1. .Introduction
Human milk (HM) feeding reduces risk for necrotizing enterocolitis (NEC), a severe inflammatory bowel disease that primarily affects preterm infants and occurs in the distal portion of the small intestine [
1]. Part of the mechanism by which NEC risk may be reduced in infants fed HM is via bioactive peptides released from milk’s proteins during digestion [
2]. Milk protein-derived bioactive peptides have an array of functions including antimicrobial, immunomodulatory, mucin-stimulatory, gut motility modulation, antioxidant and prebiotic [
2,
3]. As NEC is associated with inflammation and changes in the gut microbiome [
4], the activities of milk peptides released in the gut could impact NEC risk. Peptidomics research has revealed that thousands of unique peptides are released from milk proteins during gastric [
5,
6,
7,
8] and intestinal [
9] digestion in preterm infants, and some milk peptides survive to fecal excretion [
10]. Among these peptides found in the infant gut, many have high homology with or are identical to known bioactive peptides, and novel antimicrobial peptides have been identified in these digesta [
9,
10]. Moreover, HM peptides derived from in vitro simulated preterm digestion and in vivo preterm infant intestinal digestion modulate the response of macrophages to inflammation [
11]. Our previous finding that in vitro-digested colostrum and transitional HM (days two to eight postpartum) reduced lipopolysaccharide (LPS) and tumor necrosis factor (TNF)-induced inflammation and cytotoxicity in intestinal epithelial cells to a greater extent than undigested colostrum and transitional HM may be due to the bioactive peptides released during digestion [
12,
13].
The objectives of this study were to: 1) characterize and compare peptides present in colostrum samples before and after exposure to an in vitro model of preterm infant digestion; 2) to predict antimicrobial activities of peptides using sequence-based prediction; and 3) to test antimicrobial activity of predicted peptides against neonatal pathogens. We hypothesized that peptide abundances in colostrum increase following in vitro digestion and that by using sequence-based prediction, we can identify peptides with antimicrobial activities and demonstrate these activities in pathogenic bacterial culture models.
2. Materials and Methods
2.1. Colostrum Samples
Ten de-identified colostrum samples from different lactating parents of preterm infants were separated into two aliquots: 1) left undigested; and 2) digested using an in vitro digestion designed to simulate preterm infant digestion based on previous work [
12,
13]. In brief, the in vitro digestion protocol involved colostrum samples (200 µL colostrum + 2.2235 mL de-ionized water) being treated with pepsin (80 mg pepsin [≥250 units mg
—1] in 2 mL of de-ionized water) and titrated to pH 4.0 with 1 M hydrochloric acid (gastric digestion) and shaken at 100 rpm 30 min at 37 °C. A variable amount (10–22 μL) of 1 M sodium bicarbonate was added to increase to intestinal pH of 6.0, followed by the addition of pancreatin and bile salts (20 mg pancreatin [4X USP] + 120 mg bile salts in 10 mL of 0.1 M sodium bicarbonate) and incubated at 37 °C with 100 rpm agitation for 30 min to simulate the initial stages of intestinal digestion. The pH was further increased to 7.0 followed by incubation for 2 h at 37 °C with 100 rpm agitation to simulate distal intestinal digestion [
12,
13]. After incubation, heat treatment at 90 °C for 15 min was applied to all samples to deactivate enzymes. All “undigested” colostrum aliquots were diluted with deionized water in the same volume of digestive additives as in the “digested” counterparts and exposed to the same final heat treatment.
2.2. Peptide Extraction from the Colostrum Samples
Peptides were extracted from undigested and digested colostrum samples using centrifugation, acid precipitation using trichloroacetic acid and solid phase extraction as described previously [
5]. Briefly, all colostrum samples were centrifuged, and the liquid infranatant including proteins and peptides was collected. The collected samples were mixed with trichloroacetic acid to precipitate proteins, centrifuged and the supernatant including peptides was collected. The extracted peptides in both undigested and digested colostrum samples were further purified by solid phase extraction using C18 96-well plates (Glygen Corp, Columbia, MD) and completely dried via vacuum centrifugation.
