Submitted:
15 June 2024
Posted:
17 June 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
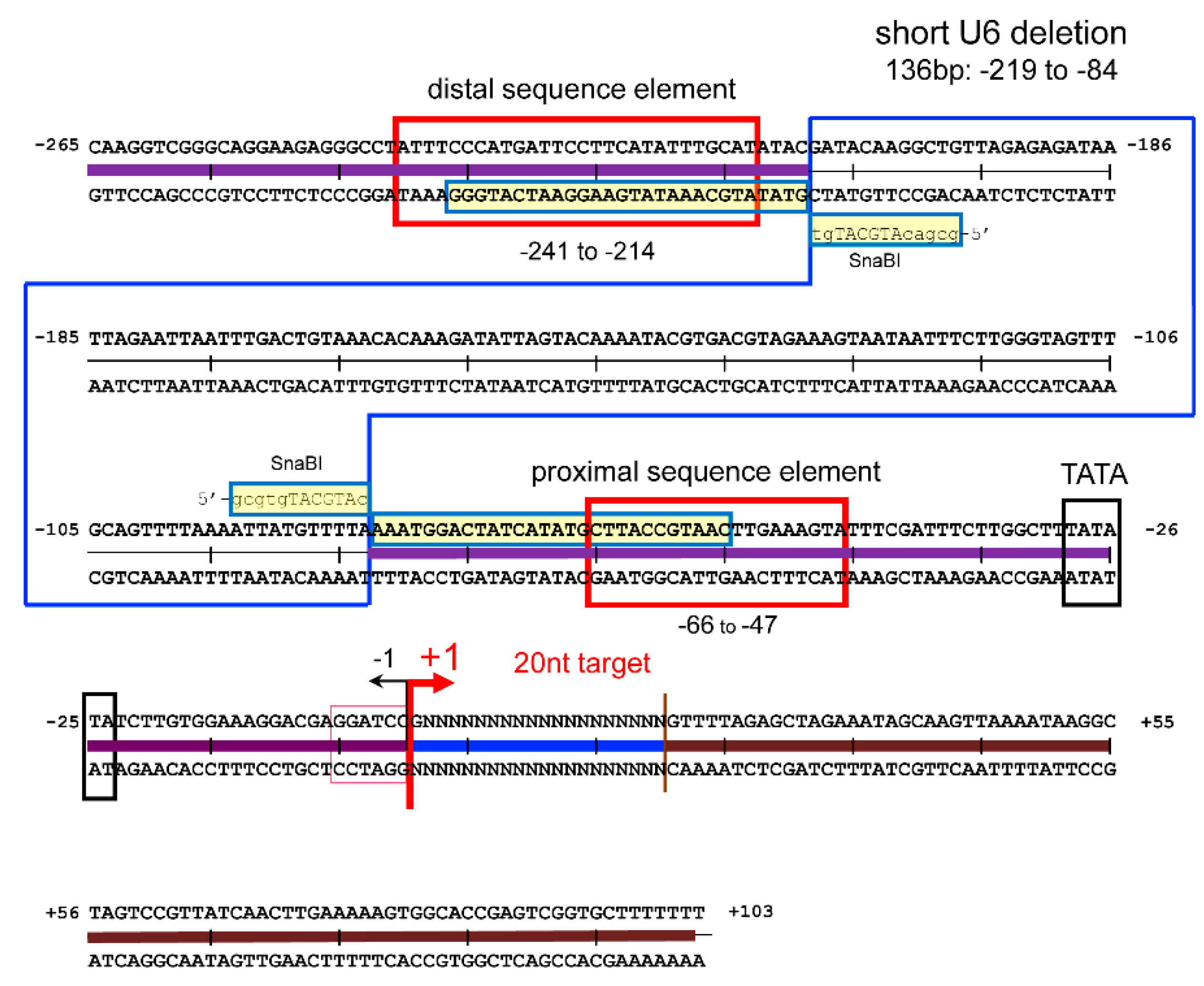
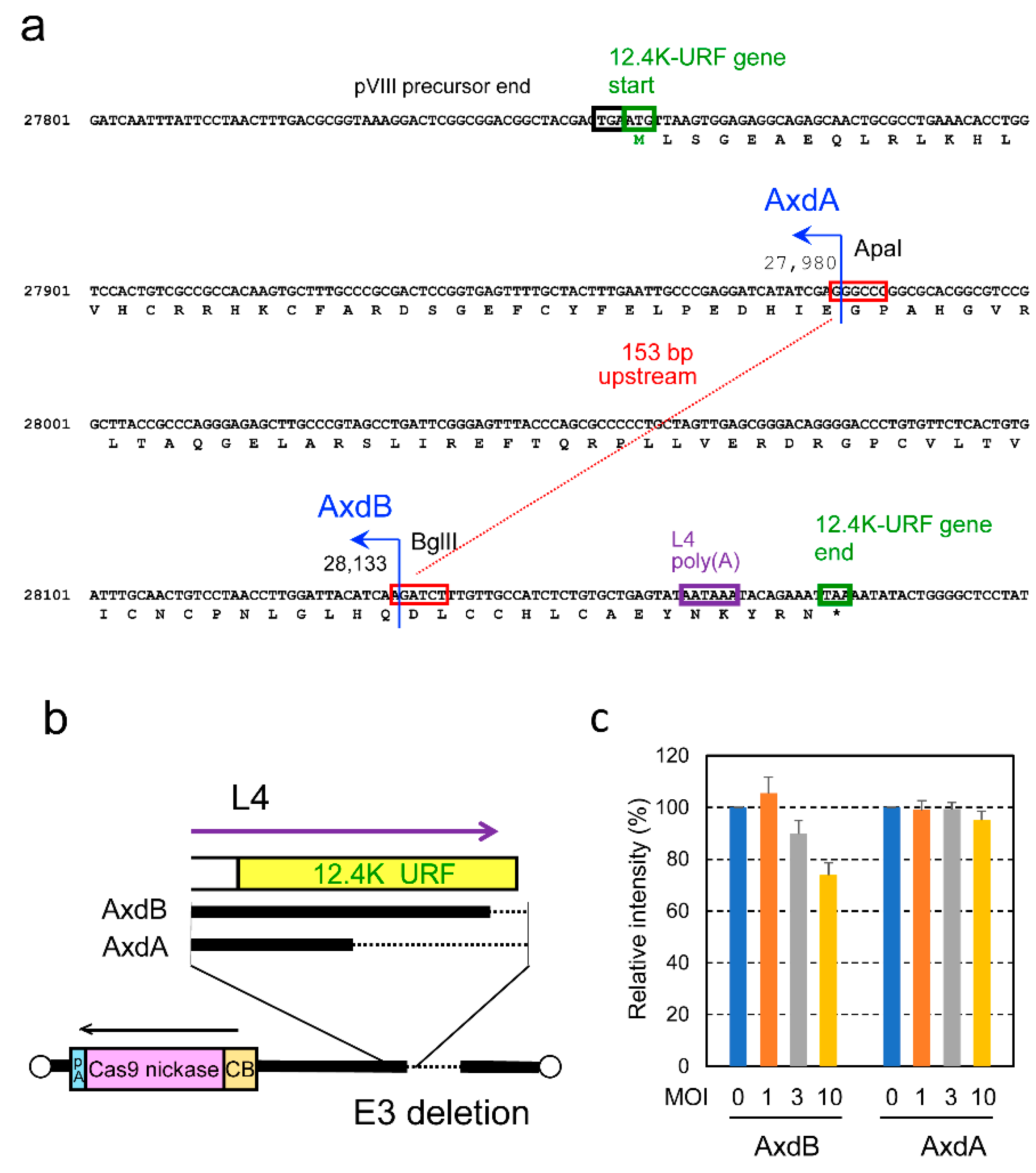
2.1. Construction of Shortened U6 Promoter and Enlargement of E3/L4 Deletion in the Vector Backbone
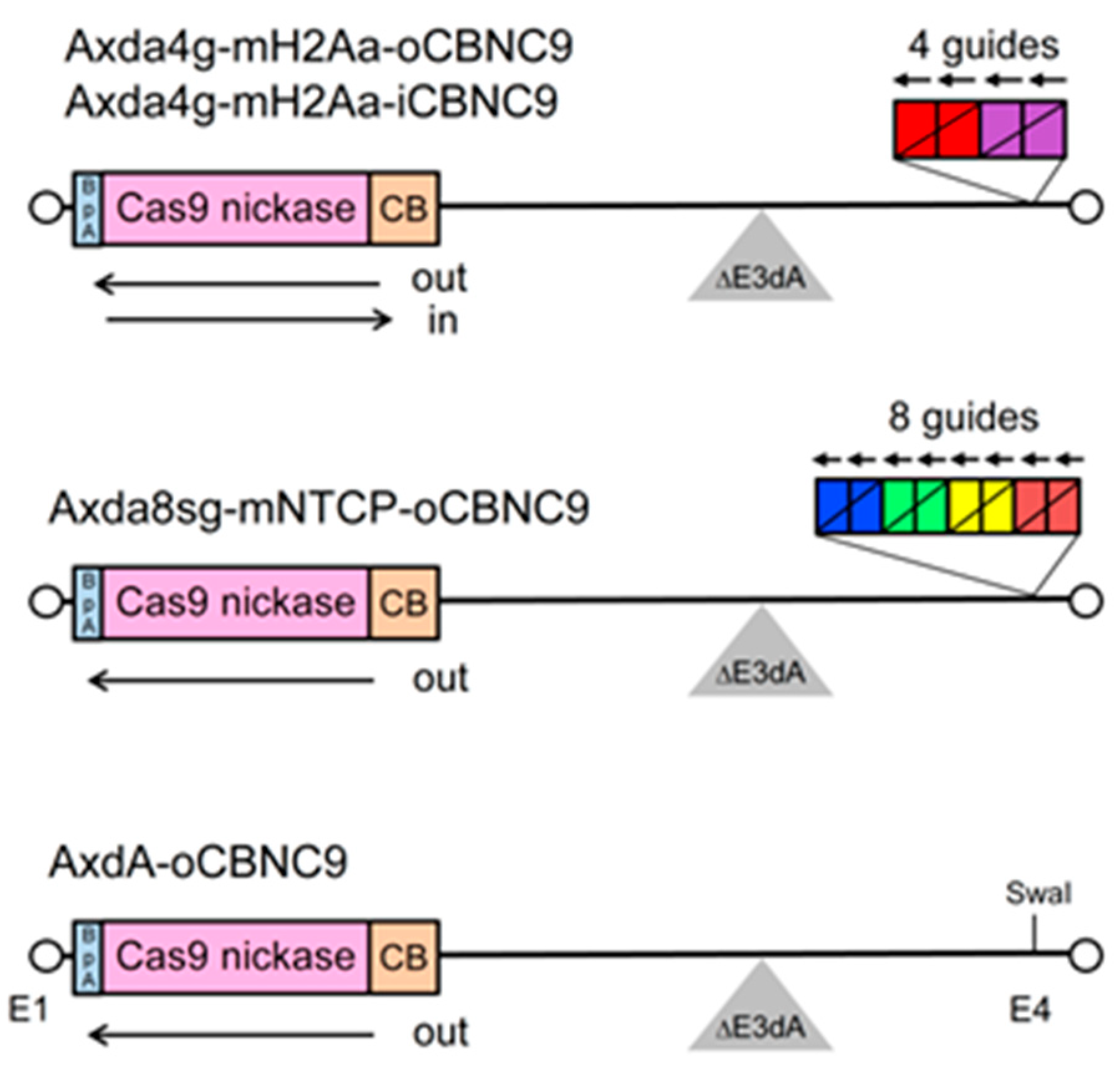
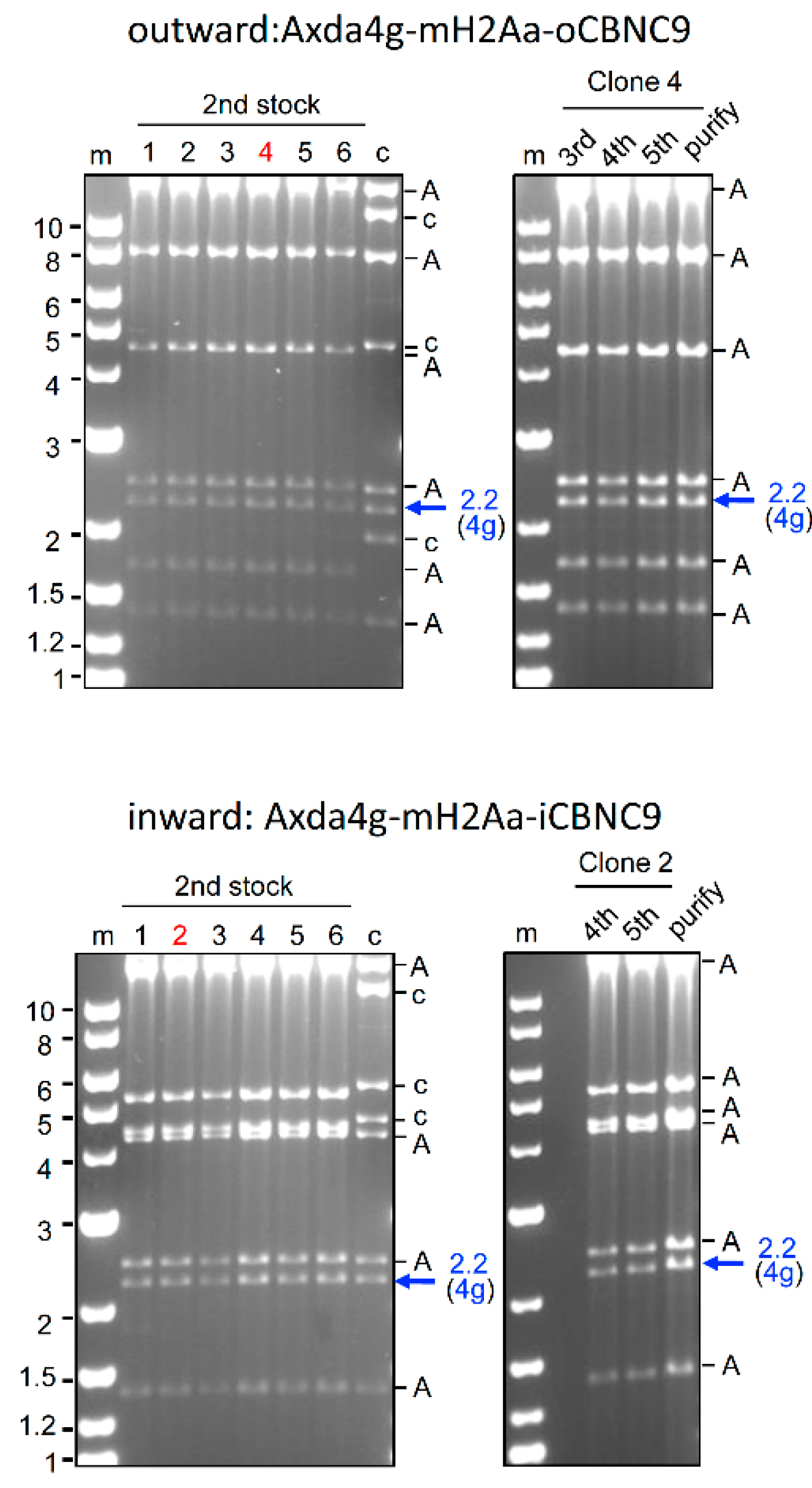
2.2. Construction of All-in-One AdVs Expressing four gRNAs
2.3. Preparation of All-in-One AdV Containing Eight gRNA Expression Units
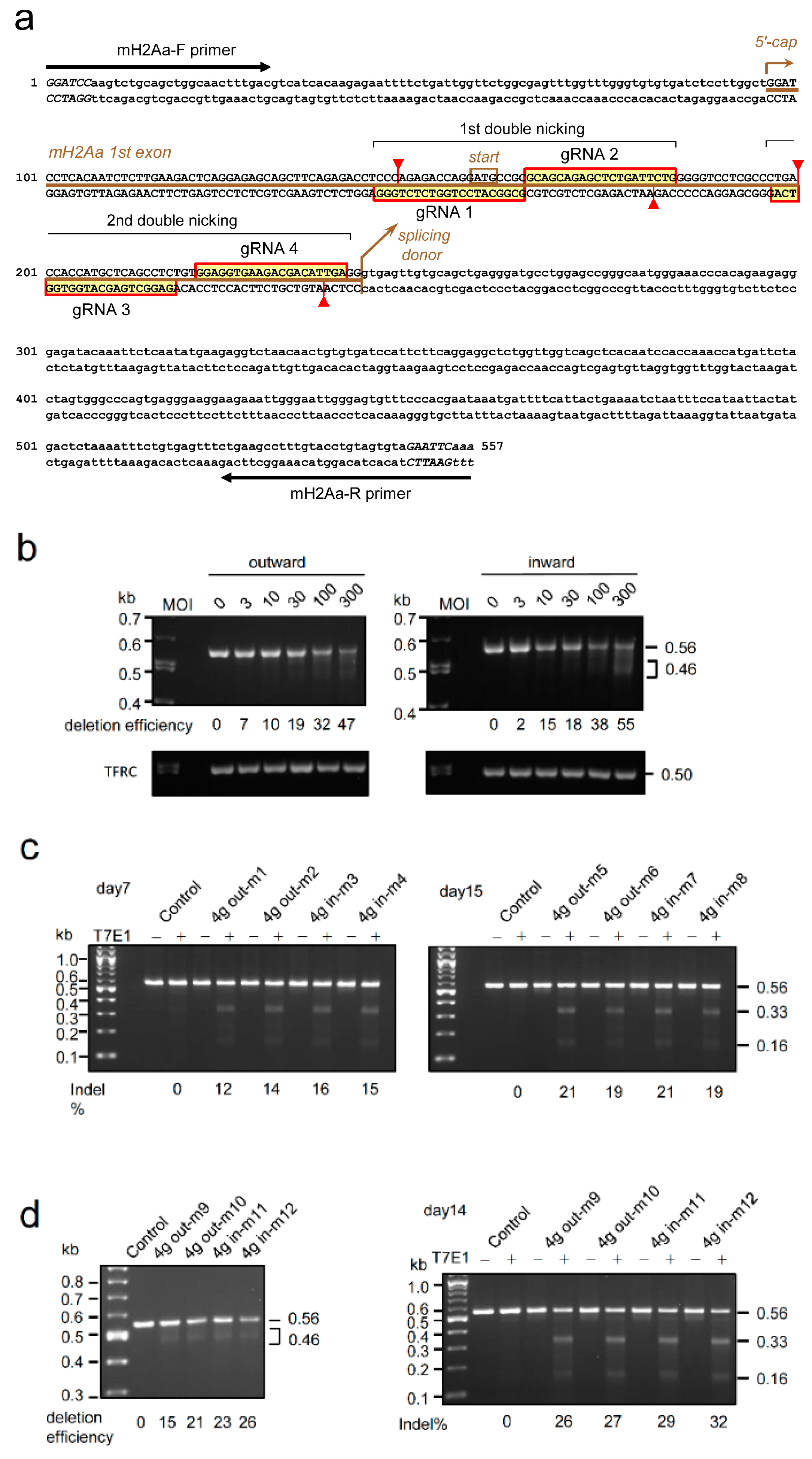
2.4. Genome editing of target gene using 4g all-in-one AdVs
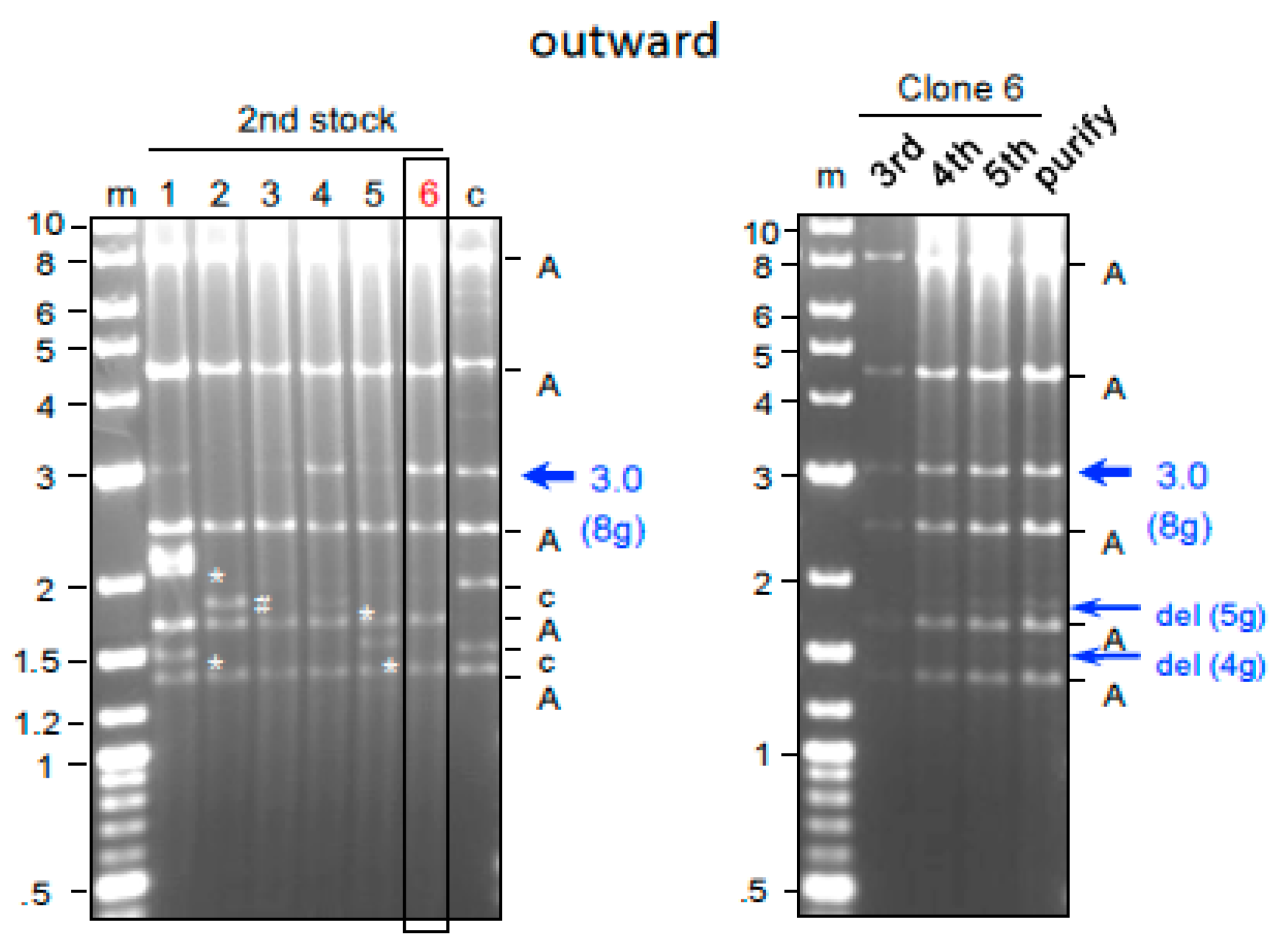
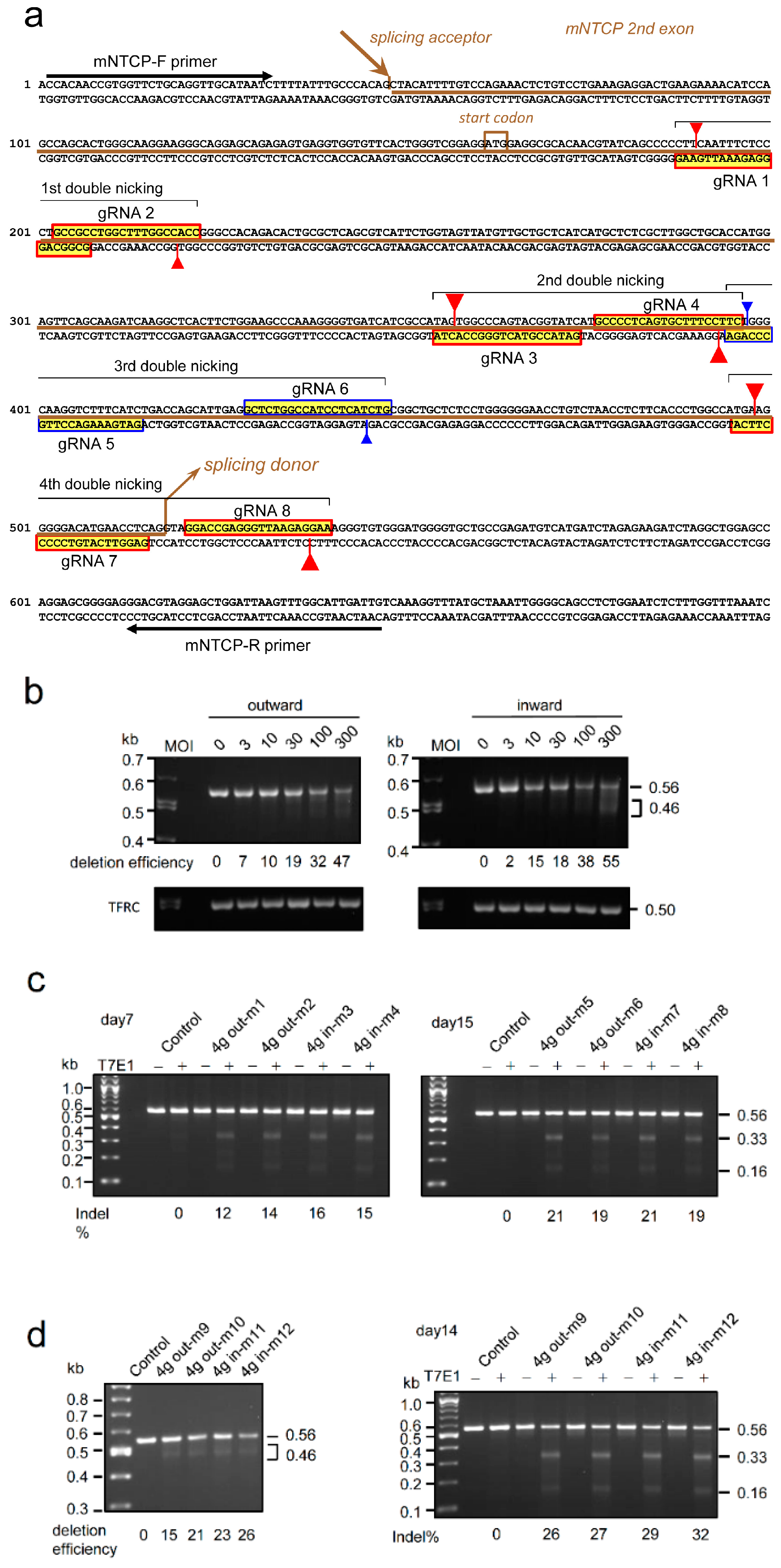
2.5. Genome Editing of Targeted Genes Using 8g All-in-One AdV
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Construction of Cosmids Containing AdV Genome Bearing Multiplex gRNA Units
4.3. Production of AdVs
4.4. Conventional PCR
4.5. T7 Endonuclease Assay
4.6. Cellular DNA Isolation
4.7. In Vitro and In Vivo AdV Infection
