncol. 2021, 13, 1210–1212. [Google Scholar] [CrossRef]
- KOLENDA, T. , GUGLAS, K., BARANOWSKI, D., SOBOCINSKA, J., KOPCZYNSKA, M., TERESIAK, A., BLIZNIAK, R. & LAMPERSKA, K. 2020. cfRNAs as biomarkers in oncology - still experimental or applied tool for personalized medicine already? Rep Pract Oncol Radiother, 25, 783-792.
- Krawczyk, N.; Meier-Stiegen, F.; Banys, M.; Neubauer, H.; Ruckhaeberle, E.; Fehm, T. Expression of Stem Cell and Epithelial-Mesenchymal Transition Markers in Circulating Tumor Cells of Breast Cancer Patients. BioMed Res. Int. 2014, 2014, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Kwan, T.T.; Bardia, A.; Spring, L.M.; Giobbie-Hurder, A.; Kalinich, M.; Dubash, T.; Sundaresan, T.; Hong, X.; LiCausi, J.A.; Ho, U.; et al. A Digital RNA Signature of Circulating Tumor Cells Predicting Early Therapeutic Response in Localized and Metastatic Breast Cancer. Cancer Discov. 2018, 8, 1286–1299. [Google Scholar] [CrossRef] [PubMed]
- Lampis, A.; Ghidini, M.; Ratti, M.; Mirchev, M.B.; Okuducu, A.F.; Valeri, N.; Hahne, J.C. Circulating Tumour DNAs and Non-Coding RNAs as Liquid Biopsies for the Management of Colorectal Cancer Patients. Gastrointest. Disord. 2020, 2, 212–235. [Google Scholar] [CrossRef]
- Li, J.; Guan, X.; Fan, Z.; Ching, L.-M.; Li, Y.; Wang, X.; Cao, W.-M.; Liu, D.-X. Non-Invasive Biomarkers for Early Detection of Breast Cancer. Cancers 2020, 12, 2767. [Google Scholar] [CrossRef]
- Liebs, S.; Nonnenmacher, A.; Klauschen, F.; Keilholz, U.; Vecchione, L. Liquid biopsy assessment of synchronous malignancies: a case report and review of the literature. ESMO Open 2019, 4, e000528. [Google Scholar] [CrossRef]
- Lim, M.; Kim, C.-J.; Sunkara, V.; Kim, M.-H.; Cho, Y.-K. Liquid Biopsy in Lung Cancer: Clinical Applications of Circulating Biomarkers (CTCs and ctDNA). Micromachines 2018, 9, 100. [Google Scholar] [CrossRef] [PubMed]
- Lin, B.; Jiang, J.; Jia, J.; Zhou, X. Recent Advances in Exosomal miRNA Biosensing for Liquid Biopsy. Molecules 2022, 27, 7145. [Google Scholar] [CrossRef]
- Lin, D.; Shen, L.; Luo, M.; Zhang, K.; Li, J.; Yang, Q.; Zhu, F.; Zhou, D.; Zheng, S.; Chen, Y.; et al. Circulating tumor cells: biology and clinical significance. Signal Transduct. Target. Ther. 2021, 6, 1–24. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, Y.; Sun, S.; Chen, Z.; Xiang, S.; Ding, Z.; Huang, Z.; Zhang, B. Understanding the versatile roles and applications of EpCAM in cancers: from bench to bedside. Exp. Hematol. Oncol. 2022, 11, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Lokhandwala, P.M.; Riel, S.L.; Haley, L.; Lu, C.; Chen, Y.; Silberstein, J.; Zhu, Y.; Zheng, G.; Lin, M.-T.; Gocke, C.D.; et al. Analytical Validation of Androgen Receptor Splice Variant 7 Detection in a Clinical Laboratory Improvement Amendments (CLIA) Laboratory Setting. J. Mol. Diagn. 2016, 19, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Lorente, D.; Olmos, D.; Mateo, J.; Bianchini, D.; Seed, G.; Fleisher, M.; Danila, D.C.; Flohr, P.; Crespo, M.; Figueiredo, I.; et al. Decline in Circulating Tumor Cell Count and Treatment Outcome in Advanced Prostate Cancer. Eur. Urol. 2016, 70, 985–992. [Google Scholar] [CrossRef] [PubMed]
