Submitted:
16 May 2024
Posted:
17 May 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. Antimicrobial Susceptibility
2.2. Genomic Analysis of Clinical K. pneumoniae Isolate 130125
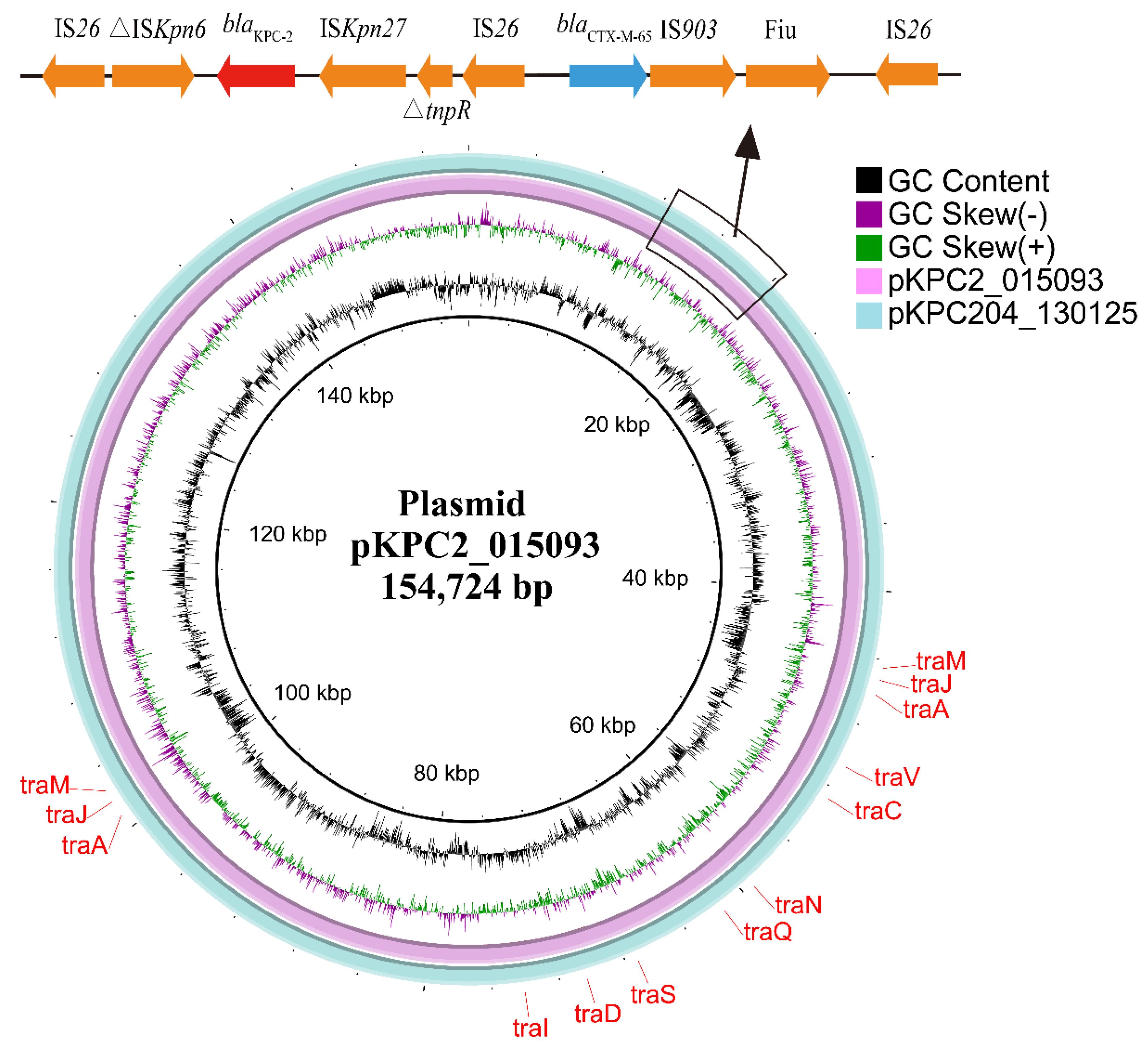
2.3. Genetic Context of blaKPC-204-Carrying Plasmid
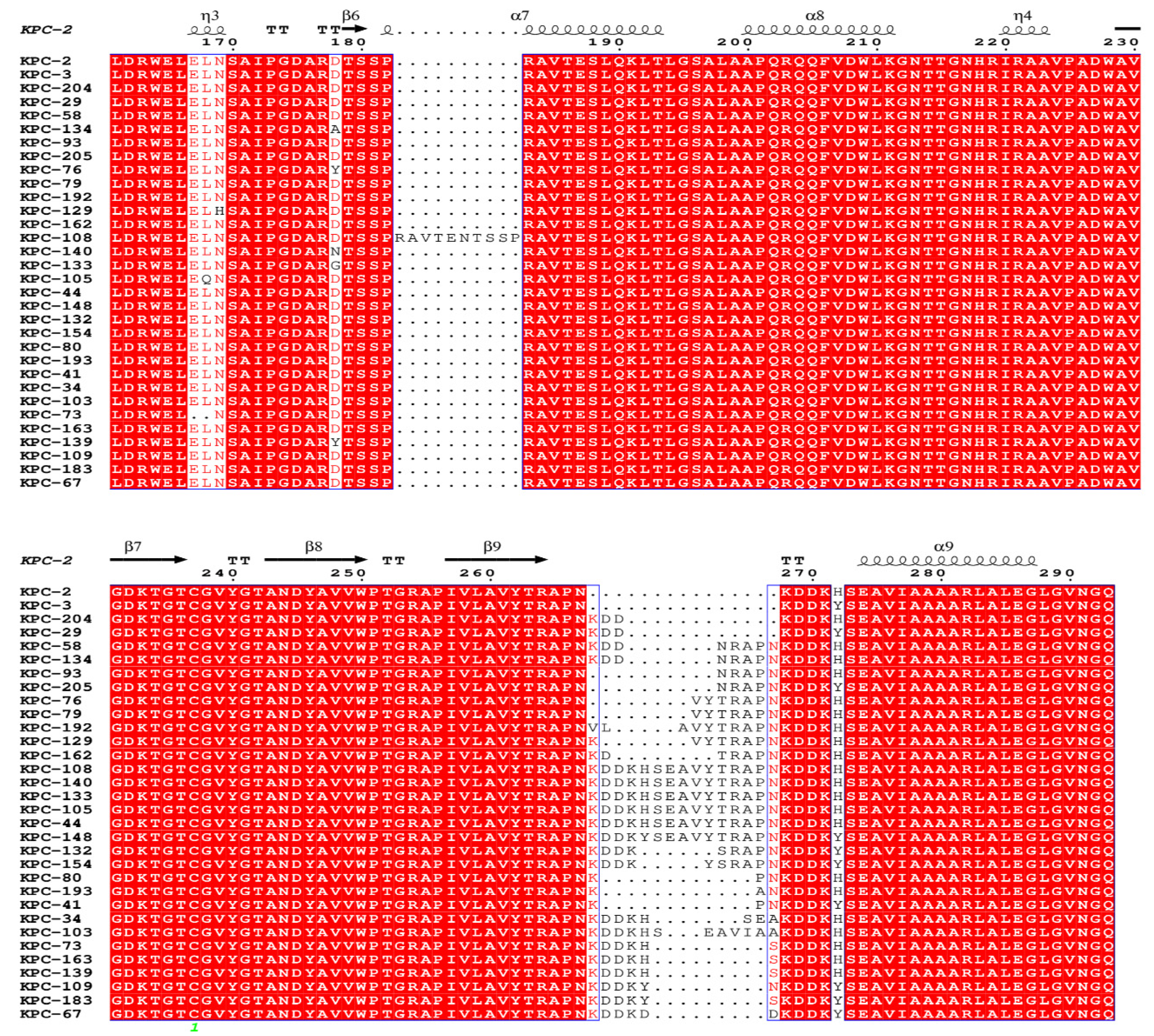
2.4. Identification of blaKPC-204 Involved in CZA Resistance
2.5. Enzyme Kinetic Parameters and IC50 Values
2.6. blaKPC-204 Was Located in a Self-Transmissible Plasmid
3. Discussion
4. Materials and Methods
4.1. The Strains and In Vitro Susceptibility
4.2. Whole Genome Sequencing and Analysis
4.3. Cloning Experiment
4.4. Kinetic Assay and Determination of IC50 Values
4.5. Mating Experiments
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ma, J.; Song, X.; Li, M.; Yu, Z.; Cheng, W.; Yu, Z.; Zhang, W.; Zhang, Y.; Shen, A.; Sun, H.; et al. Global Spread of Carbapenem-Resistant Enterobacteriaceae: Epidemiological Features, Resistance Mechanisms, Detection and Therapy. Microbiological Research 2023, 266, 127249. [Google Scholar] [CrossRef]
- Kalil, A.C.; Klompas, M. Ceftazidime-Avibactam versus Meropenem for the Treatment of Nosocomial Pneumonia. LANCET INFECTIOUS DISEASES 2018, 18, 229–231. [Google Scholar] [CrossRef]
- Bush, K.; Bradford, P.A. Interplay between β-Lactamases and New β-Lactamase Inhibitors. #N/A 2019, 17, 295–306. [Google Scholar] [CrossRef]
- Hobson, C.A.; Pierrat, G.; Tenaillon, O.; Bonacorsi, S.; Bercot, B.; Jaouen, E.; Jacquier, H.; Birgy, A. Klebsiella Pneumoniae Carbapenemase Variants Resistant to Ceftazidime-Avibactam: An Evolutionary Overview. Antimicrob Agents Chemother 2022, 66, e0044722. [Google Scholar] [CrossRef]
- Ding, L.; Shen, S.; Chen, J.; Tian, Z.; Shi, Q.; Han, R.; Guo, Y.; Hu, F. Klebsiella Pneumoniae Carbapenemase Variants: The New Threat to Global Public Health. Clin Microbiol Rev 2023, 36, e0000823. [Google Scholar] [CrossRef]
- Shields, R.K.; Nguyen, M.H.; Press, E.G.; Chen, L.; Kreiswirth, B.N.; Clancy, C.J. In Vitro Selection of Meropenem Resistance among Ceftazidime-Avibactam-Resistant, Meropenem-Susceptible Klebsiella Pneumoniae Isolates with Variant KPC-3 Carbapenemases. Antimicrob Agents Chemother 2017, 61. [Google Scholar] [CrossRef]