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- You, L.; Tong, R.; Li, M.; Liu, Y.; Xue, J.; Lu, Y. , Advancements and Obstacles of CRISPR-Cas9 Technology in Translational Research. Mol Ther Methods Clin Dev 2019, 13, 359–370. [Google Scholar] [CrossRef] [PubMed]
- Doudna, J. A. , The promise and challenge of therapeutic genome editing. Nature 2020, 578, 229–236. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Qin, C.; An, C.; Zheng, X.; Wen, S.; Chen, W.; Liu, X.; Lv, Z.; Yang, P.; Xu, W.; Gao, W.; Wu, Y. , Application of the CRISPR/Cas9-based gene editing technique in basic research, diagnosis, and therapy of cancer. Mol Cancer 2021, 20, 126. [Google Scholar] [CrossRef] [PubMed]
- Syyam, A.; Nawaz, A.; Ijaz, A.; Sajjad, U.; Fazil, A.; Irfan, S.; Muzaffar, A.; Shahid, M.; Idrees, M.; Malik, K.; Afzal, S. , Adenovirus vector system: construction, history and therapeutic applications. Biotechniques 2022, 73, 297–305. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhang, F.; Gao, G. , CRISPR-Based Therapeutic Genome Editing: Strategies and In Vivo Delivery by AAV Vectors. Cell 2020, 181, 136–150. