1. Introduction
Diffuse glioma is thought to derive from glial stem cells, and is the most common primary brain tumor. Based on their histological characteristics, gliomas are classified as astrocytes, oligodendrocytes, or ependymal tumors, and their malignancy is designated as I to IV by the World Health Organization (WHO) [
1]. Recent progress in the genetic analysis of brain tumors has demonstrated that they differ greatly with respect to morphology, location, genetic characteristics, and response to treatment. Histopathology is the gold standard for glioma grading, but histological classification and grading of diffuse glioma present many challenges for pathologists, and reliance on traditional histology to grade gliomas carries some disadvantages. First, these tumors differ greatly between observers and within observers [
2]. Because the diagnostic criteria are not accurate, this variability leads to poor consistency and repeatability of tumor grading. Moreover, diffuse gliomas often show obvious phenotypic heterogeneity and present spatial differences in cell phenotype and anaplastic degree, which leads to uneven distribution of mitotic images. This tendency leads to a high rate of variation among observers in the diagnosis of diffuse gliomas, including oligodendrocytoma (ODG) [
3]. Because of these factors, it is not possible to accurately identify the biological specificity of tumors.
In recent years, many studies have submitted pathological specimens of brain gliomas to computer algorithms for machine-aided diagnosis, such as deep convolutional neural networks. This technology has been used to detect tumor cells, classify tumor subtypes, and diagnose diseases [
4,
5]. The motivation for adopting this technology is to provide rapid, reproducible, and quantitative diagnosis [
6]. Zhou et al. established a convenient and non-invasive method for preoperative grading of glioma [
7]; Their method, which relies on MRI proteomics technology combined with enhanced T1WI, substantially improved the diagnostic accuracy. Zhang et al. used machine learning to explore the potential of MRI imaging features for distinguishing anaplastic oligodendroglioma (AO) from atypical low-grade oligodendroglioma, and established a prediction model based on T1C and FLAIR image features [
8]; Gao et al. introduced a non-invasive method for preoperative prediction of glioma grade and expression levels of various pathological biomarkers, achieving good prediction accuracy and stability [
9]. A large number of training datasets is often needed to achieve effective classification recognition, and detection efficiency is relatively low. The new (2016) WHO classification criteria for central nervous system tumors marked the end of traditional diagnostic methods solely based on histological standards and molecular biomarkers that combined morphological and molecular features from diffuse glioma [
10]. Therefore, there is an urgent need for an objective, stable, and reproducible method to accurately grade diffuse gliomas that faithfully reflects the essential attributes of tumors, thus effectively guiding clinical surgery and prognosis treatment.
Micro-hyperspectral images carry important information about biological tissues, and can be used to analyze their biochemical properties. Previous research has achieved automated detection of various cancers, including visualization enhancement of blood vessels, by combining hyperspectral imaging with deep learning [
11,
12].This research has made it possible to identify intestinal ischemia [
13] , measure the oxygen saturation of the retina [
14] , estimate the human cholesterol level [
15] and cancer testing [
16,
17,
18,
19]. Hyperspectral images carry many advantages compared with traditional medical images. Xuehu Wang [
20] An automatic detection method for predicting hyperspectral characteristics based on radiological characteristics of human computed tomography (CT) is proposed for the non-invasive detection of early liver tumors. Through image enhancement and XGBoost model construction, the mapping relationship between CT characteristics and corresponding hyperspectral imaging (HSI) data is learned, and good classification and recognition results are achieved. Wang [
21] has proposed a feature fusion network of RGB and hyperspectral dual-mode images (DuFF-Net), in which features were mined using SE-based attention modules and Pearson correlation. The results showed that DuFF-Net improved the screening accuracy of two morphologically similar gastric precancerous tissues, reaching accuracy values up to 96.15%. Jian et al[
22] introduced a spectral space transfer convolutional neural network (SST-CNN) to address the issue of limited sample size of medical hyperspectral data. This method achieved classification accuracy values of 95.46% for gastric cancer and 95.89% for thyroid cancer.
In light of the above results, we designed a multi-modal and multi-scale hyperspectral image feature extraction model (SMLME-Resnet) based on microscopic hyperspectral images of brain gliomas. In our model, the channel attention mechanism (SE module) is introduced before the traditional residual connection module, to explore interactions among spectral channels in the three-dimensional data cube of hyperspectral images of brain glioma from pathological slices. This design was chosen to realize the fusion of features between spectral channels and therefore improve the feature extraction ability of the network, while effectively reducing redundant information. Building on this architecture, we introduced the parallel multi-scale image feature method to further extract features of hyperspectral images from the three-dimensional data cube. We also improved the convolutional layer in the residual connection by introducing multi-branch modules with different image sizes. This design characteristic not only achieves deep data feature mining, but also realizes a lightweight model. Our model delivered accurate and efficient identification of diffuse glioma from pathological sections, in conformity with the classification criteria introduced by the WHO in 2016.