- Huang, X.; Shen, S.; Chang, F.; Liu, X.; Yue, J.; Xie, N.; Yin, L.; Hu, F.; Xiao, D. Emergence of KPC-134, a KPC-2 Variant Associated with Ceftazidime-Avibactam Resistance in a ST11 Klebsiella Pneumoniae Clinical Strain. Microbiol Spectr 2023, 11, e0072523. [Google Scholar] [CrossRef]
- Wu, Y.; Yang, X.; Liu, C.; Zhang, Y.; Cheung, Y.C.; Wai Chi Chan, E.; Chen, S.; Zhang, R. Identification of a KPC Variant Conferring Resistance to Ceftazidime-Avibactam from ST11 Carbapenem-Resistant Klebsiella Pneumoniae Strains. Microbiol Spectr 2022, 10, e0265521. [Google Scholar] [CrossRef]
- Li, X.; Quan, J.; Ke, H.; Wu, W.; Feng, Y.; Yu, Y.; Jiang, Y. Emergence of a KPC Variant Conferring Resistance to Ceftazidime-Avibactam in a Widespread ST11 Carbapenem-Resistant Klebsiella Pneumoniae Clone in China. Front Microbiol 2021, 12, 724272. [Google Scholar] [CrossRef]
- Li, X.; Ke, H.; Wu, W.; Tu, Y.; Zhou, H.; Yu, Y. Molecular Mechanisms Driving the In Vivo Development of KPC-71-Mediated Resistance to Ceftazidime-Avibactam during Treatment of Carbapenem-Resistant Klebsiella Pneumoniae Infections. mSphere 2021, 6, e0085921. [Google Scholar] [CrossRef]
- Cano, A.; Guzman-Puche, J.; Garcia-Gutierrez, M.; Caston, J.J.; Gracia-Ahufinger, I.; Perez-Nadales, E.; Recio, M.; Natera, A.M.; Marfil-Perez, E.; Martinez-Martinez, L.; et al. Use of Carbapenems in the Combined Treatment of Emerging Ceftazidime/Avibactam-Resistant and Carbapenem-Susceptible KPC-Producing Klebsiella Pneumoniae Infections: Report of a Case and Review of the Literature. J Glob Antimicrob Resist 2020, 22, 9–12. [Google Scholar] [CrossRef]
- Hobson, C.A.; Bonacorsi, S.; Jacquier, H.; Choudhury, A.; Magnan, M.; Cointe, A.; Bercot, B.; Tenaillon, O.; Birgy, A. KPC Beta-Lactamases Are Permissive to Insertions and Deletions Conferring Substrate Spectrum Modifications and Resistance to Ceftazidime-Avibactam. ANTIMICROBIAL AGENTS AND CHEMOTHERAPY 2020. [Google Scholar] [CrossRef]