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, H.; An, Y.; Chen, Z. , Construction and application of adenoviral vectors. Mol Ther Nucleic Acids 2023, 34, 102027. [Google Scholar] [CrossRef] [PubMed]
- Kuzmin, D. A.; Shutova, M. V.; Johnston, N. R.; Smith, O. P.; Fedorin, V. V.; Kukushkin, Y. S.; van der Loo, J. C. M.; Johnstone, E. C. , The clinical landscape for AAV gene therapies. Nat Rev Drug Discov 2021, 20, 173–174. [Google Scholar] [CrossRef] [PubMed]
- Haeussler, M.; Concordet, J. P. , Genome Editing with CRISPR-Cas9: Can It Get Any Better? J Genet Genomics 2016, 43, 239–50. [Google Scholar] [CrossRef] [PubMed]
- Ran, F. A.; Cong, L.; Yan, W. X.; Scott, D. A.; Gootenberg, J. S.; Kriz, A. J.; Zetsche, B.; Shalem, O.; Wu, X.; Makarova, K. S.; Koonin, E. V.; Sharp, P. A.; Zhang, F. , In vivo genome editing using Staphylococcus aureus Cas9. Nature 2015, 520, 186–91. [Google Scholar] [CrossRef]
- Ran, F. A.; Hsu, P. D.; Lin, C. Y.; Gootenberg, J. S.; Konermann, S.; Trevino, A. E.; Scott, D. A.; Inoue, A.; Matoba, S.; Zhang, Y.; Zhang, F. , Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 2013, 154, 1380–9. [Google Scholar] [CrossRef]
- Hryhorowicz, M.; Lipinski, D.; Zeyland, J.; Slomski, R. , CRISPR/Cas9 Immune System as a Tool for Genome Engineering. Arch Immunol Ther Exp (Warsz) 2017, 65, 233–240. [Google Scholar] [CrossRef] [PubMed]
- Kato, Y.; Tabata, H.; Sato, K.; Nakamura, M.; Saito, I.; Nakanishi, T. , Adenovirus Vectors Expressing Eight Multiplex Guide RNAs of CRISPR/Cas9 Efficiently Disrupted Diverse Hepatitis B Virus Gene Derived from Heterogeneous Patient. Int J Mol Sci 2021, 22. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, T.; Maekawa, A.; Suzuki, M.; Tabata, H.; Sato, K.; Mori, M.; Saito, I. , Construction of adenovirus vectors simultaneously expressing four multiplex, double-nicking guide RNAs of CRISPR/Cas9 and in vivo genome editing. Sci Rep 2021, 11, 3961. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, T.; Maekawa, A.; Tabata, H.; Yoshioka, T.; Pei, Z.; Sato, K.; Mori, M.; Kato, M.; Saito, I. , Highly multiplex guide RNA expression units of CRISPR/Cas9 were completely stable using cosmid amplification in a novel polygonal structure. J Gene Med 2019, 21, e3115. [Google Scholar] [CrossRef] [PubMed]
- Maggio, I.; Liu, J.; Janssen, J. M.; Chen, X.; Goncalves, M. A. , Adenoviral vectors encoding CRISPR/Cas9 multiplexes rescue dystrophin synthesis in unselected populations of DMD muscle cells. Sci Rep 2016, 6, 37051. [Google Scholar] [CrossRef] [PubMed]
- Stephens, C. J.; Lauron, E. J.; Kashentseva, E.; Lu, Z. H.; Yokoyama, W. M.; Curiel, D. T. , Long-term correction of hemophilia B using adenoviral delivery of CRISPR/Cas9. J Control Release 2019, 298, 128–141. [Google Scholar] [CrossRef] [PubMed]
- Palmer, D. J.; Turner, D. L.; Ng, P. , Production of CRISPR/Cas9-Mediated Self-Cleaving Helper-Dependent Adenoviruses. Mol Ther Methods Clin Dev 2019, 13, 432–439. [Google Scholar] [CrossRef]