2.3. Peptide Analysis Using Liquid Chromatography and Mass Spectrometry (LC-MS)
Dried samples were reconstituted with 100 μL of 3% acetonitrile, 0.1% formic acid and diluted tenfold. Peptides were measured using an Orbitrap Fusion™ Lumos™ Tribrid™ mass spectrometer (Thermo Fisher Scientific, Waltham, MA) combined with a NanoAcquity Ultra High-Performance Liquid Chromatography (UPLC) instrument (Waters Corporation, Milford, MA). One microliter of peptides was loaded onto a C18 180 μm × 20 mm, 5-μm bead nanoAcquity UPLC trap column (Waters) and separated with a 100 μm × 100 mm, 1.7-μm bead Acquity UPLC Peptide BEH C18 column (Waters). Peptides were separated and eluted for 60 min at a flowrate of 0.5 µL min—1 using the following gradient conditions: 3% to 11.5% B, 0 min to 10 min; 11.5% to 20% B, 10 min to 31 min; 20% to 30% B, 31 min to 36 min; 30% to 95% B, 36 min to 45 min; 95% B, 45 min to 54.5 min, 95 to 3% B over 0.5 min then finally the column was re-equilibrated with 97% A for 5 min. Mobile phase A was 100% ultrapure water, 0.1% formic acid and mobile phase B was 100% acetonitrile, 0.1% formic acid. Peptides were ionized with a nanospray ion source with an electrospray voltage of 2,320 V and an ion transfer tube temperature of 300 °C. MS spectra were acquired in positive ionization mode over an m/z range of 300–2,000 with the Orbitrap at a resolution of 60,000. The automatic gain control (AGC) target was set to 4.0 × 105, with a maximum injection time of 50 ms. The MS cycle time was set to 3 s. Precursor ions were automatically selected by the acquisition software for a data-dependent MS/MS scan based on the following criteria: ion-intensity threshold of 5.0 × 104, charge state of 2–8 and exclusion duration of 60 s and mass tolerance of 10 ppm. The fragmentation of precursor ions was performed by electron-transfer/higher-energy collision dissociation (EThcD) with optimized electron-transfer dissociation (ETD) reaction times depending on charge state (2+, 130 ms; 3+, 70 ms; 4+, 50 ms; 5+, 40 ms; 6+ to 8+, 20 ms) and supplemental higher-energy collision-induced dissociation (HCD) activation (25% of collision energy). MS/MS spectra were acquired in the positive ionization mode over an m/z range of 300–2,000 with the Orbitrap at a resolution of 30,000. The AGC target was set to 5.0 × 104.
2.4. LC-MS Data Analysis
Peptide identification was performed using Thermo Proteome Discoverer (v2.2) with a SequestHT search engine from the LC-MS raw files based on database searching using an in-house HM protein sequence database (n=378). Cleavage sites were set to “No-Enzyme (Unspecific).” The precursor mass tolerance and fragment mass tolerance were set to 10 ppm and 0.5 Da, respectively. Allowed dynamic modifications included phosphorylation of serine and threonine, oxidation of methionine, acetylation at the N-terminus of the protein and amidation at the C-terminus of the protein. Peptide sequences of all proteins identified in digested colostrum samples that were not identified in undigested colostrum were examined to rule out porcine origin (digestive enzymes used for the in vitro digestion protocol were of porcine origin) using the Protein Basic Local Alignment Search Tool (BLASTp®) of the National Library of Medicine.
2.5. Homology Search against Known Bioactive Peptides
Identified HM peptides were examined for homology with known bioactive peptides using the Milk Bioactive Peptide Database (MBPDB;
https://mbpdb.nws.oregonstate.edu/) [
2,
3]. The search parameters were set to “Sequence” for Search type, “80%” for Similarity threshold”, “Identity” for Scoring matrix and “human (homo sapiens)” for Species.
2.6. Sequence-Based Prediction of Antimicrobial Peptides
The amino acid sequences of peptides found from digested samples were used as an input to AmPEP tool to predict their antimicrobial activity potential [
14]. This tool is a sequence-based predictor, which recognizes distribution patterns of amino acid properties using a random forest algorithm to predict antimicrobial activity. Peptides with predicted probability values greater than 0.5 were classified as potentially antimicrobial. Four HM peptides with varying predicted antimicrobial probability values were chosen for empirical testing.
2.7. Bacterial Strains and Culture Conditions
Bacteria commonly observed in neonates with culture-positive sepsis were used for antimicrobial testing [
15].
Citrobacter freundii ATCC 13316 and
Serratia marcescens subsp.
marcescens Bizio were obtained from American Type Culture Collection (ATCC).
Klebsiella aerogenes and
Escherichia coli were obtained from Ward’s Science (strain not provided; Rochester, NY). All bacterial strains were cultured on nutrient agar at 37 ˚C for 24–48 h. Single colonies were isolated for liquid culture.
E. coli was grown in Luria-Bertani (LB) broth containing 1% (w/v) tryptone, 1% (w/v) sodium chloride and 0.5% (w/v) yeast extract; other strains were grown in nutrient broth containing 0.3% (w/v) beef extract and 0.5% (w/v) peptone.
2.8. Antimicrobial Activity of Predicted Peptides
The top four candidate peptides (α
S1-casein [157 – 163] QYVPFPP, α
S1-casein [157 – 165] QYVPFPPFS, β-casein [153 – 159] VPQPIPQ and plasminogen [591 – 597] SWPWQVS) were synthesized by GenScript (Piscataway, NJ) with ≥98% purity. Peptides were reconstituted with ultrapure water to 5 mg mL
—1. Minimal inhibitory concentration (MIC) determination was performed based on a published method [
16]. The antimicrobial assay was performed in a 96-well microtiter plate with two-fold dilution of each peptide ranging from 4.9 µg mL
—1 to 2,500 µg mL
—1. Bacterial culture with a density equal to McFarland standard 0.5 was inoculated into each well in a 1:1 ratio with the peptide solution. Controls included a sterility control without inoculum and a growth control without peptides. MIC test with varying peptide concentrations of each peptide on bacterial growth was measured through readings at the optical density of 600 nm by a microplate reader at baseline and after incubation at 37 ˚C for 18–24 h. Promotion or inhibition on bacterial growth was identified by calculating the percentage change of optical density at 600 nm against the growth control. The data presented are the mean values obtained from two independent experiments. To test whether peptides had bactericidal effects, bacterial cultures from the wells that had been spiked with 2,500 µg mL
—1 of each peptide were spread on nutrient agar plates without peptides, and growth of bacteria was monitored over 48 h.