- Lozano, R.; Lorente, D.; Aragon, I.M.; Romero-Laorden, N.; Nombela, P.; Mateo, J.; Reid, A.H.M.; Cendón, Y.; Bianchini, D.; Llacer, C.; et al. Value of Early Circulating Tumor Cells Dynamics to Estimate Docetaxel Benefit in Metastatic Castration-Resistant Prostate Cancer (mCRPC) Patients. Cancers 2021, 13, 2334. [Google Scholar] [CrossRef] [PubMed]
- Lu, D.; Krupa, R.; Harvey, M.; Graf, R.P.; Schreiber, N.; Barnette, E.; Carbone, E.; Jendrisak, A.; Gill, A.; Orr, S.; et al. Development of an immunofluorescent AR-V7 circulating tumor cell assay – A blood-based test for men with metastatic prostate cancer. J. Circ. Biomarkers 2020, 9, 13–19. [Google Scholar] [CrossRef]
- Lu, S.; Kong, H.; Hou, Y.; Ge, D.; Huang, W.; Ou, J.; Yang, D.; Zhang, L.; Wu, G.; Song, Y.; et al. Two plasma microRNA panels for diagnosis and subtype discrimination of lung cancer. Lung Cancer 2018, 123, 44–51. [Google Scholar] [CrossRef] [PubMed]
- LU, Y. T. , DELIJANI, K., MECUM, A. & GOLDKORN, A. 2019. ≪p>Current Status of Liquid Biopsies for the Detection and Management of Prostate Cancer. Cancer Management and Research, Volume 11, 5271-5291.
- Markou, .; Londra, D.; Tserpeli, V.; Kollias, .; Tsaroucha, E.; Vamvakaris, I.; Potaris, K.; Pateras, I.; Kotsakis, .; Georgoulias, V.; et al. DNA methylation analysis of tumor suppressor genes in liquid biopsy components of early stage NSCLC: a promising tool for early detection. Clin. Epigenetics 2022, 14, 1–9. [Google Scholar] [CrossRef]
- Martins, I.; Ribeiro, I.P.; Jorge, J.; Gonçalves, A.C.; Sarmento-Ribeiro, A.B.; Melo, J.B.; Carreira, I.M. Liquid Biopsies: Applications for Cancer Diagnosis and Monitoring. Genes 2021, 12, 349. [Google Scholar] [CrossRef] [PubMed]
- Mathios, D.; Johansen, J.S.; Cristiano, S.; Medina, J.E.; Phallen, J.; Larsen, K.R.; Bruhm, D.C.; Niknafs, N.; Ferreira, L.; Adleff, V.; et al. Detection and characterization of lung cancer using cell-free DNA fragmentomes. Nat. Commun. 2021, 12, 1–14. [Google Scholar] [CrossRef]
- MAZARD, T., CAYREFOURCQ, L., PERRIARD, F., SENELLART, H., LINOT, B., DE LA FOUCHARDIèRE, C., TERREBONNE, E., FRANçOIS, E., OBLED, S., GUIMBAUD, R., MINEUR, L., FONCK, M., DAURèS, J. P., YCHOU, M., ASSENAT, E. & ALIX-PANABIèRES, C. 2021. Clinical Relevance of Viable Circulating Tumor Cells in Patients with Metastatic Colorectal Cancer: The COLOSPOT Prospective Study. Cancers (Basel), 13.
- Mazzitelli, C.; Santini, D.; Corradini, A.G.; Zamagni, C.; Trerè, D.; Montanaro, L.; Taffurelli, M. Liquid Biopsy in the Management of Breast Cancer Patients: Where Are We Now and Where Are We Going. Diagnostics 2023, 13, 1241. [Google Scholar] [CrossRef]
- MEDINA DIAZ, I. , NOCON, A., MEHNERT, D. H., FREDEBOHM, J., DIEHL, F. & HOLTRUP, F. 2016. Performance of Streck cfDNA Blood Collection Tubes for Liquid Biopsy Testing. PLOS ONE, 11, e0166354.