- Shen, Z.; Ding, B.; Ye, M.; Wang, P.; Bi, Y.; Wu, S.; Xu, X.; Guo, Q.; Wang, M. High Ceftazidime Hydrolysis Activity and Porin OmpK35 Deficiency Contribute to the Decreased Susceptibility to Ceftazidime/Avibactam in KPC-Producing Klebsiella Pneumoniae. JOURNAL OF ANTIMICROBIAL CHEMOTHERAPY 2017, 72, 1930–1936. [Google Scholar] [CrossRef]
- Tian, X.; Wang, Q.; Perlaza-Jiménez, L.; Zheng, X.; Zhao, Y.; Dhanasekaran, V.; Fang, R.; Li, J.; Wang, C.; Liu, H.; et al. First Description of Antimicrobial Resistance in Carbapenem-Susceptible Klebsiella Pneumoniae after Imipenem Treatment, Driven by Outer Membrane Remodeling. BMC Microbiol 2020, 20, 218. [Google Scholar] [CrossRef]
- Robert, X.; Gouet, P. Deciphering Key Features in Protein Structures with the New ENDscript Server. Nucleic Acids Res 2014, 42, W320–W324. [Google Scholar] [CrossRef]
- Alikhan, N.-F.; Petty, N.K.; Ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple Prokaryote Genome Comparisons. BMC Genomics 2011, 12, 402. [Google Scholar] [CrossRef]
- Virolle, C.; Goldlust, K.; Djermoun, S.; Bigot, S.; Lesterlin, C. Plasmid Transfer by Conjugation in Gram-Negative Bacteria: From the Cellular to the Community Level. Genes (Basel) 2020, 11, 1239. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, Q.; Yin, Y.; Chen, H.; Jin, L.; Gu, B.; Xie, L.; Yang, C.; Ma, X.; Li, H.; et al. Epidemiology of Carbapenem-Resistant Enterobacteriaceae Infections: Report from the China CRE Network. Antimicrob Agents Chemother 2018, 62, e01882-17. [Google Scholar] [CrossRef]
- Zhang, R.; Liu, L.; Zhou, H.; Chan, E.W.; Li, J.; Fang, Y.; Li, Y.; Liao, K.; Chen, S. Nationwide Surveillance of Clinical Carbapenem-Resistant Enterobacteriaceae (CRE) Strains in China. EBioMedicine 2017, 19, 98–106. [Google Scholar] [CrossRef]
- Cui, X.; Shan, B.; Zhang, X.; Qu, F.; Jia, W.; Huang, B.; Yu, H.; Tang, Y.W.; Chen, L.; Du, H. Reduced Ceftazidime-Avibactam Susceptibility in KPC-Producing Klebsiella Pneumoniae From Patients Without Ceftazidime-Avibactam Use History - A Multicenter Study in China. Front. Microbiol. 2020, 11, 1365. [Google Scholar] [CrossRef]
- Carattoli, A.; Arcari, G.; Bibbolino, G.; Sacco, F.; Tomolillo, D.; Di Lella, F.M.; Trancassini, M.; Faino, L.; Venditti, M.; Antonelli, G.; et al. Evolutionary Trajectories toward Ceftazidime-Avibactam Resistance in Klebsiella Pneumoniae Clinical Isolates. Antimicrob Agents Chemother 2021, 65, e0057421. [Google Scholar] [CrossRef]