2.9. Statistical Analysis
Peptidomic data were entered into SPSS (IBM SPSS Statistics for Windows, Version 25.0. Armonk, NY: IBM Corp.), and descriptive analyses were performed to assess normality. Peptide abundances were deemed non-normal; thus, a related-samples Wilcoxon Signed Rank test was conducted to compare peptide abundances between undigested and digested samples. Heat mapping was conducted via Heatmapper to evaluate differences of peptide profiles between undigested and digested colostrum samples [
17].
3. Results
3.1. Comparison of Peptides Identified in Undigested and Digested Samples
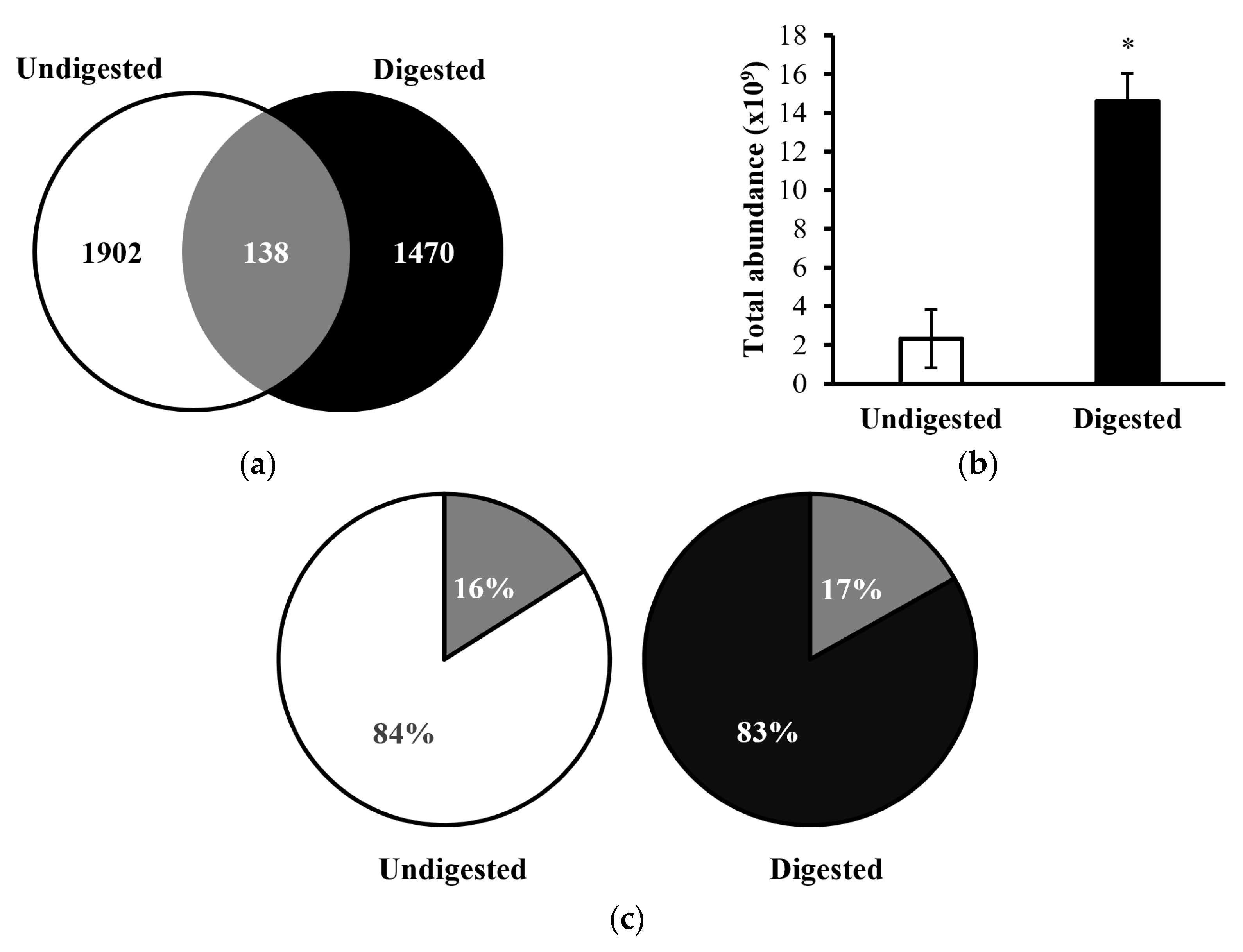
A total of 2,040 peptides were identified from the ten undigested colostrum samples, and a total of 1,608 peptides were identified in the ten in vitro-digested colostrum samples (
Figure 1a,
Supplementary Tables S1 and S2). Among these peptides, 138 sequences were shared between the undigested and digested samples (making up 6.8% of peptides in the undigested samples and 8.6% of the peptides in digested