2. Materials and Methods
2.1. Dataset Structure
All sections were digitized and magnified using an Olybas 40× objective lens. They were manually annotated by a pathologist with professional certification. We used cancer and normal regions as marked on digital histological images as ground truth for data selection and classification. For each slide, we selected at least three ROIs within the cancer region, which were marked by pathologists specializing in relevant research. In
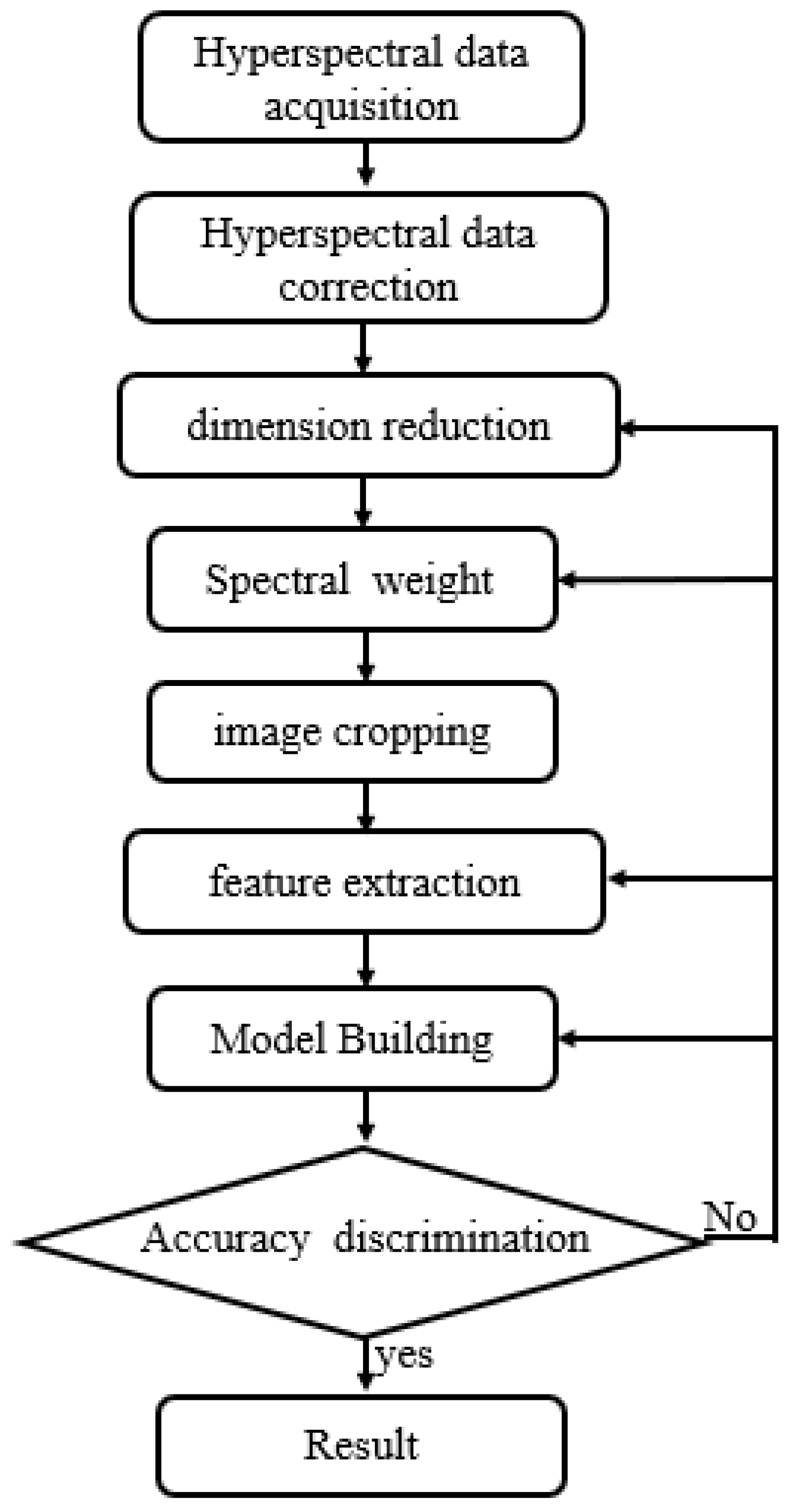
Figure 1, We collaborated with staff at the Third Hospital of Jilin University to complete formalin fixation 1, HE staining, IHC staining, and sample labeling of brain gliomas. We had access to 196 cases of brain gliomas spanning different sexes and ages (14 cases of grade 1, 64 cases of grade 2, 46 cases of grade 3, and 72 cases of grade 4), together with 22 normal brain tissues.The processing process is shown in
Figure 2.
