Submitted:
10 May 2024
Posted:
15 May 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
3. Results
4. Discussion
5. Conclusions
References
- Abd Rashid, S.A.; Yaacob, M.F.; Raihanah, N.; Anuar, T.; Johari, N.; Kamaruzzaman, A.N.A.; Yahya, M.F.Z. R.; et al. A combination of in silico subtractive and reverse vaccinology approaches reveals potential vaccine targets in Corynebacterium pseudotuberculosis. Journal of Sustainability Science and Management 2022, 17, 99–109. [Google Scholar] [CrossRef]
- Armon, R.; Starosvetzky, J.; Arbel, T.; Green, M. Survival of Legionella pneumophila and Salmonella typhimurium in biofilm systems. Water Science and Technology 1997, 35, 293–300. [Google Scholar] [CrossRef]
- Attwood, M.M.; Schiöth, H.B. Characterization of five transmembrane proteins: With focus on the Tweety, sideroflexin, and yip1 domain families. Frontiers in Cell and Developmental Biology 2021, 9. [Google Scholar] [CrossRef] [PubMed]
- Bajire, S.K.; Ghate, S.D.; Shetty, S.; Banerjee, S.; Rao RS, P.; Shetty, V.; Shastry, R.P. Unveiling the role of hub proteins in controlling quorum sensing regulated virulence through analogues in Pseudomonas aeruginosa PAO1: A functional protein-protein network biology approach. Biochemical and Biophysical Research Communications 2023, 660, 13–20. [Google Scholar] [CrossRef]
- Fernandes, L.; Centeno, M.M.; Couto, N.; Nunes, T.; Almeida, V.; Alban, L.; Pomba, C. Longitudinal characterization of monophasic salmonella typhimurium throughout the pig’s life cycle. Veterinary Microbiology 2016, 192, 231–237. [Google Scholar] [CrossRef] [PubMed]
- Hapfelmeier, S.; Hardt, W.-D. A mouse model for S. typhimurium-induced enterocolitis. Trends in Microbiology 2005, 13, 497–503. [Google Scholar] [CrossRef]