samples by count). Total abundance of the in vitro-digested colostrum samples was significantly higher than the undigested samples (p=.005, nearly five times higher,
Figure 1b). The 138 peptides present in both undigested and digested samples made up 16% of the total peptide abundance in the undigested samples and 17% of the total peptide abundance in the digested samples (
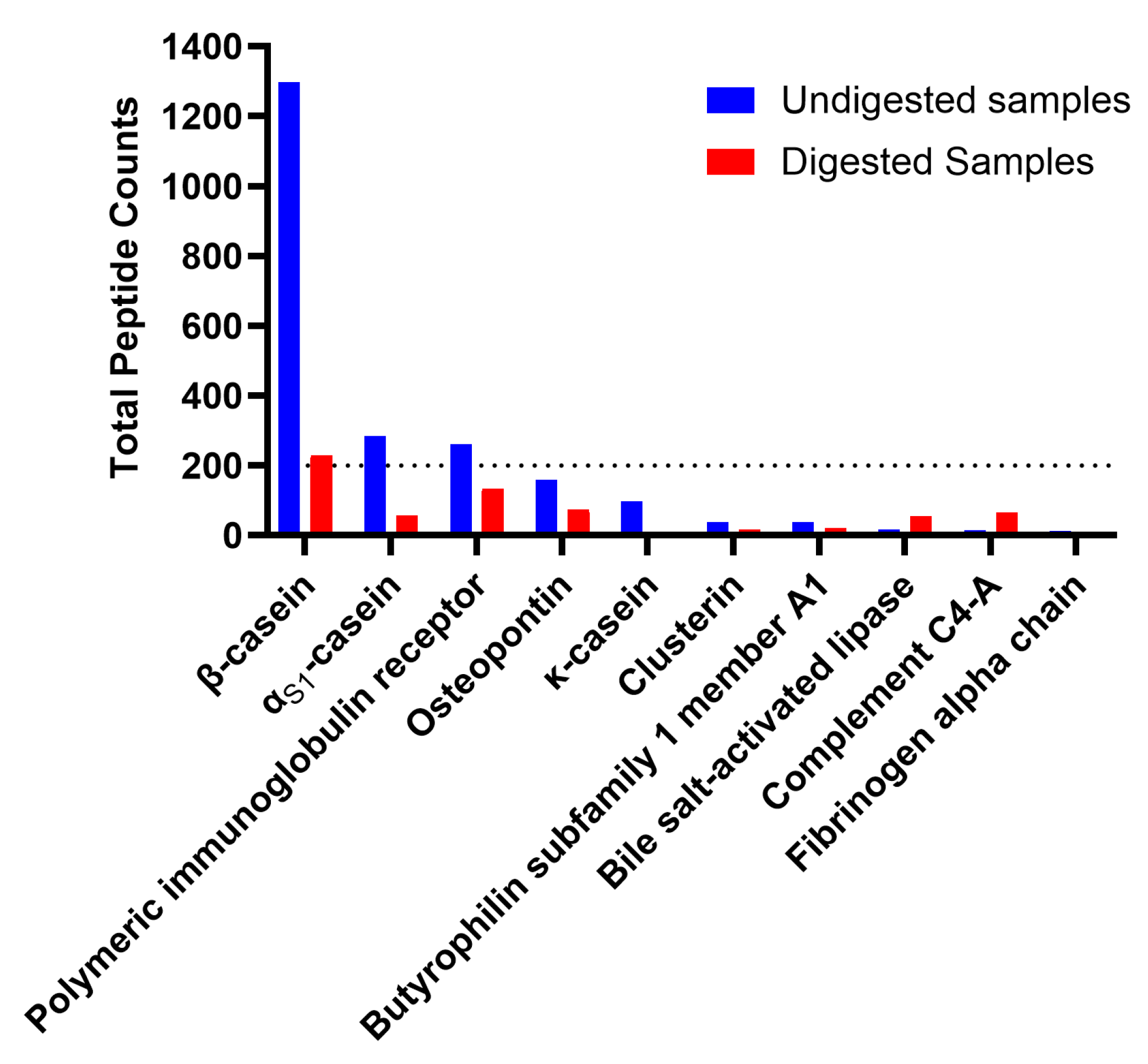
Figure 1c). Among the top ten parent proteins from which peptides derived in the undigested and digested samples, β-casein, α
S1-casein, polymeric immunoglobulin receptor, osteopontin, bile salt-activated lipase and complement C4-A were present in both undigested and digested samples (
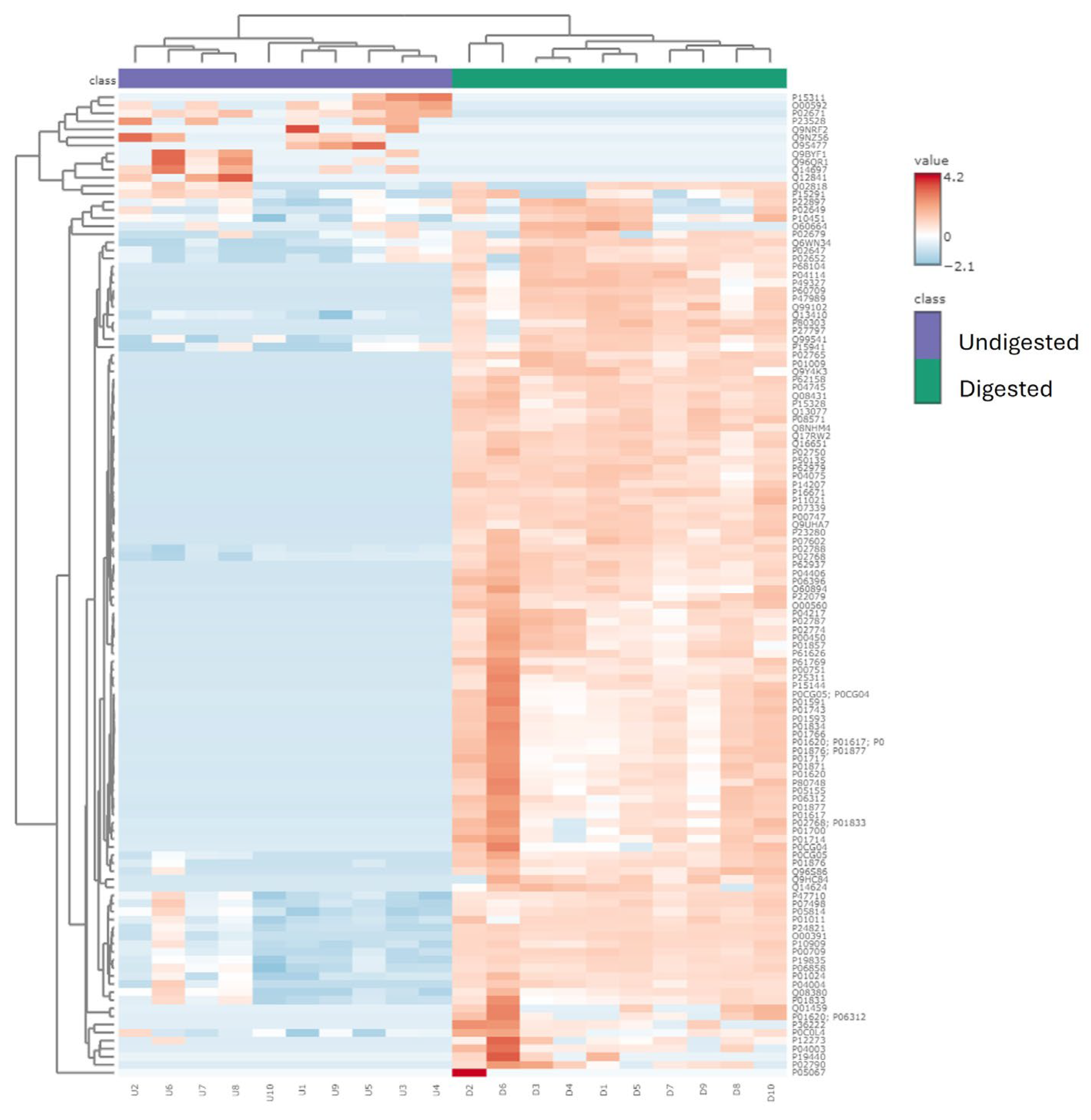
Figure 2). Heatmapping and hierarchical clustering showed a clear separation of the peptide profile patterns between undigested colostrum and in vitro-digested colostrum (
Figure 3).
Homology searching of all identified peptides against the Milk Bioactive Peptide Database [
2,
3] revealed 121 peptides in the undigested samples and 64 peptides in the digested samples that had over 80% homology to known bioactive peptides (
Table 1;
Supplementary Tables S3 and S4). Among identified peptide bioactivities from the undigested samples, the three most common functional groups were: angiotensin-converting enzyme (ACE)-inhibitory (35 peptides); antimicrobial (45 peptides); and increased cellular growth (52 peptides) (
Table 1). The profile of peptide bioactivities in the digested samples differed from the undigested peptides: most bioactive peptides in the digested samples were ACE-inhibitory (43 peptides) and only one peptide in the digested samples had >80% homology with a known antimicrobial peptide (
Table 1).
3.2. Sequence-Based Prediction of Bioactive Peptides
The AmPEP algorithm classified 96 of the identified peptides in the digested samples as potentially antimicrobial, with a prediction probability range of 0.50–0.82. Four peptides with varying predicted antimicrobial probabilities (0.55–0.73) were randomly selected for empirical testing.