2.2. Micro-Hyperspectral Data Collection
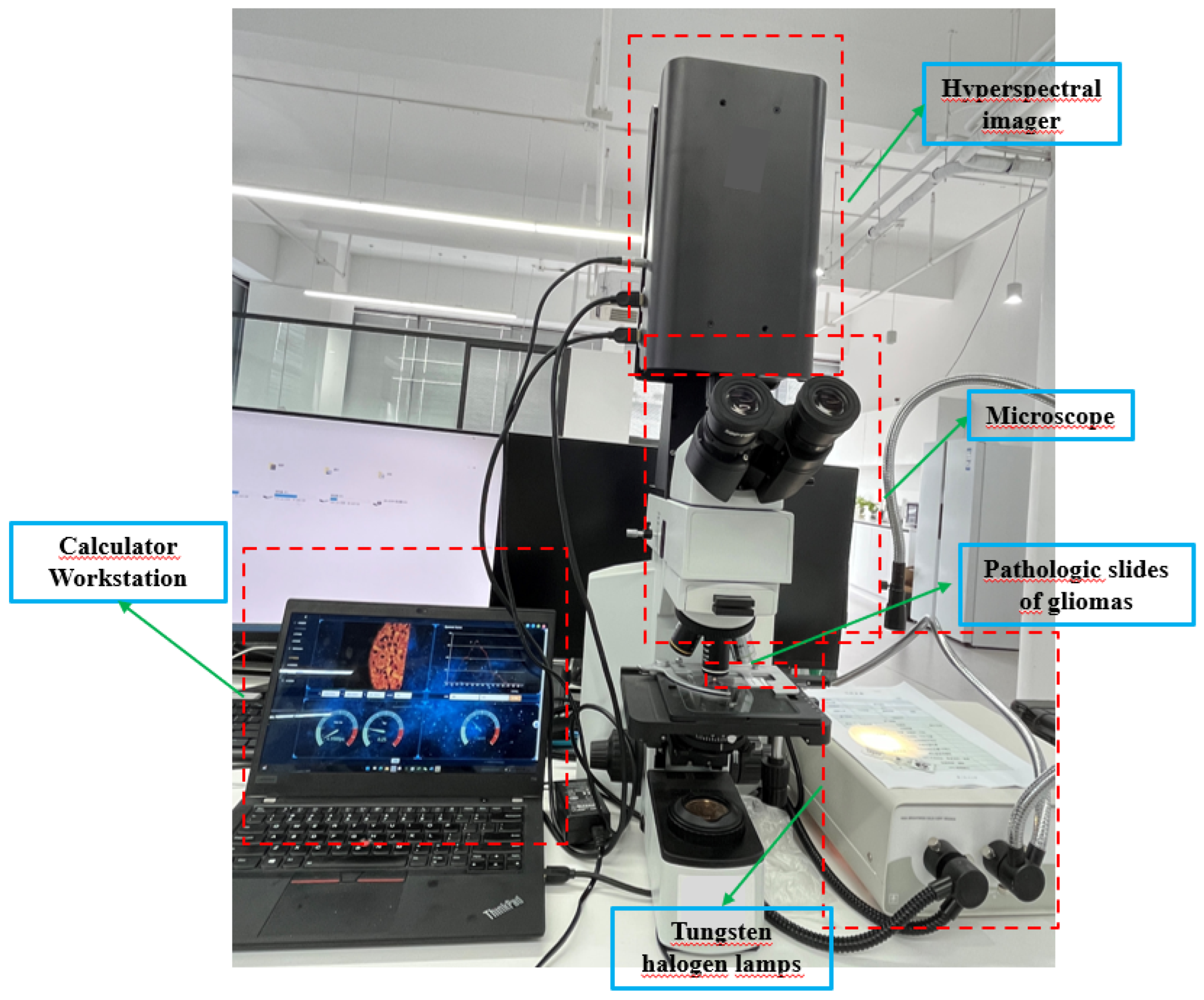
We obtained HSIs from slides of brain glioma using the micro hyperspectral imaging system integrated by Hangzhou Hyperspectral Imaging Technology Co., Ltd. in China. The hyperspectral imaging module was independently developed by the research group of Changchun Institute of Optics, Fine Mechanics and Physics (CIOMP), Chinese Academy of Sciences , as shown in
Figure 3. The acquisition system consists of an imaging system, an illumination system, and a computer. In the spectral imaging system, the objective lens is 40 times that of Olympus, and the detector is a CMOS two-dimensional area array camera of imperx. Target size is 1920×1080 pixels, spectral range of the system covers 400–1000 nm, number of spectral channels is 270, and spectral resolution is >2.5 nm. After push-scan imaging, the system can capture a three-dimensional data cube of the specimen. The lighting system consists of a cold light source of optical fiber halogen tungsten lamp with power of 150W, and the spectral range of continuous spectrum light source is 400–2200 nm.