- Räisänen, K.; Koivula, I.; Ilmavirta, H.; Puranen, S.; Kallonen, T.; Lyytikäinen, O.; Jalava, J. Emergence of Ceftazidime-Avibactam-Resistant Klebsiella Pneumoniae during Treatment, Finland, December 2018. Euro Surveill 2019, 24, 1900256. [Google Scholar] [CrossRef] [PubMed]
- Arcari, G.; Cecilia, F.; Oliva, A.; Polani, R.; Raponi, G.; Sacco, F.; De Francesco, A.; Pugliese, F.; Carattoli, A. Genotypic Evolution of Klebsiella Pneumoniae Sequence Type 512 during Ceftazidime/Avibactam, Meropenem/Vaborbactam, and Cefiderocol Treatment, Italy. Emerg Infect Dis 2023, 29, 2266–2274. [Google Scholar] [CrossRef]
- Shi, Q.; Han, R.; Guo, Y.; Yang, Y.; Wu, S.; Ding, L.; Zhang, R.; Yin, D.; Hu, F. Multiple Novel Ceftazidime-Avibactam-Resistant Variants of blaKPC-2-Positive Klebsiella Pneumoniae in Two Patients. Microbiol Spectr 2022, 10, e0171421. [Google Scholar] [CrossRef]
- Mueller, L.; Masseron, A.; Prod’Hom, G.; Galperine, T.; Greub, G.; Poirel, L.; Nordmann, P. Phenotypic, Biochemical, and Genetic Analysis of KPC-41, a KPC-3 Variant Conferring Resistance to Ceftazidime-Avibactam and Exhibiting Reduced Carbapenemase Activity. Antimicrobial Agents and Chemotherapy 2019, 63, 10.1128/aac.01111-19. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Feng, Y.; Wei, L.; Xiao, Y.; Zong, Z. KPC-2-Producing Carbapenem-Resistant Klebsiella Pneumoniae of the Uncommon ST29 Type Carrying OXA-926, a Novel Narrow-Spectrum OXA β-Lactamase. Front. Microbiol. 2021, 12, 701513. [Google Scholar] [CrossRef]
- Qin, J.; Feng, Y.; Lü, X.; Zong, Z. KPC-12 with a L169M Substitution in the Ω Loop Has Reduced Carbapenemase Activity. EUROPEAN JOURNAL OF CLINICAL MICROBIOLOGY & INFECTIOUS DISEASES 2021, 40, 1761–1766. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Shen, W.; Zhang, R.; Cai, J. Identification of a Novel Ceftazidime-Avibactam-Resistant KPC-2 Variant, KPC-123, in Citrobacter Koseri Following Ceftazidime-Avibactam Treatment. Front Microbiol 2022, 13, 930777. [Google Scholar] [CrossRef]