3.3. Antimicrobial Activity of the Peptides with Predicted Antimicrobial Activities
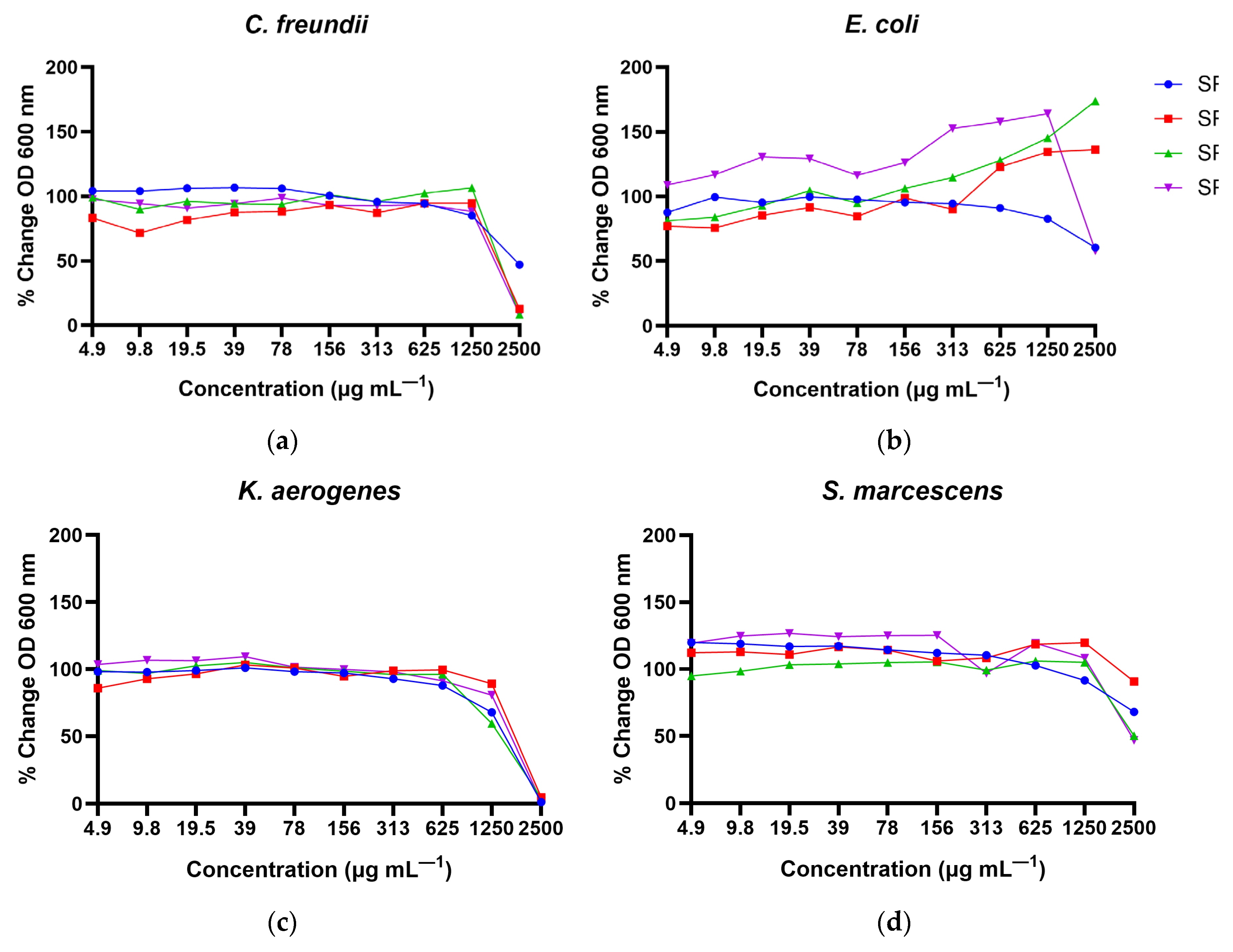
The growth of
C. freundii was inhibited by α
S1-casein [157–165], β-casein [153–159] and plasminogen [591–597] at the MIC of 2,500 µg mL
—1 (
Table 2). Though the MIC for α
S1-casein [157-163] against
C. freundii was >2,500 µg mL
—1 (
Table 2), the 2,500 µg mL
—1 dose somewhat reduced growth (
Figure 4a). The growth of the
E. coli strain was not inhibited by any of the four peptides. However, a slight growth reduction of
E. coli was observed when incubated with α
S1-casein [157–163] and plasminogen [591–597] at concentrations of 1,250 µg mL
—1 and higher, whereas α
S1-casein [157–165] and β-casein [153–159] enhanced growth at concentrations higher than 625 µg mL
—1 (
Figure 4b). All four peptides reduced the growth of
K. aerogenes starting at 1,250 µg mL
—1 and completely inhibited its growth with an MIC of 2,500 µg mL
—1 (
Figure 4c). The growth of
S. marcescens was reduced to various extents when incubated with 2,500 µg mL
—1 of all four peptides (
Figure 4d); however, the MIC was >2,500 µg mL
—1. In the bactericidal tests, growth of all the four bacterial strains were recovered in 24–48 h, indicating no bactericidal effects against
K. aerogenes,
C. freundii or
S. marcescens for any of the tested peptides. This finding indicates the observed reductions in bacterial growth were due to a bacteriostatic effect.