- Mikilps-Mikgelbs, R.; Pūpola, D.; Antone, E.; Kiršners, A.; Luguzis, A.; Salna, E.; Krams, A.; Ērglis, A. Liquid Biopsy — A Novel Diagnostic Tool for Management of Early-Stage Peripheral Lung Cancer. Proc. Latv. Acad. Sci. Sect. B. Nat. Exact, Appl. Sci. 2022, 76, 325–332. [Google Scholar] [CrossRef]
- Miller, M.C.; Doyle, G.V.; Terstappen, L.W.M.M. Significance of Circulating Tumor Cells Detected by the CellSearch System in Patients with Metastatic Breast Colorectal and Prostate Cancer. J. Oncol. 2009, 2010, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.; Dubash, T.D.; F., J.; Jewett, M.K.; Garre, S.G.; Karabacak, N.M.; Rabe, D.C.; Mutlu, B.R.; Walsh, J.R.; Kapur, R.; et al. Ultrahigh-throughput magnetic sorting of large blood volumes for epitope-agnostic isolation of circulating tumor cells. Proc. Natl. Acad. Sci. USA 2020, 117, 16839–16847. [Google Scholar] [CrossRef] [PubMed]
- Müller, I.; Beeger, C.; Alix-Panabières, C.; Rebillard, X.; Pantel, K.; Schwarzenbach, H. Identification of Loss of Heterozygosity on Circulating Free DNA in Peripheral Blood of Prostate Cancer Patients: Potential and Technical Improvements. Clin. Chem. 2008, 54, 688–696. [Google Scholar] [CrossRef] [PubMed]
- Nagasaka, M.; Uddin, M.H.; Al-Hallak, M.N.; Rahman, S.; Balasubramanian, S.; Sukari, A.; Azmi, A.S. Liquid biopsy for therapy monitoring in early-stage non-small cell lung cancer. Mol. Cancer 2021, 20, 1–16. [Google Scholar] [CrossRef] [PubMed]
- NETWORK, C. G. A. R. 2015. The Molecular Taxonomy of Primary Prostate Cancer. Cell, 163, 1011-25.
- Nordgård, O.; Forthun, R.B.; Lapin, M.; Grønberg, B.H.; Kalland, K.H.; Kopperud, R.K.; Thomsen, L.C.V.; Tjensvoll, K.; Gilje, B.; Gjertsen, B.T.; et al. Liquid Biopsies in Solid Cancers: Implementation in a Nordic Healthcare System. Cancers 2021, 13, 1861. [Google Scholar] [CrossRef] [PubMed]
- Nosaki, K.; Satouchi, M.; Kurata, T.; Yoshida, T.; Okamoto, I.; Katakami, N.; Imamura, F.; Tanaka, K.; Yamane, Y.; Yamamoto, N.; et al. Re-biopsy status among non-small cell lung cancer patients in Japan: A retrospective study. Lung Cancer 2016, 101, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Oliva, S.; D'Agostino, M.; Boccadoro, M.; Larocca, A. Clinical Applications and Future Directions of Minimal Residual Disease Testing in Multiple Myeloma. Front. Oncol. 2020, 10, 1. [Google Scholar] [CrossRef]
- Palacín-Aliana, I.; García-Romero, N.; Asensi-Puig, A.; Carrión-Navarro, J.; González-Rumayor, V.; Ayuso-Sacido. Clinical Utility of Liquid Biopsy-Based Actionable Mutations Detected via ddPCR. Biomedicines 2021, 9, 906. [Google Scholar] [CrossRef]
- Pantel, K.; Alix-Panabières, C. The clinical significance of circulating tumor cells. Nat. Clin. Pr. Oncol. 2007, 4, 62–63. [Google Scholar] [CrossRef]
- Pantel, K.; Alix-Panabières, C. Circulating tumour cells in cancer patients: challenges and perspectives. Trends Mol. Med. 2010, 16, 398–406. [Google Scholar] [CrossRef] [PubMed]
- PANTEL, K. , ALIX-PANABIèRES, C. & RIETHDORF, S. 2009. Cancer micrometastases. Nature Reviews Clinical Oncology, 6, 339-351.
