Submitted:
16 May 2024
Posted:
17 May 2024
You are already at the latest version
Abstract
Keywords:
Introduction
Methods
Results
Discussion
Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Statement of Ethics
References
- O'Mahony, A.M.; Lynn, E.; Murphy, D.J.; Fabre, A.; McCarthy, C. Lymphangioleiomyomatosis: a clinical review. Breathe 2020, 16, 200007. [Google Scholar] [CrossRef] [PubMed]
- Koslow, M.; Lynch, D.A.; Cool, C.D.; Groshong, S.D.; Downey, G.P. Lymphangioleiomyomatosis and Other Cystic Lung Diseases. Immunol. Allergy Clin. North Am. 2023, 43, 359–377. [Google Scholar] [CrossRef]
- Kundu, N.; Holz, M.K. Lymphangioleiomyomatosis: a metastatic lung disease. Am. J. Physiol. Physiol. 2023, 324, C320–C326. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.J.; Robb, V.A.; Morrison, T.A.; Ariazi, E.A.; Karbowniczek, M.; Astrinidis, A.; Wang, C.; Hernandez-Cuebas, L.; Seeholzer, L.F.; Nicolas, E.; et al. Estrogen promotes the survival and pulmonary metastasis of tuberin-null cells. Proc. Natl. Acad. Sci. 2009, 106, 2635–2640. [Google Scholar] [CrossRef] [PubMed]
- Prizant, H.; Sen, A.; Light, A.; Cho, S.-N.; DeMayo, F.J.; Lydon, J.P.; Hammes, S.R. Uterine-Specific Loss of Tsc2 Leads to Myometrial Tumors in Both the Uterus and Lungs. Mol. Endocrinol. 2013, 27, 1403–1414. [Google Scholar] [CrossRef] [PubMed]
- Avecilla, V.; Doke, M.; Das, M.; Alcazar, O.; Appunni, S.; Tondin, A.R.; Watts, B.; Ramamoorthy, V.; Rubens, M.; Das, J.K. Integrative Bioinformatics–Gene Network Approach Reveals Linkage between Estrogenic Endocrine Disruptors and Vascular Remodeling in Peripheral Arterial Disease. Int. J. Mol. Sci. 2024, 25, 4502. [Google Scholar] [CrossRef] [PubMed]
- Avecilla, V. Effect of Transcriptional Regulator ID3 on Pulmonary Arterial Hypertension and Hereditary Hemorrhagic Telangiectasia. Int. J. Vasc. Med. 2019, 2019, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Avecilla, V.; Doke, M.; Felty, Q. Contribution of Inhibitor of DNA Binding/Differentiation-3 and Endocrine Disrupting Chemicals to Pathophysiological Aspects of Chronic Disease. BioMed Res. Int. 2017, 2017, 6307109–22. [Google Scholar] [CrossRef] [PubMed]
- Doke, M.; Avecilla, V.; Felty, Q. Inhibitor of Differentiation-3 and Estrogenic Endocrine Disruptors: Implications for Susceptibility to Obesity and Metabolic Disorders. BioMed Res. Int. 2018, 2018, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Avecilla, V.E. ID3, Estrogenic Chemicals, and the Pathogenesis of Tumor-Like Proliferative Vascular Lesions. (2017). FIU Electronic Theses and Dissertations. 3519. Available online: https://digitalcommons.fiu.edu/etd/3519.