4. Discussion
The relatively limited number of peptides that were similar between undigested and digested samples (138, making up 6.8% of undigested samples and 8.6% of digested samples) indicates that digestion caused at least partial degradation of most native milk peptides and the release of novel peptides from milk protein or native milk peptide precursors. Similarly, the limited percent of total abundance in the undigested (16%) and digested colostrum (17%) made up by peptides present in both sample types indicates that digestion induced major shifts in the overall peptide profile. The large increase in total peptide abundance from undigested to digested colostrum samples also demonstrates that digestion caused the release of a large amount of peptides from intact milk proteins. The heatmap also indicates that the abundance of peptides from each protein differs between undigested and digested colostrum samples.
Homology search of the peptides identified against the MBPDB revealed 45 peptides in the undigested samples had >80% homology with a previously identified antimicrobial peptide, but only one peptide met that criterion in the digested samples. Sequence-based prediction of antimicrobial activities yielded 96 peptides from the digested samples that were candidates for antimicrobial activity. From these, we selected four peptides (α
S1-casein [157–163], α
S1-casein [157–165], β-casein [153–159] and plasminogen [591–597]) with varying prediction scores to synthesize and test for antimicrobial action against four strains of bacteria previously observed in culture-positive blood samples of preterm infants in neonatal intensive care units with sepsis and/or NEC. Each of the four peptides inhibited
C. freundii,
K. aerogenes and
S. marcescens, but only two peptides inhibited
E. coli, and only to a small degree at the concentrations tested. The inhibitory effects of these peptides were overall strongest against
C. freundii and
K. aerogenes. The pathogen inhibitory effects of these peptides suggested a potential role of these digestion-released HM peptides in inhibiting the growth of pathogenic bacteria that are often associated with sepsis and NEC in premature infants. The results also indicate that MIC of the same peptide differed depending on the bacterial strain and that some peptides appeared to promote the growth of
E. coli, similar to the findings from Beverly et al. [
9]. Though all the tested peptides were able to inhibit microbial growth of
C. freundii,
K. aerogenes and
S. marcescens, and two peptides inhibited
E. coli, the MIC results did not differ based on the predictive scores, suggesting the need to refine the prediction tools. Though milk casein proteins are precursors for a large number of known antimicrobial peptides, previous studies have not identified antimicrobial peptides from plasminogen. Therefore, the identification of plasminogen [591-597] as an antimicrobial peptide is particularly novel. Plasminogen and plasmin are abundant in HM [
18].
We suspect that milk peptides may be supporting microbiome development in the neonate by reducing potentially pathogenic bacteria while promoting commensal bacteria. Future work should examine whether these peptides can increase the growth of beneficial infant gut microbes, such as Bifidobacterium, as previously reported [
9]. HM antimicrobial peptides likely represent one component of a system of milk components (e.g., milk oligosaccharides [
19], immunoglobulins [
20]) that help guide infant gut microbial development.
5. Conclusions
By combining peptidomic analyses of digestion-released peptides from human colostrum with a machine-learning based modeling tool followed by synthesis and testing of predicted antimicrobial peptides, we were able to identify four peptides that can suppress certain common pathogenic bacteria that are associated with NEC and sepsis in the premature infant. Our findings provide insights into the potential capacity of HM peptides to inhibit the growth of pathogenic virulent bacteria in the infant gut and exert potential protection against infection.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org. Supplementary Tables S1–S4.
Author Contributions
Conceptualization, Y.C. and D.C.D.; Methodology, Y.L., Y.C., B.J.K., J.S.P. and D.C.D.; Formal Analysis, Y.L., Y.C. and B.J.K.; Writing – Original Draft Preparation, Y.C.; Writing – Review & Editing, Y.L., Y.C., B.J.K., J.S.P. and D.C.D.; Visualization, Y.L., B.J.K.; Supervision, Y.C. and D.C.D.; Funding Acquisition, Y.C. and D.C.D.
Funding
This research was funded by the National Institutes of Health (R01HD097367 and R01HD109193) (D.C.D.), the United States Department of Agriculture National Institute of Food and Agriculture (2018-67017-27521) (D.C.D.), the Gerber Foundation (2017-1586) (D.C.D.), the United States Department of Agriculture Multistate Workgroup W4002 and the International Society for Research on Human Milk and Lactation–Family Larsson-Rosenquist Foundation Trainee Expansion Program. JSP was supported by the Institute for Modeling Collaboration and Innovation sponsored by the NIGMS under award number National Institutes of Health P20GM104420.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data available upon request.