- Paoletti, C.; Larios, J.M.; Muñiz, M.C.; Aung, K.; Cannell, E.M.; Darga, E.P.; Kidwell, K.M.; Thomas, D.G.; Tokudome, N.; Brown, M.E.; et al. Heterogeneous estrogen receptor expression in circulating tumor cells suggests diverse mechanisms of fulvestrant resistance. Mol. Oncol. 2016, 10, 1078–1085. [Google Scholar] [CrossRef] [PubMed]
- Paracchini, L.; D’incalci, M.; Marchini, S. Liquid Biopsy in the Clinical Management of High-Grade Serous Epithelial Ovarian Cancer—Current Use and Future Opportunities. Cancers 2021, 13, 2386. [Google Scholar] [CrossRef] [PubMed]
- Pardini, B.; Sabo, A.A.; Birolo, G.; Calin, G.A. Noncoding RNAs in Extracellular Fluids as Cancer Biomarkers: The New Frontier of Liquid Biopsies. Cancers 2019, 11, 1170. [Google Scholar] [CrossRef] [PubMed]
- E Payne, R.; Wang, F.; Su, N.; Krell, J.; Zebrowski, A.; Yagüe, E.; Ma, X.-J.; Luo, Y.; Coombes, R.C. Viable circulating tumour cell detection using multiplex RNA in situ hybridisation predicts progression-free survival in metastatic breast cancer patients. Br. J. Cancer 2012, 106, 1790–1797. [Google Scholar] [CrossRef] [PubMed]
- van der Pol, Y.; Moldovan, N.; Verkuijlen, S.; Ramaker, J.; Boers, D.; Onstenk, W.; de Rooij, J.; Bahce, I.; Pegtel, D.M.; Mouliere, F. The Effect of Preanalytical and Physiological Variables on Cell-Free DNA Fragmentation. Clin. Chem. 2022, 68, 803–813. [Google Scholar] [CrossRef] [PubMed]
- Ponti, G.; Manfredini, M.; Tomasi, A. Non-blood sources of cell-free DNA for cancer molecular profiling in clinical pathology and oncology. Crit. Rev. Oncol. 2019, 141, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Powell, A.A.; Talasaz, A.H.; Zhang, H.; Coram, M.A.; Reddy, A.; Deng, G.; Telli, M.L.; Advani, R.H.; Carlson, R.W.; Mollick, J.A.; et al. Single Cell Profiling of Circulating Tumor Cells: Transcriptional Heterogeneity and Diversity from Breast Cancer Cell Lines. PLOS ONE 2012, 7, e33788. [Google Scholar] [CrossRef] [PubMed]
- Preethi, K.A.; Selvakumar, S.C.; Ross, K.; Jayaraman, S.; Tusubira, D.; Sekar, D. Liquid biopsy: Exosomal microRNAs as novel diagnostic and prognostic biomarkers in cancer. Mol. Cancer 2022, 21, 1–15. [Google Scholar] [CrossRef]
- Ramirez-Garrastacho, M.; Bajo-Santos, C.; Line, A.; Martens-Uzunova, E.S.; de la Fuente, J.M.; Moros, M.; Soekmadji, C.; Tasken, K.A.; Llorente, A. Extracellular vesicles as a source of prostate cancer biomarkers in liquid biopsies: a decade of research. Br. J. Cancer 2021, 126, 331–350. [Google Scholar] [CrossRef]
- Ramirez, J.-M.; Fehm, T.; Orsini, M.; Cayrefourcq, L.; Maudelonde, T.; Pantel, K.; Alix-Panabières, C. Prognostic Relevance of Viable Circulating Tumor Cells Detected by EPISPOT in Metastatic Breast Cancer Patients. Clin. Chem. 2014, 60, 214–221. [Google Scholar] [CrossRef] [PubMed]
- Rastogi, A. Changing role of histopathology in the diagnosis and management of hepatocellular carcinoma. World J. Gastroenterol. 2018, 24, 4000–4013. [Google Scholar] [CrossRef] [PubMed]