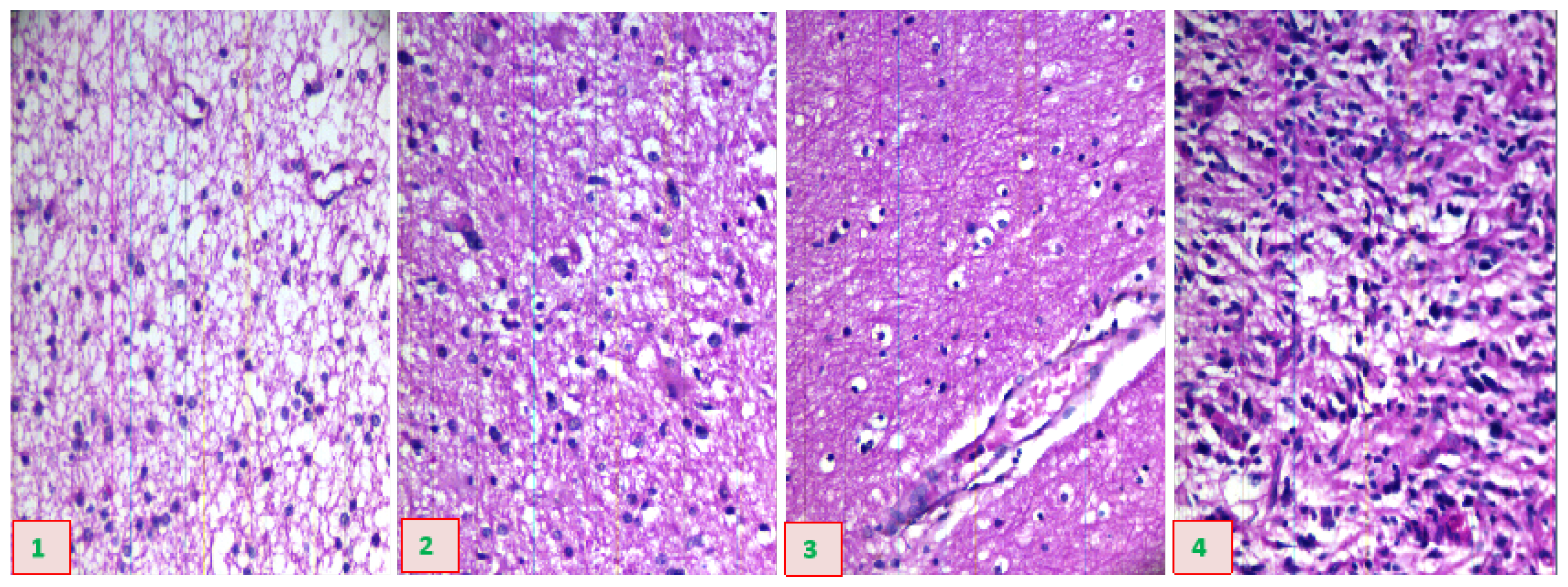
Figure 4 provides a schematic diagram of hyperspectral data images from glioma pathological sections of different grades, collected using the system just described.
2.3. Spectral Data Preprocessing
We used hyperspectral images of brain glioma tissue array chip to create the database used in this study. In order to reduce system noise, the radiation intensity difference of the pixel response, and the influence of light source instability, while at the same time preserving spectrum information as much as possible, we correct system transmittance based on Lambert-Beer law [
23]. We implemented normalization and SG filtering as detailed in Formula (
1), where
T is the transmittance data after spectral correction, representing both the original sample data and the blank slide data under the same experimental conditions.