- Isa SF, M.; Abdul Hamid, U.M.; Zaman Raja Yahya, M.F. Treatment with the combined antimicrobials triggers proteomic changes in P. aeruginosa-C. albicans polyspecies biofilms. ScienceAsia 2022, 48. [Google Scholar]
- Jalal, K.; Khan, K.; Hassam, M.; Abbas, M.N.; Uddin, R.; Khusro, A.; Gajdács, M.; et al. Identification of a novel therapeutic target against XDR Salmonella Typhi H58 using genomics driven approach followed up by natural products virtual screening. Microorganisms 2021, 9, 2512. [Google Scholar] [CrossRef] [PubMed]
- Johari, N.A.; Aazmi, M.S.; Yahya, M.F.Z.R. FTIR Spectroscopic Study of Inhibition of Chloroxylenol-Based Disinfectant Against Salmonella enterica serovar Thyphimurium Biofilm. Malaysian Applied Biology 2023, 52, 97–107. [Google Scholar] [CrossRef]
- Kamaruzzaman, A.N.A.; Mulok, T.E.T.Z.; Nor, N.H.M.; Yahya, M.F.Z.R. FTIR spectral changes in Candida albicans biofilm following exposure to antifungals. Malaysian Applied Biology 2022, 51, 57–66. [Google Scholar] [CrossRef]
- Khan, K.; Uddin, R. Integrated bioinformatics based subtractive genomics approach to decipher the therapeutic function of hypothetical proteins from Salmonella typhi XDR H-58 strain. Biotechnology Letters 2022, 1–20. [Google Scholar] [CrossRef]
- Lapidot, A.; Romling, U.; Yaron, S. Biofilm formation and the survival of Salmonella Typhimurium on parsley. International journal of food microbiology 2006, 109, 229–233. [Google Scholar] [CrossRef]
- Mohammed, B.T. Identification and bioinformatic analysis of invA gene of Salmonella in free range chicken. Brazilian Journal of Biology 2022, 84, e263363. [Google Scholar] [CrossRef] [PubMed]