- Ratti, M.; Lampis, A.; Ghidini, M.; Salati, M.; Mirchev, M.B.; Valeri, N.; Hahne, J.C. MicroRNAs (miRNAs) and Long Non-Coding RNAs (lncRNAs) as New Tools for Cancer Therapy: First Steps from Bench to Bedside. Target. Oncol. 2020, 15, 261–278. [Google Scholar] [CrossRef] [PubMed]
- Rink, M.; Party, O.B.O.T.E.Y.A.U.C.W.; Schwarzenbach, H.; Vetterlein, M.W.; Riethdorf, S.; Soave, A. The current role of circulating biomarkers in non-muscle invasive bladder cancer. Transl. Androl. Urol. 2019, 8, 61–75. [Google Scholar] [CrossRef] [PubMed]
- RUSSANO, M. , NAPOLITANO, A., RIBELLI, G., IULIANI, M., SIMONETTI, S., CITARELLA, F., PANTANO, F., DELL'AQUILA, E., ANESI, C., SILVESTRIS, N., ARGENTIERO, A., SOLIMANDO, A. G., VINCENZI, B., TONINI, G. & SANTINI, D. 2020. Liquid biopsy and tumor heterogeneity in metastatic solid tumors: the potentiality of blood samples. J Exp Clin Cancer Res, 39, 95.
- SACHER, A. G. , PAWELETZ, C., DAHLBERG, S. E., ALDEN, R. S., O'CONNELL, A., FEENEY, N., MACH, S. L., JäNNE, P. A. & OXNARD, G. R. 2016. Prospective Validation of Rapid Plasma Genotyping for the Detection of EGFR and KRAS Mutations in Advanced Lung Cancer. JAMA Oncol, 2, 1014-22.
- SAHA, S. , ARAF, Y. & PROMON, S. K. 2022. Circulating tumor DNA in cancer diagnosis, monitoring, and prognosis. Journal of the Egyptian National Cancer Institute, 34, 8.
- Sanches, S.M.; Braun, A.C.; Calsavara, V.F.; Barbosa, P.N.V.P.; Chinen, L.T.D. Comparison of hormonal receptor expression and HER2 status between circulating tumor cells and breast cancer metastases. Clinics 2021, 76. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, B.; Weickmann, S.; Witt, C.; Fleischhacker, M. Improved Method for Isolating Cell-Free DNA. Clin. Chem. 2005, 51, 1561–1563. [Google Scholar] [CrossRef] [PubMed]
- Sequist, L.V.; Nagrath, S.; Toner, M.; Haber, D.A.; Lynch, T.J. The CTC-Chip: An Exciting New Tool to Detect Circulating Tumor Cells in Lung Cancer Patients. J. Thorac. Oncol. 2009, 4, 281–283. [Google Scholar] [CrossRef] [PubMed]
- Serratì, S.; Palazzo, A.; Lapenna, A.; Mateos, H.; Mallardi, A.; Marsano, R.M.; Quarta, A.; Del Rosso, M.; Azzariti, A. Salting-Out Approach Is Worthy of Comparison with Ultracentrifugation for Extracellular Vesicle Isolation from Tumor and Healthy Models. Biomolecules 2021, 11, 1857. [Google Scholar] [CrossRef] [PubMed]
- Shegekar, T.; Vodithala, S.; Juganavar, A. The Emerging Role of Liquid Biopsies in Revolutionising Cancer Diagnosis and Therapy. Cureus 2023, 15, e43650. [Google Scholar] [CrossRef]
- Shih, J.; Gow, C.; Yu, C.; Yang, C.; Chang, Y.; Tsai, M.; Hsu, Y.; Chen, K.; Su, W.; Yang, P. Epidermal growth factor receptor mutations in needle biopsy/aspiration samples predict response to gefitinib therapy and survival of patients with advanced nonsmall cell lung cancer. Int. J. Cancer 2005, 118, 963–969. [Google Scholar] [CrossRef]
- Shoukry, M.; Broccard, S.; Kaplan, J.; Gabriel, E. The Emerging Role of Circulating Tumor DNA in the Management of Breast Cancer. Cancers 2021, 13, 3813. [Google Scholar] [CrossRef] [PubMed]