represents the radiant intensity transmitted by the blank slide, and
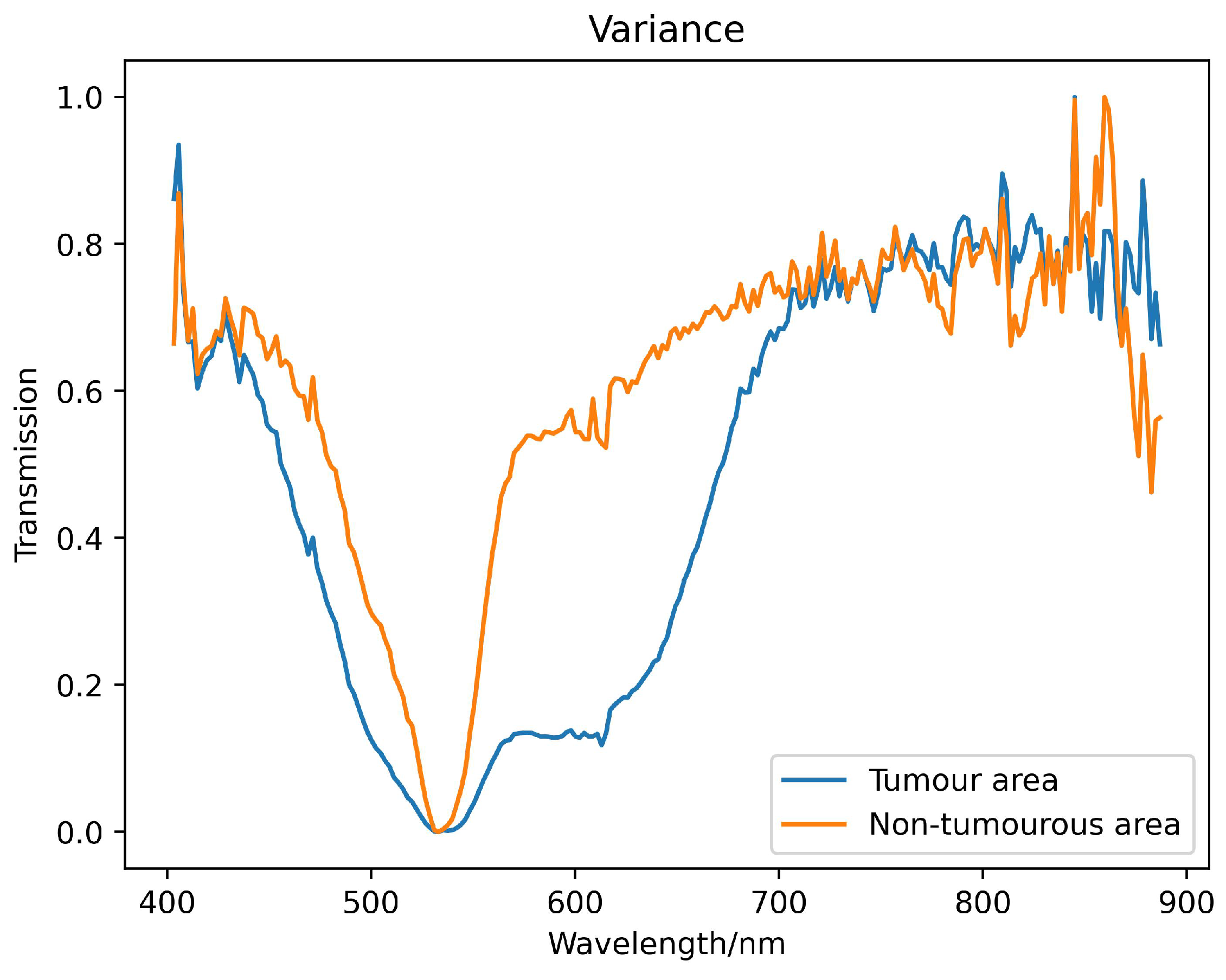
represents the radiant intensity transmitted by the glioma slides. This process ensures data consistency and repeatability of algorithm results. The spectral transmittance curve obtained after preprocessing is shown in
Figure 5.
2.4. Experimental Setup
Our hardware consisted of a GPU (NVIDIA Geforce RTX208Ti) . We used Windows 11 as the operating system, python 3.7 as the programming language, and Pytorch as the deep-learning framework. We adopted the following parameters: parametric reLU activation function, batch size of 32, epoch equal to 100, and and the ratio of training set, validation set and test set is set to 4:1:5. The study utilized Adam as the optimizer. Additionally, due to the intricacy of the medical sample production process and the requirement for professional doctors to annotate the images, there were limited samples available for training. Therefore, regularization was implemented to decrease the network’s complexity, prevent overfitting, and enhance the model’s generalization ability. To improve classification accuracy and avoid overfitting, we utilize a layer-specific regularization and normalization classification mechanism. This mechanism includes a fully connected layer, a sigmoid activation function, and an L2 regularization function. We evaluated classification performance using overall accuracy (OA), average accuracy (AA), and Kappa coefficient. OA represents the ratio between the total number of correctly classified samples and all tested samples. AA represents the average of classification accuracy across all categories. Kappa coefficient is used to evaluate classification consistency across all categories. To eliminate the influence of random factors, all experimental results in this study are obtained from an average of 10 experiments.