- Nithya, C.; Kiran, M.; Nagarajaram, H.A. Dissection of hubs and bottlenecks in a protein-protein interaction network. Computational Biology and Chemistry 2023, 102, 107802. [Google Scholar] [CrossRef] [PubMed]
- Nogueira, W.G.; Jaiswal, A.K.; Tiwari, S.; Ramos, R.T.; Ghosh, P.; Barh, D.; Soares, S.C.; et al. Computational identification of putative common genomic drug and vaccine targets in Mycoplasma genitalium. Genomics 2021, 113, 2730–2743. [Google Scholar] [CrossRef]
- Okoro, C.K.; Barquist, L.; Connor, T.R.; Harris, S.R.; Clare, S.; Stevens, M.P.; Arends, M.J.; Hale, C.; Kane, L.; Pickard, D.J.; Hill, J.; Harcourt, K.; Parkhill, J.; Dougan, G.; Kingsley, R.A. Signatures of adaptation in human invasive salmonella typhimurium ST313 populations from Sub-Saharan africa. PLOS Neglected Tropical Diseases 2015, 9. [Google Scholar]
- Othman, N.A.; Yahya, M.F.Z.R. In silico analysis of essential gene and non-homologous proteins in Salmonella typhimurium biofilm. Journal of Physics: Conference Series 2019, 1349, 012133. [Google Scholar]
- Ryu, H.; Fuwad, A.; Yoon, S.; Jang, H.; Lee, J.; Kim, S.; Jeon, T.-J. Biomimetic membranes with transmembrane proteins: State-of-the-art in transmembrane protein applications. International Journal of Molecular Sciences 2019, 20, 1437. [Google Scholar] [CrossRef]
- Sanches, R.C.; Tiwari, S.; Ferreira, L.C.; Oliveira, F.M.; Lopes, M.D.; Passos, M.J.; Lopes, D.O.; et al. Immunoinformatics design of multi-epitope peptide-based vaccine against Schistosoma mansoni using transmembrane proteins as a target. Frontiers in immunology 2021, 12, 621706. [Google Scholar] [CrossRef]