- Stergiopoulou, D.; Markou, A.; Strati, A.; Zavridou, M.; Tzanikou, E.; Mastoraki, S.; Kallergi, G.; Georgoulias, V.; Lianidou, E. Comprehensive liquid biopsy analysis as a tool for the early detection of minimal residual disease in breast cancer. Sci. Rep. 2023, 13, 1–13. [Google Scholar] [CrossRef]
- Stevic, I.; Buescher, G.; Ricklefs, F.L. Monitoring Therapy Efficiency in Cancer through Extracellular Vesicles. Cells 2020, 9, 130. [Google Scholar] [CrossRef]
- Stott, S.L.; Hsu, C.-H.; Tsukrov, D.I.; Yu, M.; Miyamoto, D.T.; Waltman, B.A.; Rothenberg, S.M.; Shah, A.M.; Smas, M.E.; Korir, G.K.; et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc. Natl. Acad. Sci. USA 2010, 107, 18392–18397. [Google Scholar] [CrossRef]
- Stranska, R.; Gysbrechts, L.; Wouters, J.; Vermeersch, P.; Bloch, K.; Dierickx, D.; Andrei, G.; Snoeck, R. Comparison of membrane affinity-based method with size-exclusion chromatography for isolation of exosome-like vesicles from human plasma. J. Transl. Med. 2018, 16, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Sun, N.; Li, X.; Wang, Z.; Li, Y.; Pei, R. High-purity capture of CTCs based on micro-beads enhanced isolation by size of epithelial tumor cells (ISET) method. Biosens. Bioelectron. 2018, 102, 157–163. [Google Scholar] [CrossRef]
- Sundaresan, T.K.; Sequist, L.V.; Heymach, J.V.; Riely, G.J.; Jänne, P.A.; Koch, W.H.; Sullivan, J.P.; Fox, D.B.; Maher, R.; Muzikansky, A.; et al. Detection of T790M, the Acquired Resistance EGFR Mutation, by Tumor Biopsy versus Noninvasive Blood-Based Analyses. Clin. Cancer Res. 2016, 22, 1103–1110. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, T.; Mori, K.; Sunayama, H.; Takano, E.; Kitayama, Y.; Shimizu, T.; Hirose, Y.; Inubushi, S.; Sasaki, R.; Tanino, H. Antibody-Conjugated Signaling Nanocavities Fabricated by Dynamic Molding for Detecting Cancers Using Small Extracellular Vesicle Markers from Tears. J. Am. Chem. Soc. 2020, 142, 6617–6624. [Google Scholar] [CrossRef]
- Temilola, D.O.; Wium, M.; Coulidiati, T.H.; Adeola, H.A.; Carbone, G.M.; Catapano, C.V.; Zerbini, L.F. The Prospect and Challenges to the Flow of Liquid Biopsy in Africa. Cells 2019, 8, 862. [Google Scholar] [CrossRef]
- Tesovnik, T.; Bizjan, B.J.; Šket, R.; Debeljak, M.; Battelino, T.; Kovač, J. Technological Approaches in the Analysis of Extracellular Vesicle Nucleotide Sequences. Front. Bioeng. Biotechnol. 2021, 9, 787551. [Google Scholar] [CrossRef]
- Toro, P.V.; Erlanger, B.; Beaver, J.A.; Cochran, R.L.; VanDenBerg, D.A.; Yakim, E.; Cravero, K.; Chu, D.; Zabransky, D.J.; Wong, H.Y.; et al. Comparison of cell stabilizing blood collection tubes for circulating plasma tumor DNA. Clin. Biochem. 2015, 48, 993–998. [Google Scholar] [CrossRef] [PubMed]
- Tran, L.S.; Pham, H.-A.T.; Tran, V.-U.; Tran, T.-T.; Dang, A.-T.H.; Le, D.-T.; Nguyen, S.-L.; Nguyen, N.-V.; Nguyen, T.-V.; Vo, B.T.; et al. Ultra-deep massively parallel sequencing with unique molecular identifier tagging achieves comparable performance to droplet digital PCR for detection and quantification of circulating tumor DNA from lung cancer patients. PLOS ONE 2019, 14, e0226193. [Google Scholar] [CrossRef]