- Palmer, D. J.; Turner, D. L.; Ng, P. , A Single "All-in-One" Helper-Dependent Adenovirus to Deliver Donor DNA and CRISPR/Cas9 for Efficient Homology-Directed Repair. Mol Ther Methods Clin Dev 2020, 17, 441–447. [Google Scholar] [CrossRef]
- Schiwon, M.; Ehrke-Schulz, E.; Oswald, A.; Bergmann, T.; Michler, T.; Protzer, U.; Ehrhardt, A. , One-Vector System for Multiplexed CRISPR/Cas9 against Hepatitis B Virus cccDNA Utilizing High-Capacity Adenoviral Vectors. Mol Ther Nucleic Acids 2018, 12, 242–253. [Google Scholar] [CrossRef]
- Miyake, S.; Makimura, M.; Kanegae, Y.; Harada, S.; Sato, Y.; Takamori, K.; Tokuda, C.; Saito, I. , Efficient generation of recombinant adenoviruses using adenovirus DNA-terminal protein complex and a cosmid bearing the full-length virus genome. Proc Natl Acad Sci U S A 1996, 93, 1320–4. [Google Scholar] [CrossRef]
- Bett, A. J.; Prevec, L.; Graham, F. L. , Packaging capacity and stability of human adenovirus type 5 vectors. J Virol 1993, 67, 5911–21. [Google Scholar] [CrossRef] [PubMed]