- Avecilla, A.; Doke, M.; Jovellanos, J.; Avecilla, V. Contribution of Inhibitor of Differentiation and Estrogenic Endocrine Disruptors to Neurocognitive Disorders. Med Sci. 2018, 6, 61. [Google Scholar] [CrossRef] [PubMed]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res. 2012, 41, D991–D995. [Google Scholar] [CrossRef] [PubMed]
- Pacheco-Rodriguez, G.; Kumaki, F.; Steagall, W.K.; Zhang, Y.; Ikeda, Y.; Lin, J.-P.; Billings, E.M.; Moss, J. Chemokine-Enhanced Chemotaxis of Lymphangioleiomyomatosis Cells with Mutations in the Tumor Suppressor TSC2 Gene. J. Immunol. 2009, 182, 1270–1277. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Law, C.W.; Chen, Y.; Shi, W.; Smyth, G.K. Voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014, 15, R29. [Google Scholar] [CrossRef] [PubMed]
- Phipson, B.; Lee, S.; Majewski, I.J.; Alexander, W.S.; Smyth, G.K. Robust hyperparameter estimation protects against hypervariable genes and improves power to detect differential expression. Ann. Appl. Stat. 2016, 10, 946–963. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef] [PubMed]
- Keenan, A.B.; Torre, D.; Lachmann, A.; Leong, A.K.; Wojciechowicz, M.L.; Utti, V.; Jagodnik, K.M.; Kropiwnicki, E.; Wang, Z.; Ma’ayan, A. ChEA3: transcription factor enrichment analysis by orthogonal omics integration. Nucleic Acids Res. 2019, 47, W212–W224. [Google Scholar] [CrossRef] [PubMed]
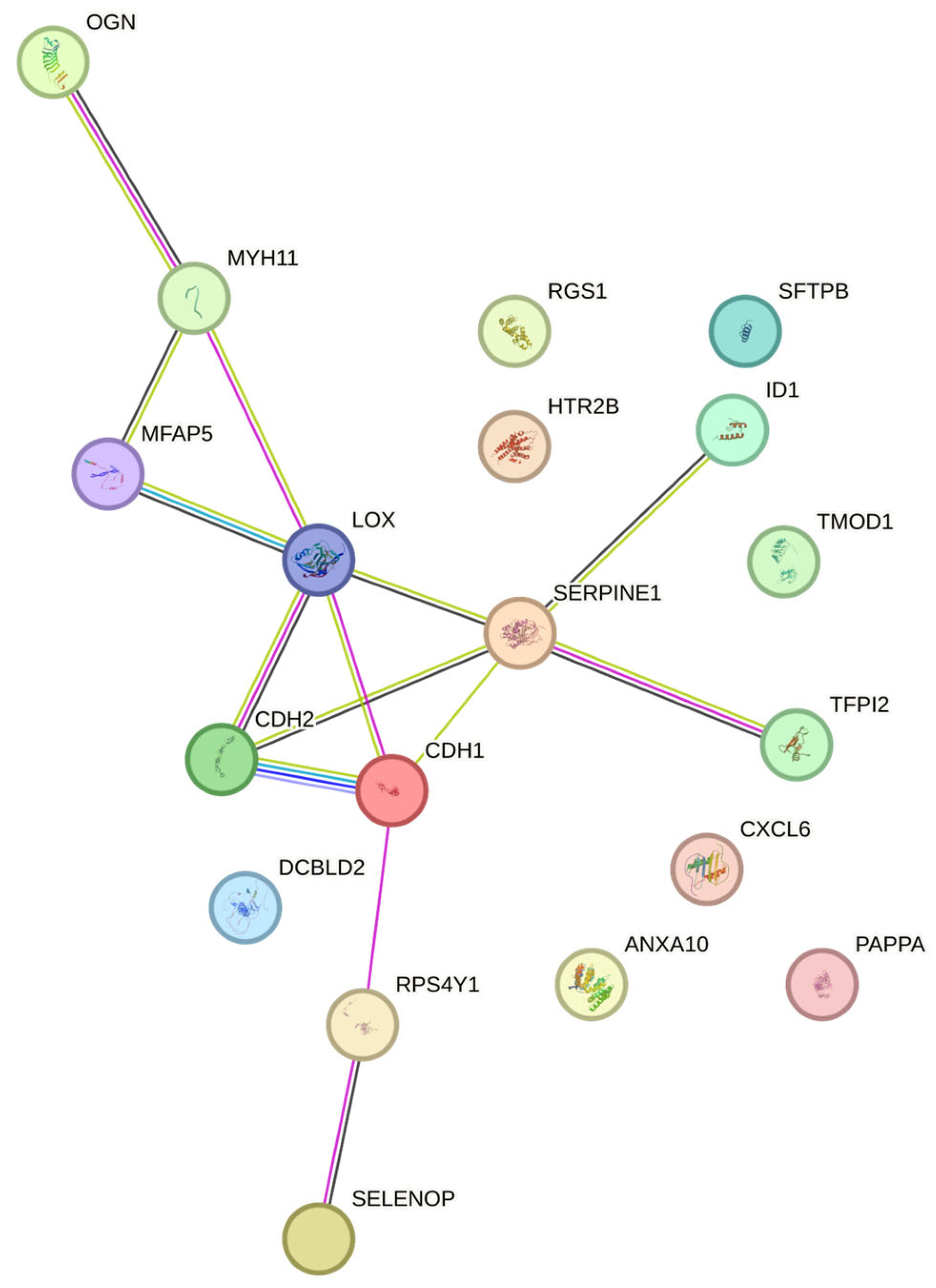

| Gene Symbol | Gene Title | Log2 (Fold Change) | p – value |
|---|---|---|---|
| CDH2 | cadherin 2 | 7.6295995 | 3.73E-12 |
| CXCL6 | C-X-C motif chemokine ligand 6 | 7.2061993 | 3.26E-14 |
| ANXA10 | annexin A10 | 6.7952473 | 5.57E-13 |
| MFAP5 | microfibrillar associated protein 5 | 5.7810303 | 5.57E-13 |
| RPS4Y1 | ribosomal protein S4, Y-linked 1 | 5.6284047 | 1.57E-15 |
| DCBLD2 | discoidin, CUB and LCCL domain containing 2 | 5.5517299 | 9.99E-13 |
| PAPPA | pappalysin 1 | 5.5260289 | 1.43E-11 |
| TFPI2 | tissue factor pathway inhibitor 2 | 5.4304952 | 2.63E-13 |
| SERPINE1 | serpin family E member 1 | 5.3529649 | 3.70E-11 |
| LOX | lysyl oxidase | 5.0978044 | 2.85E-12 |
| MYH11 | myosin heavy chain 11 | -12.0887684 | 1.11E-17 |
| RGS1 | regulator of G-protein signaling 1 | -9.9510615 | 4.09E-13 |
| SEPP1 | selenoprotein P, plasma, 1 | -9.5071713 | 2.18E-14 |
| TMOD1 | tropomodulin 1 | -9.2792733 | 1.57E-15 |
| ID1 | Inhibitor of differentiation - 1 | -9.126583 | 1.32E-06 |
| SFTPB | surfactant protein B | -8.471653 | 4.91E-08 |
| CDH1 | cadherin 1 | -8.3548182 | 2.27E-13 |
| HTR2B | 5-hydroxytryptamine receptor 2B | -7.6577741 | 4.34E-13 |
| OGN | osteoglycin | -7.6373228 | 1.53E-12 |
| IGLC1 | immunoglobulin lambda constant 1 | -7.6012966 | 1.12E-07 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