Acknowledgments
The Mass Spectrometry Center at Oregon State University, which is supported in part by the National Institute of Health grant (NIH # 1S10OD020111-01)
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Altobelli, E.; Angeletti, P.M.; Verrotti, A.; Petrocelli, R. The impact of human milk on necrotizing enterocolitis: A systematic review and meta-analysis. Nutrients. 2020, 12, 1322. [CrossRef]
- Nielsen, S.D.; Beverly, R.L.; Qu, Y.; Dallas, D.C. Milk bioactive peptide database: A comprehensive database of milk protein-derived bioactive peptides and novel visualization. Food Chem. 2017, 232, 673-682. [CrossRef]
- Nielsen, S.D.; Liang, N.; Rathish, H.; Kim, B.J.; Lueangsakulthai, J.; Koh, J.; Qu, Y.; Schulz, H.J.; Dallas, D.C. Bioactive milk peptides: An updated comprehensive overview and database. Crit Rev Food Sci Nutr. 2023, 1-20. [CrossRef]
- Bazacliu, C.; Neu, J. Pathophysiology of necrotizing enterocolitis: An update. Curr Pediatr Rev. 2019, 15, 68-87. [CrossRef]
- Nielsen, S.D.; Beverly, R.L.; Underwood, M.A.; Dallas, D.C. Differences and similarities in the peptide profile of preterm and term mother’s milk, and preterm and term infant gastric samples. Nutrients. 2020, 12, 2825. [CrossRef]
- Beverly, R.L.; Huston, R.K.; Markell, A.M.; McCulley, E.A.; Martin, R.L.; Dallas, D.C. Differences in human milk peptide release along the gastrointestinal tract between preterm and term infants. Clin Nutr. 2021, 40, 1214-1223. [CrossRef]
- Beverly, R.L.; Underwood, M.A.; Dallas, D.C. Peptidomics analysis of milk protein-derived peptides released over time in the preterm infant stomach. J Proteome Res. 2019, 18, 912-922. [CrossRef]
- Nielsen, S.D.; Beverly, R.L.; Underwood, M.A.; Dallas, D.C. Release of functional peptides from mother’s milk and fortifier proteins in the premature infant stomach. PLoS One. 2018, 13, e0208204. [CrossRef]
- Beverly, R.L.; Woonnimani, P.; Scottoline, B.P.; Lueangsakulthai, J.; Dallas, D.C. Peptides from the intestinal tract of breast milk-fed infants have antimicrobial and bifidogenic activity. Int J Mol Sci. 2021, 22. [CrossRef]
- Beverly, R.L.; Huston, R.K.; Markell, A.M.; McCulley, E.A.; Martin, R.L.; Dallas, D.C. Milk peptides survive in vivo gastrointestinal digestion and are excreted in the stool of infants. J Nutr. 2020, 150, 712-721. [CrossRef]
- Liang, N.; Beverly, R.L.; Scottoline, B.P.; Dallas, D.C. Peptides derived from in vitro and in vivo digestion of human milk are immunomodulatory in THP-1 human macrophages. J Nutr. 2022, 152, 331-342. [CrossRef]
- Chen, Y.; Patel, A.; Meier, P.P.; Fantuzzi, G. Digested early preterm human milk suppresses tumor necrosis factor-induced inflammation and cytotoxicity in intestinal epithelial cells. J Pediatr Gastroenterol Nutr. 2018, 66, e153-e157. [CrossRef]
- Lyu, Y.; Chen, Y. Digested human colostrum reduces interleukin-8 production in induced human intestinal epithelial cells. Nutrients. 2022, 14, 2787. [CrossRef]
- Bhadra, P.; Yan, J.; Li, J.; Fong, S.; Siu, S.W. AmPEP: Sequence-based prediction of antimicrobial peptides using distribution patterns of amino acid properties and random forest. Sci Rep. 2018, 8, 1697. [CrossRef]
- Hornik, C.P.; Fort, P.; Clark, R.H.; Watt, K.; Benjamin, D.K., Jr.; Smith, P.B.; Manzoni, P.; Jacqz-Aigrain, E.; Kaguelidou, F.; Cohen-Wolkowiez, M. Early and late onset sepsis in very-low-birth-weight infants from a large group of neonatal intensive care units. Early Hum Dev. 2012, 88 Suppl 2, S69-74. [CrossRef]
- Wiegand, I.; Hilpert, K.; Hancock, R.E. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc. 2008, 3, 163-175. [CrossRef]
- Babicki, S.; Arndt, D.; Marcu, A.; Liang, Y.; Grant, J.R.; Maciejewski, A.; Wishart, D.S. Heatmapper: Web-enabled heat mapping for all. Nucleic Acids Res. 2016, 44, W147-W153. [CrossRef]
- Heegaard, C.W.; Larsen, L.B.; Rasmussen, L.K.; Højberg, K.E.; Petersen, T.E.; Andreasen, P.A. Plasminogen activation system in human milk. J Pediatr Gastroenterol Nutr. 1997, 25, 159-166. [CrossRef]
- He, Y.; Liu, S.; Leone, S.; Newburg, D.S. Human colostrum oligosaccharides modulate major immunologic pathways of immature human intestine. Mucosal Immunol. 2014, 7, 1326-1339. [CrossRef]
- Mantis, N.J.; Rol, N.; Corthésy, B. Secretory IgA’s complex roles in immunity and mucosal homeostasis in the gut. Mucosal Immunol. 2011, 4, 603-611. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).