- Stunkel, W.; Kober, I.; Seifart, K. H. , A nucleosome positioned in the distal promoter region activates transcription of the human U6 gene. Mol Cell Biol 1997, 17, 4397–405. [Google Scholar] [CrossRef] [PubMed]
- Bett, A. J.; Haddara, W.; Prevec, L.; Graham, F. L. , An efficient and flexible system for construction of adenovirus vectors with insertions or deletions in early regions 1 and 3. Proc Natl Acad Sci U S A 1994, 91, 8802–6. [Google Scholar] [CrossRef]
- Shintani, Y.; Yotsuyanagi, H.; Moriya, K.; Fujie, H.; Tsutsumi, T.; Kanegae, Y.; Kimura, S.; Saito, I.; Koike, K. , Induction of apoptosis after switch-on of the hepatitis B virus X gene mediated by the Cre/loxP recombination system. J Gen Virol 1999, 80 (Pt 12) Pt 12, 3257–65. [Google Scholar] [CrossRef]
- Suzuki, M.; Kondo, S.; Yamasaki, M.; Matsuda, N.; Nomoto, A.; Suzuki, T.; Saito, I.; Kanegae, Y. , Efficient genome replication of hepatitis B virus using adenovirus vector: a compact pregenomic RNA-expression unit. Sci Rep 2017, 7, 41851. [Google Scholar] [CrossRef] [PubMed]