- Jiang, Y.; Yu, D.; Wei, Z.; Shen, P.; Zhou, Z.; Yu, Y. Complete Nucleotide Sequence of Klebsiella Pneumoniae Multidrug Resistance Plasmid pKP048, Carrying blaKPC-2, blaDHA-1, qnrB4, and armA. Antimicrob Agents Chemother 2010, 54, 3967–3969. [Google Scholar] [CrossRef]
- Cuzon, G.; Naas, T.; Nordmann, P. Functional Characterization of Tn4401, a Tn3-Based Transposon Involved in blaKPC Gene Mobilization. Antimicrob. Agents Chemother. 2011, 55, 5370–5373. [Google Scholar] [CrossRef]
- Wayne, PA CLSI. Performance Standards for Antimicrobial Susceptibility Testing32nd Informational Supplement: M100-S32. 2022.
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving Bacterial Genome Assemblies from Short and Long Sequencing Reads. PLoS Comput Biol 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Jain, C.; Rodriguez-R, L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High Throughput ANI Analysis of 90K Prokaryotic Genomes Reveals Clear Species Boundaries. Nat Commun 2018, 9, 5114. [Google Scholar] [CrossRef]
- Richter, M.; Rosselló-Móra, R. Shifting the Genomic Gold Standard for the Prokaryotic Species Definition. Proc Natl Acad Sci U S A 2009, 106, 19126–19131. [Google Scholar] [CrossRef]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-Access Bacterial Population Genomics: BIGSdb Software, the PubMLST.Org Website and Their Applications. Wellcome Open Res 2018, 3, 124. [Google Scholar] [CrossRef]
- Wyres, K.L.; Wick, R.R.; Gorrie, C.; Jenney, A.; Follador, R.; Thomson, N.R.; Holt, K.E. Identification of Klebsiella Capsule Synthesis Loci from Whole Genome Data. Microbial Genomics 2016, 2. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Møller Aarestrup, F.; Hasman, H. In Silico Detection and Typing of Plasmids Using PlasmidFinder and Plasmid Multilocus Sequence Typing. Antimicrob Agents Chemother 2014, 58, 3895–3903. [Google Scholar] [CrossRef]
- Siguier, P.; Perochon, J.; Lestrade, L.; Mahillon, J.; Chandler, M. ISfinder: The Reference Centre for Bacterial Insertion Sequences. Nucleic Acids Res 2006, 34, D32–D36. [Google Scholar] [CrossRef]
- Sambrook J, Russell D. 2001. Molecular Cloning: A Laboratory Manual, 3rd Ed. Cold Spring Harbor, (NY): Cold Spring Harbor Laboratory Press.