- Yaacob, M.F.; Murata, A.; Nor, N.H.M.; Jesse, F.F.A.; Yahya, M.F.Z.R. Biochemical composition, morphology and antimicrobial susceptibility pattern of Corynebacterium pseudotuberculosis biofilm. Journal of King Saud University-Science 2021, 33, 101225. [Google Scholar] [CrossRef]
- Yahya, M.F.Z.R.; Hamid, U.M.A.; Norfatimah, M.Y.; Kambol, R. In silico analysis of essential tricarboxylic acid cycle enzymes from biofilm-forming bacteria. Trends in Bioinformatics 2014, 7, 19–26. [Google Scholar] [CrossRef]
- Yahya, M.F.; Alias, Z.; Karsani, S.A. Subtractive protein profiling of Salmonella typhimurium biofilm treated with DMSO. The Protein Journal 2017, 36, 286–298. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R. DEG: A database of essential genes. Nucleic Acids Research 2004, 32. [Google Scholar] [CrossRef]
- Zulkiply, N.; Ramli, M.E.; Yahya, M.F.Z.R. In silico identification of antigenic proteins in Staphylococcus aureus. Journal of Sustainability Science and Management 2022, 17, 18–26. [Google Scholar] [CrossRef]
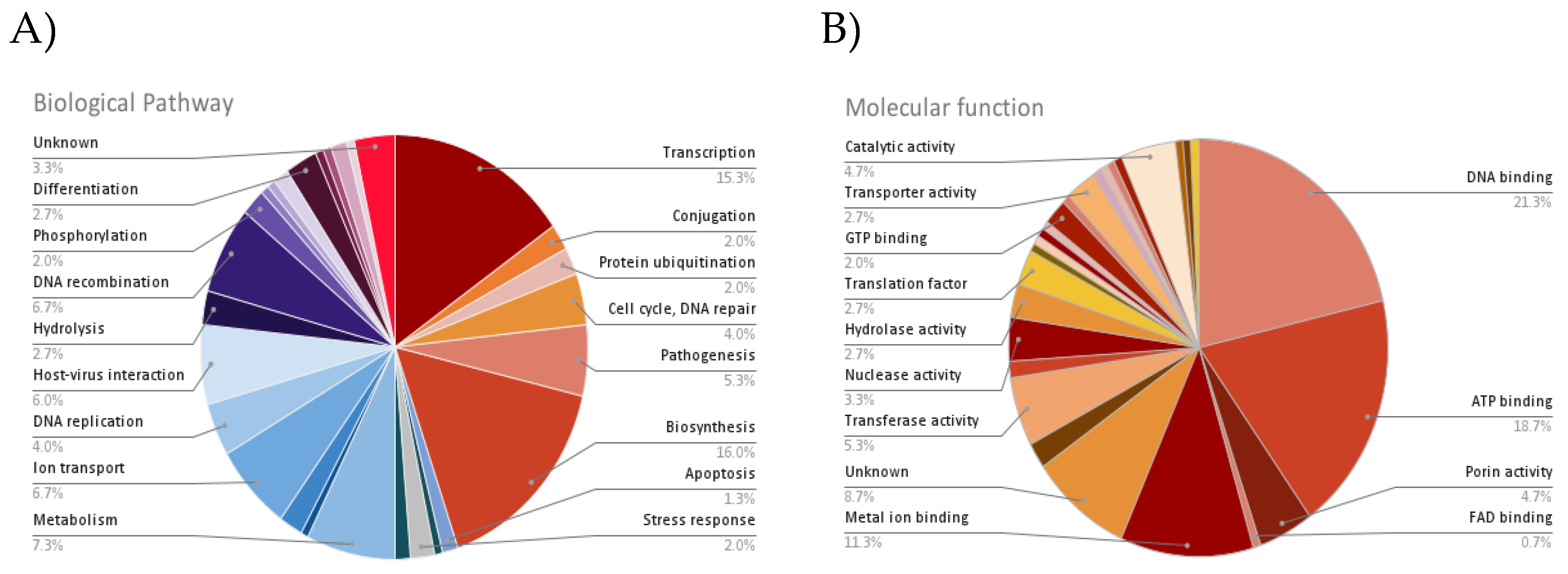
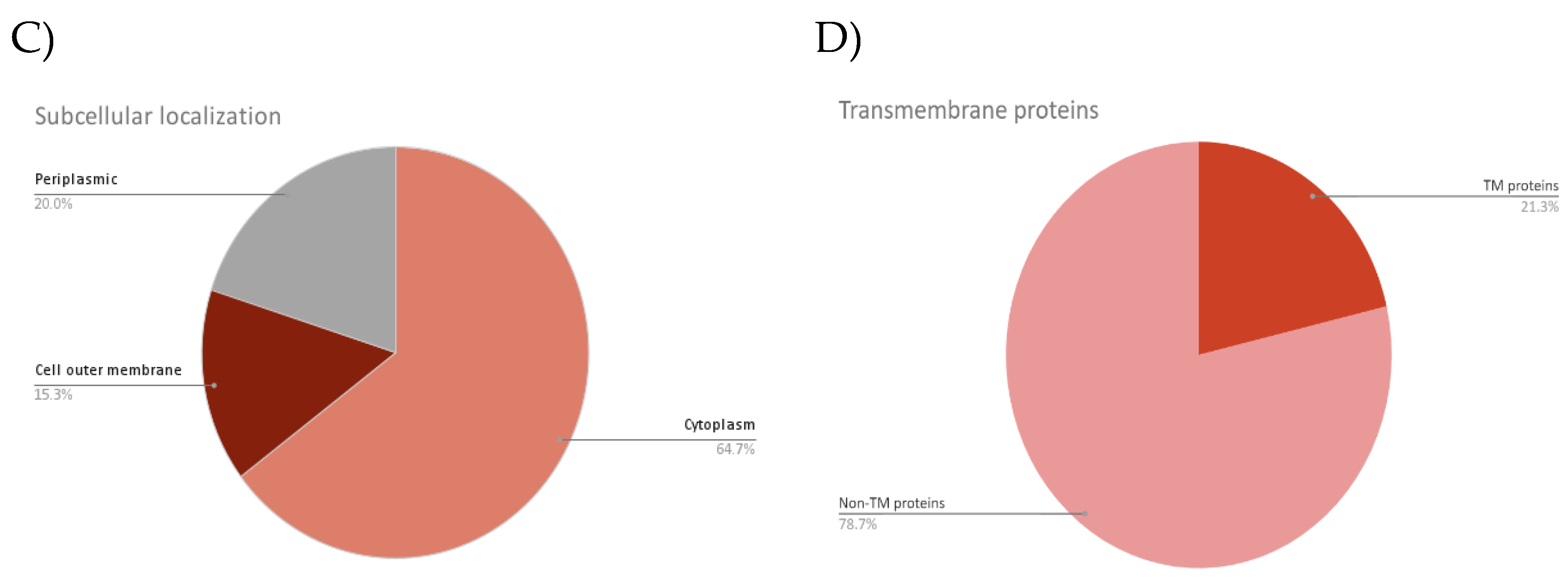
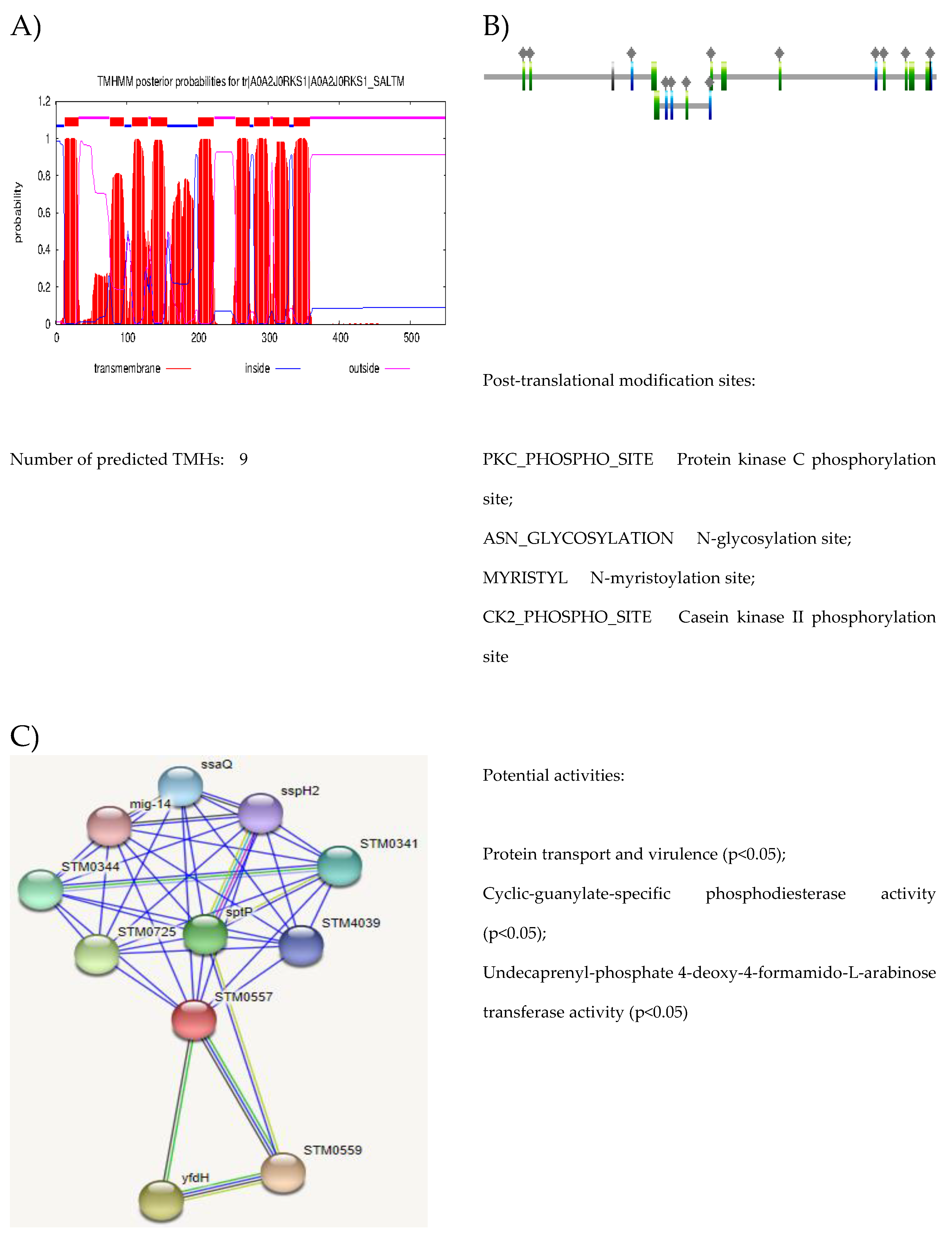
| Accession | No. of TM domain | Biological pathway | Molecular function | Subcellular localization |
|---|---|---|---|---|
| A0A2J0RKS1 | 9 | Conjugation | Porin activity | Cell outer membrane |
| A0A1P8DMP4 | 1 | Unknown | ATP binding | Plasma membrane |
| A0A3Z7JEV9 | 1 | DNA repair | DNA ligase activity | Cytosol |
| A0A2J0RFX8 | 1 | Catabolic process | D-aminoacyl-tRNA deacylase activity | Cytoplasm |
| A0A2J0RDC1 | 1 | Translocation | Unknown | Nucleus |
| A0A3T3ZZV4 | 1 | tRNA processing | ATP binding | Cytoplasm |
| A0A3V6H1D5 | 1 | Pathogenesis | Unknown | Plasma membrane |