- Ghosh-Choudhury, G.; Haj-Ahmad, Y.; Graham, F. L. , Protein IX, a minor component of the human adenovirus capsid, is essential for the packaging of full length genomes. EMBO J 1987, 6, 1733–9. [Google Scholar] [CrossRef] [PubMed]
- Mizuguchi, H.; Kay, M. A.; Hayakawa, T. , In vitro ligation-based cloning of foreign DNAs into the E3 and E1 deletion regions for generation of recombinant adenovirus vectors. Biotechniques 2001, 30, 1112–4. [Google Scholar] [CrossRef] [PubMed]
- Koike, K. , Hepatitis B virus X gene is implicated in liver carcinogenesis. Cancer Lett 2009, 286, 60–8. [Google Scholar] [CrossRef]
- Kanegae, Y.; Makimura, M.; Saito, I. , A simple and efficient method for purification of infectious recombinant adenovirus. Jpn J Med Sci Biol 1994, 47, 157–66. [Google Scholar] [CrossRef]
- Suzuki, M.; Kondo, S.; Pei, Z.; Maekawa, A.; Saito, I.; Kanegae, Y. , Preferable sites and orientations of transgene inserted in the adenovirus vector genome: The E3 site may be unfavorable for transgene position. Gene Ther 2015, 22, 421–429. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).