- Tulpule, V.; Morrison, G.J.; Falcone, M.; Quinn, D.I.; Goldkorn, A. Integration of Liquid Biopsies in Clinical Management of Metastatic Prostate Cancer. Curr. Oncol. Rep. 2022, 24, 1287–1298. [Google Scholar] [CrossRef] [PubMed]
- Uchiyama, K.; Yagi, N.; Mizushima, K.; Higashimura, Y.; Hirai, Y.; Okayama, T.; Yoshida, N.; Kamada, K.; Handa, O.; Ishikawa, T.; et al. Serum metabolomics analysis for early detection of colorectal cancer. J. Gastroenterol. 2016, 52, 677–694. [Google Scholar] [CrossRef] [PubMed]
- Vacante, M.; Ciuni, R.; Basile, F.; Biondi, A. The Liquid Biopsy in the Management of Colorectal Cancer: An Overview. Biomedicines 2020, 8, 308. [Google Scholar] [CrossRef]
- van der Pol, Y.; Tantyo, N.A.; Evander, N.; E Hentschel, A.; Wever, B.M.; Ramaker, J.; Bootsma, S.; Fransen, M.F.; Lenos, K.J.; Vermeulen, L.; et al. Real-time analysis of the cancer genome and fragmentome from plasma and urine cell-free DNA using nanopore sequencing. EMBO Mol. Med. 2023, 15, e17282. [Google Scholar] [CrossRef] [PubMed]
- van der Toom, E.E.; Verdone, J.E.; Gorin, M.A.; Pienta, K.J. Technical challenges in the isolation and analysis of circulating tumor cells. Oncotarget 2016, 7, 62754–62766. [Google Scholar] [CrossRef] [PubMed]
- Venetis, K.; Cursano, G.; Pescia, C.; D'Ercole, M.; Porta, F.M.; Blanco, M.C.; Frascarelli, C.; Ivanova, M.; Rocco, E.G.; Fusco, N. Liquid biopsy: Cell-free DNA based analysis in breast cancer. J. Liq. Biopsy 2023, 1. [Google Scholar] [CrossRef]
- Vidlarova, M.; Rehulkova, A.; Stejskal, P.; Prokopova, A.; Slavik, H.; Hajduch, M.; Srovnal, J. Recent Advances in Methods for Circulating Tumor Cell Detection. Int. J. Mol. Sci. 2023, 24, 3902. [Google Scholar] [CrossRef]
- Wan, J.C.M.; Massie, C.; Garcia-Corbacho, J.; Mouliere, F.; Brenton, J.D.; Caldas, C.; Pacey, S.; Baird, R.; Rosenfeld, N. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat. Rev. Cancer 2017, 17, 223–238. [Google Scholar] [CrossRef]
- Wang, H.; Tan, Z.; Hu, H.; Liu, H.; Wu, T.; Zheng, C.; Wang, X.; Luo, Z.; Wang, J.; Liu, S.; et al. microRNA-21 promotes breast cancer proliferation and metastasis by targeting LZTFL1. BMC Cancer 2019, 19, 1–13. [Google Scholar] [CrossRef]
- Wang, W.; Li, X.; Liu, C.; Zhang, X.; Wu, Y.; Diao, M.; Tan, S.; Huang, S.; Cheng, Y.; You, T. MicroRNA-21 as a diagnostic and prognostic biomarker of lung cancer: a systematic review and meta-analysis. Biosci. Rep. 2022, 42. [Google Scholar] [CrossRef] [PubMed]
- Watson, P.A.; Arora, V.K.; Sawyers, C.L. Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat. Rev. Cancer 2015, 15, 701–711. [Google Scholar] [CrossRef] [PubMed]
- WOODHOUSE, R. , LI, M., HUGHES, J., DELFOSSE, D., SKOLETSKY, J., MA, P., MENG, W., DEWAL, N., MILBURY, C., CLARK, T., DONAHUE, A., STOVER, D., KENNEDY, M., DACPANO-KOMANSKY, J., BURNS, C., VIETZ, C., ALEXANDER, B., HEGDE, P. & DENNIS, L. 2020. Clinical and analytical validation of FoundationOne Liquid CDx, a novel 324-Gene cfDNA-based comprehensive genomic profiling assay for cancers of solid tumor origin. PLoS One, 15, e0237802.