- Liu, S.; Jing, L.; Yu, Z.-J.; Wu, C.; Zheng, Y.; Zhang, E.; Chen, Q.; Yu, Y.; Guo, L.; Wu, Y.; et al. ((S)-3-Mercapto-2-Methylpropanamido)Acetic Acid Derivatives as Metallo-β-Lactamase Inhibitors: Synthesis, Kinetic and Crystallographic Studies. Eur J Med Chem 2018, 145, 649–660. [Google Scholar] [CrossRef] [PubMed]
- Coque, T.M.; Oliver, A.; Pérez-Díaz, J.C.; Baquero, F.; Cantón, R. Genes Encoding TEM-4, SHV-2, and CTX-M-10 Extended-Spectrum Beta-Lactamases Are Carried by Multiple Klebsiella Pneumoniae Clones in a Single Hospital (Madrid, 1989 to 2000). Antimicrob Agents Chemother 2002, 46, 500–510. [Google Scholar] [CrossRef] [PubMed]
| MICs (mg/L)a | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Strains | PIP | TZP | FOX | FEP | ATM | CAZ | CZA | IPM | IMR | MEM | MEV | ETP |
| 130125 | >512 | 256 | >512 | >512 | 512 | >512 | 256 | 64 | 0.25 | 64 | 0.5 | 64 |
| 015093 | >512 | >512 | >512 | >512 | 512 | >512 | 0.5 | 128 | 0.25 | 256 | 0.06 | 64 |
| DH5α::pEKPC-2 | >512 | >512 | >512 | >512 | 512 | 128 | 0.5 | 16 | 0.125 | 8 | 0.03 | 8 |
| DH5α::pEKPC-204 | >512 | 256 | >512 | 512 | 256 | 128 | 64 | 16 | 0.125 | 16 | 0.125 | 8 |
| DH5α::pET28a | 1 | 1 | 2 | 0.06 | 0.125 | 0.25 | 0.25 | 0.25 | 0.06 | ≤0.015 | ≤0.015 | ≤0.015 |
| E. coli J53 | 1 | 1 | 1 | 0.06 | 0.125 | 0.25 | 0.125 | 0.125 | 0.06 | ≤0.015 | ≤0.015 | ≤0.015 |
| J53::pKPC2_015093 | >512 | >512 | >512 | 512 | 512 | 512 | 0.5 | 32 | 0.25 | 32 | 0.03 | 32 |
| J53::KPC204_130125 | >512 | 256 | >512 | 512 | 512 | 512 | 64 | 32 | 0.25 | 64 | 0.25 | 32 |
| Accession no. | Size,bp | Replicon type | Resistance genes | ||
|---|---|---|---|---|---|
| β-Lactam | Other | ||||
| 130125_chr | CP148996 | 5,462,753 | - | blaSHV-158 | aadA2, fosA6 |
| pKPC204_130125 | CP148997 | 154,728 | IncR, IncFII | blaKPC-204, blaTEM-1, blaCTX-M-65 | rmtB1 |
| p1_130125 | CP148998 | 10,060 | ColRNAI | ||
| p2_130125 | CP148999 | 5,596 | - | ||
| KPC-2 | KPC-204 | ||||||
|---|---|---|---|---|---|---|---|
| β-Lactam | Km (μM) | kcat(s-1) | kcat/Km (μM-1·s-1) | Km (μM) | kcat(s-1) | kcat/Km (μM-1·s-1) | |
| Nitrocefin | 22.124 | 97.589 | 4.411 | 31.178 | 116.419 | 3.734 | |
| Ceftazidime | 870.413 | 5.226 | 0.006 | 975.154 | 7.801 | 0.008 | |
| Meropenem | 15.283 | 5.194 | 0.34 | 14.157 | 8.325 | 0.588 |
| IC50 (μM) | ||
|---|---|---|
| Inhibitor | KPC-2 | KPC-204 |
| Avibactam | 0.045 | 0.569 |
| Tazobactam | 1.782 | 0.083 |
| Clavulanic acid | 0.887 | 0.124 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