2.5. Data Dimensionality Reduction
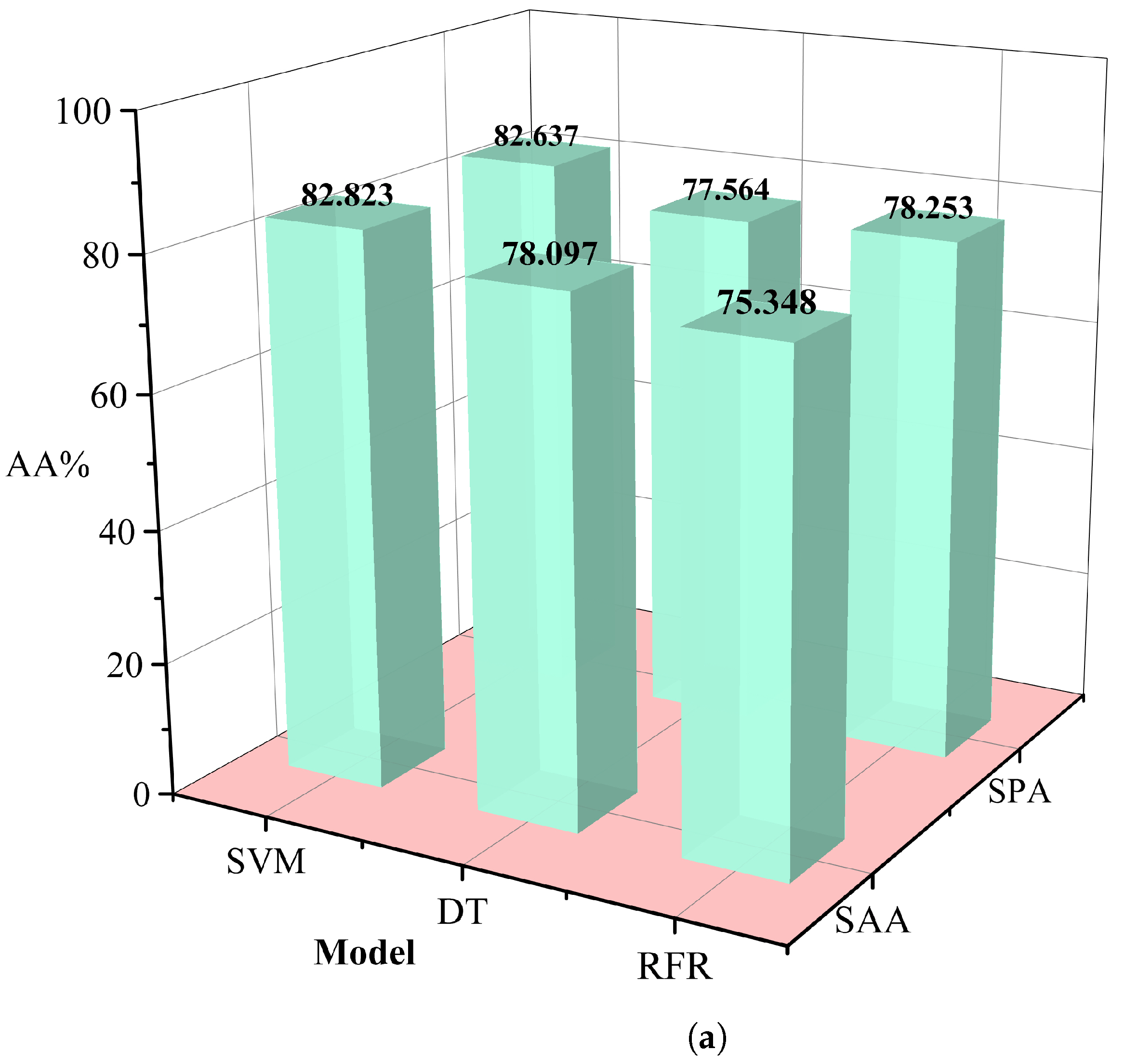
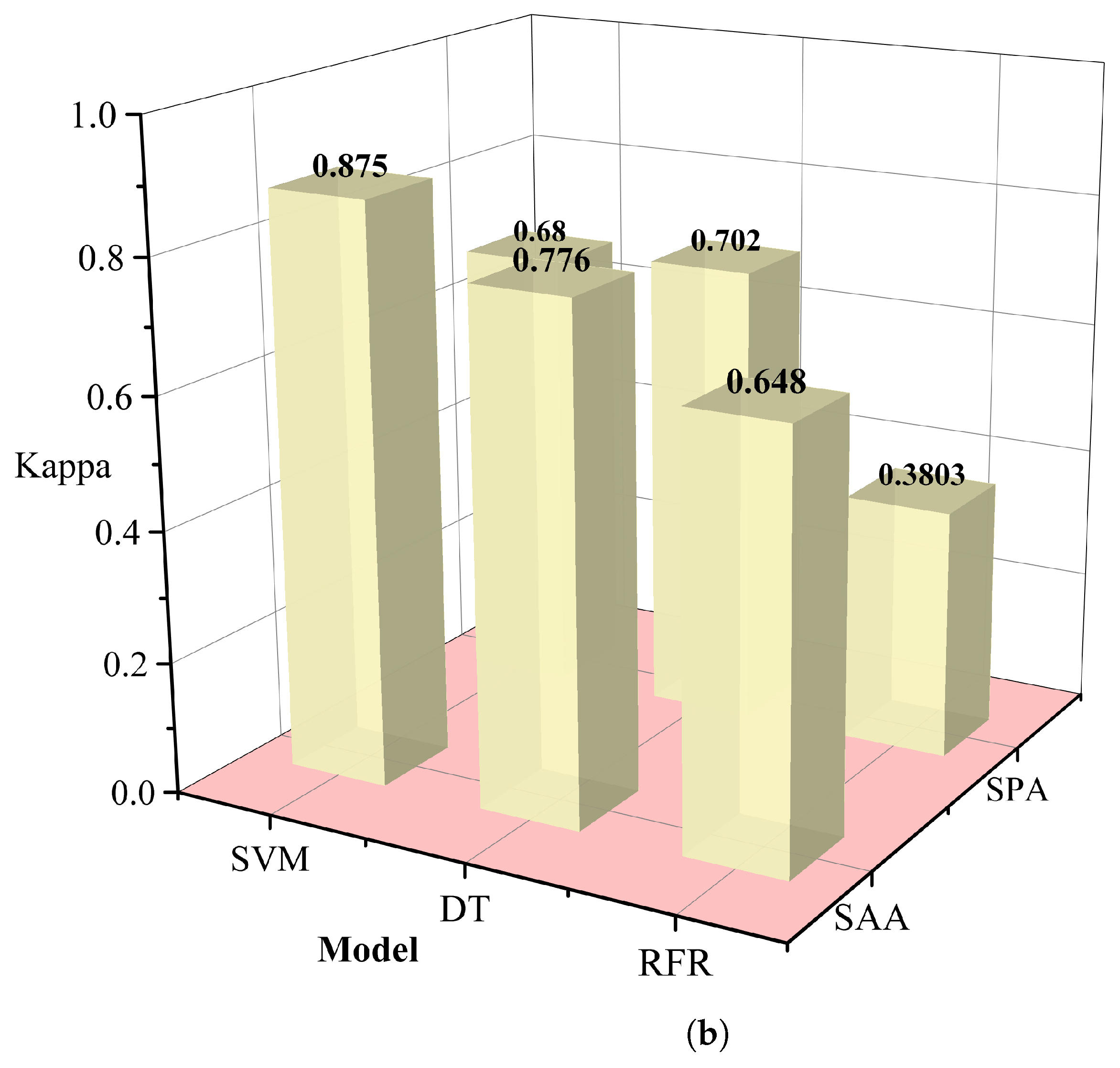
The redundancy associated with hyperspectral data may lead to a decrease in the classification efficiency of malignant grades. In this study, we used the simulated annealing algorithm and principal component analysis to select the appropriate wavelength. In this procedure, the annealing algorithm is simulated by defining an appropriate energy function, that is an objective function. The new wavelength subset can be accepted or rejected according to the probability of energy change, and the most suitable wavelength subset can be gradually optimized to achieve the purpose of wavelength dimensionality reduction. Successive projections algorithm(SPA)[
24] utilizes projection analysis of vectors by projecting wavelengths onto other wavelengths, comparing the magnitude of the projection vectors, taking the wavelength with the largest projection vector as the wavelength to be selected, and then selecting the final feature wavelength based on the correction model. SPA selects the combination of variables that contains the least amount of redundancy information and the least amount of covariance. We used several different machine learning methods to control variables, including support vector machines (SVM) [
25], decision tree classifier (RT) [
26], random forest (RFR) [
27], and selected the data dimensionality reduction method that was most suitable for this study. Relevant results are shown in
Table 1. The simulated annealing algorithm is clearly superior to the PCA algorithm for different classification models. Based on these results, we selected the simulated annealing algorithm as a dimensionality reduction algorithm for hyperspectral data in
Figure 6.