- Wu, L.-L.; Tang, M.; Zhang, Z.-L.; Qi, C.-B.; Hu, J.; Ma, X.-Y.; Pang, D.-W. Chip-Assisted Single-Cell Biomarker Profiling of Heterogeneous Circulating Tumor Cells Using Multifunctional Nanospheres. Anal. Chem. 2018, 90, 10518–10526. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.-M.; Liu, J.-B.; Liu, Y.; Shi, Y.; Li, W.; Wang, G.-R.; Ma, Y.-S.; Fu, D. Power and Promise of Next-Generation Sequencing in Liquid Biopsies and Cancer Control. Cancer Control. 2020, 27. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Yang, Z.; Dai, Y.; Zhu, Q.; Chen, L.-A. Update on liquid biopsy in clinical management of non-small cell lung cancer. OncoTargets Ther. 2019, ume 12, 5097–5109. [Google Scholar] [CrossRef]
- Xue, V.W.; Yang, C.; Wong, S.C.C.; Cho, W.C.S. Proteomic profiling in extracellular vesicles for cancer detection and monitoring. Proteomics 2021, 21, 2000094. [Google Scholar] [CrossRef] [PubMed]
- Yan, T.-B.; Huang, J.-Q.; Huang, S.-Y.; Ahir, B.K.; Li, L.-M.; Mo, Z.-N.; Zhong, J.-H. Advances in the Detection of Pancreatic Cancer Through Liquid Biopsy. Front. Oncol. 2021, 11, 801173. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.-C.; Hu, J.-J.; Li, Y.-X.; Luo, W.; Liu, J.-Z.; Ye, D.-W. Clinical Applications of Liquid Biopsy in Hepatocellular Carcinoma. Front. Oncol. 2022, 12, 781820. [Google Scholar] [CrossRef]
- Ye, S.; You, Q.; Song, S.; Wang, H.; Wang, C.; Zhu, L.; Yang, Y. Nanostructures and Nanotechnologies for the Detection of Extracellular Vesicle. Adv. Biol. 2022, 7, e2200201. [Google Scholar] [CrossRef]
- You, C.; Jin, L.; Xu, Q.; Shen, B.; Jiao, X.; Huang, X. Expression of miR-21 and miR-138 in colon cancer and its effect on cell proliferation and prognosis. Oncol. Lett. 2018, 17, 2271–2277. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Sun, C.; Zhao, Y.; Wang, Q.; Guo, J.; Ye, B.; Yu, G. Overview of MicroRNAs as Diagnostic and Prognostic Biomarkers for High-Incidence Cancers in 2021. Int. J. Mol. Sci. 2022, 23, 11389. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Kang, G.; Jiang, P.; Qiao, R.; Lam, W.K.J.; Yu, S.C.Y.; Ma, M.-J.L.; Ji, L.; Cheng, S.H.; Gai, W.; et al. Epigenetic analysis of cell-free DNA by fragmentomic profiling. Proc. Natl. Acad. Sci. 2022, 119. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Fang, Y.; Guo, K.; Ni, Z.; Xiang, N. Next-generation liquid biopsy instruments: Challenges and opportunities. Electrophoresis 2023, 44, 775–783. [Google Scholar] [CrossRef]
- Tamkovich, S.; Tupikin, A.; Kozyakov, A.; Laktionov, P. Size and Methylation Index of Cell-Free and Cell-Surface-Bound DNA in Blood of Breast Cancer Patients in the Contest of Liquid Biopsy. Int. J. Mol. Sci. 2022, 23, 8919. [Google Scholar] [CrossRef]
 |
 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