| A0A0D6HCE5 | 2 | Lipoprotein biosynthesis | phosphatidylglycerol-prolipoprotein diacylglyceryl transferase activity | Plasma membrane |
| A0A2J0RDS6 | 1 | Host-virus interaction | Unknown | Host membrane |
| A0A3V7XF93 | 1 | Carbohydrate metabolic process | Carbohydrate binding | Extracellular region or secreted |
| A0A3V7X7Z5 | 2 | Host-virus interaction | ATP binding | Cell membrane |
| A0A5Z7LRR5 | 4 | Proteolysis | Metal ion binding | Cell membrane |
| A0A5Z7LRA6 | 1 | Cell cycle | ATP binding | Nucleus |
| A0A0F7J9G5 | 1 | DNA packaging | Metal ion binding | viral terminase complex |
| A0A0F7JGQ1 | 1 | Cell adhesion | Protein-containing complex binding | Extracellular region or secreted |
| A0A0F7JFI2 | 1 | Differentiation | Zinc ion binding | Extracellular region or secreted |
| A0A0F7JDX1 | 2 | Establishment of competence for transformation | Unknown | Plasma membrane |
| A0A5Y2HQY6 | 1 | Amino acid biosynthesis | ATP binding | Cytoplasm |
| A0A0F7J982 | 1 | Protein biosynthesis | GTP binding | Cytoplasm |
| A0A6C8WQJ4 | 3 | inorganic anion transport | Chloride channel activity | Plasma membrane |
| A0A735ZTN8 | 7 | Transmembrane transport | Porin activity | Plasma membrane |
| A0A731QU75 | 2 | Chemical synaptic transmission | G-protein coupled receptor activity [serotonin] | Plasma membrane |
| A0A607WTJ3 | 6 | Phospholipid biosynthetic process | Transferase activity | Plasma membrane |
| A0A705WX69 | 1 | Oxidative phosphorylation | Translocase | Plasma membrane |
| A0A707YZC5 | 1 | Cytochrome complex assembly | heme transmembrane transporter activity | Plasma membrane |
| A0A706T8K4 | 2 | Transmembrane transport | transmembrane transporter activity | Vacuole |
| A0A717VZE3 | 1 | DNA replication | DNA binding | Plasma membrane |
| A0A736JL85 | 2 | Folate biosynthesis | metal ion binding | Inner membrane |
| A0A610AT56 | 3 | Phospholipid biosynthetic process | Transferase activity | Plasma membrane |
| A0A705WZ57 | 1 | Transmembrane transport | Transmembrane transporter activity | Plasma membrane |
| A0A701H8E2 | 4 | Cytochrome c-type biogenesis | Heme binding | Plasma membrane |
| A0A7G2DIQ3 | 2 | Viral tail assembly | Unknown | Host cell cytoplasm |
| Protein | Human | Cattle | Sheep | Goat | Horses |
|---|---|---|---|---|---|
| A0A2J0RKS1 | 0 | 0 | 0 | 0 | 0 |
| A0A1P8DMP4 | 0 | 0 | 0 | 0 | 0 |
| A0A3Z7JEV9 | 0 | 0 | 0 | 0 | 0 |
| A0A2J0RFX8 | 0 | 0 | 0 | 0 | 0 |
| A0A2J0RDC1 | 0 | 0 | 0 | 0 | 0 |
| A0A3T3ZZV4 | 0 | 0 | 0 | 0 | 0 |
| A0A3V6H1D5 | 0 | 0 | 0 | 0 | 0 |
| A0A0D6HCE5 | 0 | 0 | 0 | 0 | 0 |
| A0A2J0RDS6 | 0 | 0 | 0 | 0 | 0 |
| A0A3V7XF93 | 0 | 0 | 0 | 0 | 0 |
| A0A3V7X7Z5 | 0 | 0 | 0 | 0 | 0 |
| A0A5Z7LRR5 | 0 | 0 | 0 | 0 | 0 |
| A0A5Z7LRA6 | 0 | 0 | 0 | 0 | 0 |
| A0A0F7J9G5 | 0 | 0 | 0 | 0 | 0 |
| A0A0F7JGQ1 | 0 | 0 | 0 | 0 | 0 |
| A0A0F7JFI2 | 0 | 0 | 0 | 0 | 0 |
| A0A0F7JDX1 | 0 | 0 | 1 | 0 | 0 |
| A0A5Y2HQY6 | 0 | 0 | 0 | 0 | 0 |
| A0A0F7J982 | 0 | 0 | 0 | 0 | 0 |
| A0A6C8WQJ4 | 0 | 0 | 0 | 0 | 0 |
| A0A735ZTN8 | 0 | 0 | 0 | 0 | 0 |
| A0A731QU75 | 0 | 0 | 0 | 0 | 0 |
| A0A607WTJ3 | 0 | 0 | 0 | 0 | 0 |
| A0A705WX69 | 0 | 0 | 0 | 0 | 0 |
| A0A707YZC5 | 0 | 0 | 0 | 0 | 0 |
| A0A706T8K4 | 0 | 0 | 0 | 0 | 0 |
| A0A717VZE3 | 1 | 0 | 0 | 0 | 0 |
| A0A736JL85 | 0 | 0 | 0 | 0 | 0 |
| A0A610AT56 * | 0 | 0 | 0 | 0 | 0 |
| A0A705WZ57 | 0 | 0 | 0 | 0 | 0 |
| A0A701H8E2 | 0 | 0 | 0 | 0 | 0 |
| A0A7G2DIQ3 | 0 | 0 | 0 | 0 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