2.6. Identify Model
Based on the results from the previous section, we performed partial Decision Tree(DT), SVM classification, RFR, one-dimensional convolutional neural network (CNN) and MSE-ResNet [
28]. We used the improved algorithm to establish the classification model for grading pathological sections of glioma with different malignant grades, which we term SMLME-Resnet. We used support vector classification (SVC) to establish models for classifying pathological sections of gliomas with different malignant grades. SVC is a typical nonlinear classification method, which maps low-dimensional data to high-dimensional space and realizes the classification of samples by finding the optimal hyperplane. In SVC, the appropriate choice of the penalty coefficient
C and kernel function
G ensures strong generalization ability [
29]. We used a genetic algorithm to optimize the penalty coefficient
C and kernel function parameter
G. Decision Tree ( Decision Tree) is a predictive analytics model expressed in the form of a tree structure (including binary and multinomial trees). Each non-leaf node represents a test on a feature attribute, each branch represents the output of this feature attribute on a certain value domain, and each leaf node stores a category. The random forest algorithm consists of several decision trees, with no correlation between decision trees in the forest. The final output of the model is determined by all decision trees, which carries strong anti-interference ability and anti-over-fitting ability. Jian [
30] used spectral information from microscopic hyperspectral images to classify gastric cancer tissue via 1D-CNN. The network model included one input layer, two convolution layers, two pooling layers, one fully connected layer, and one output layer. Chongxuan et al. [
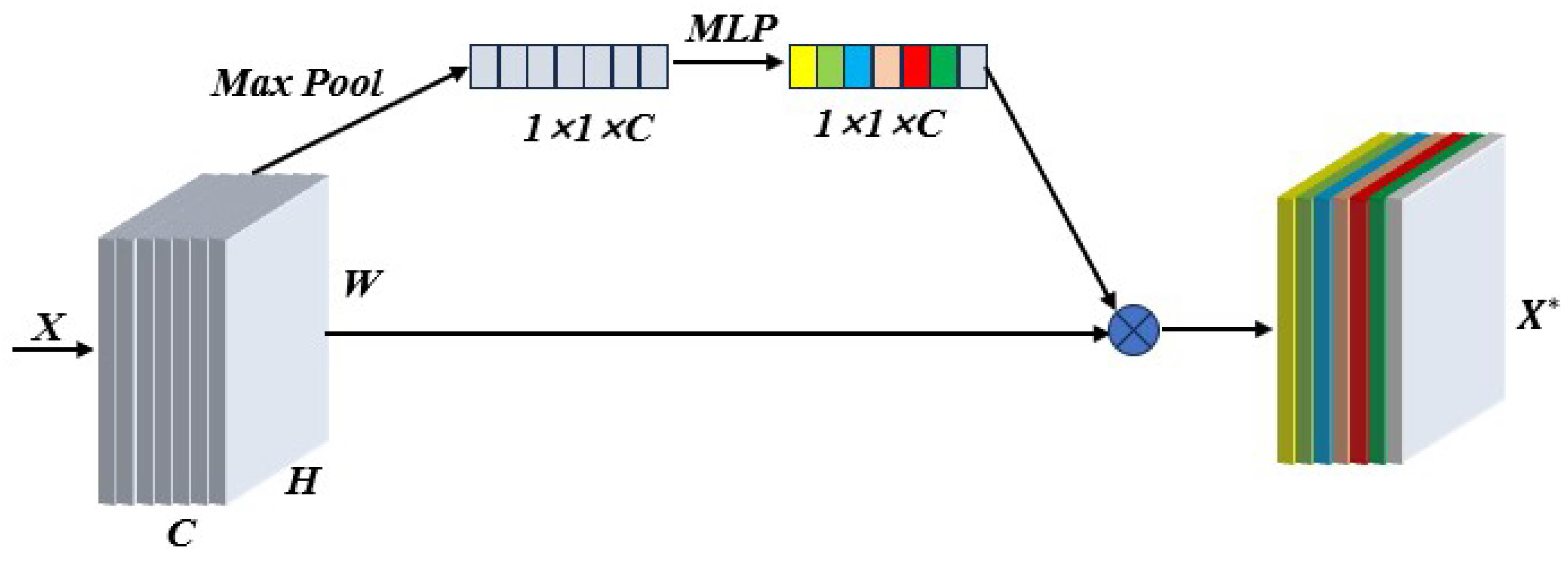
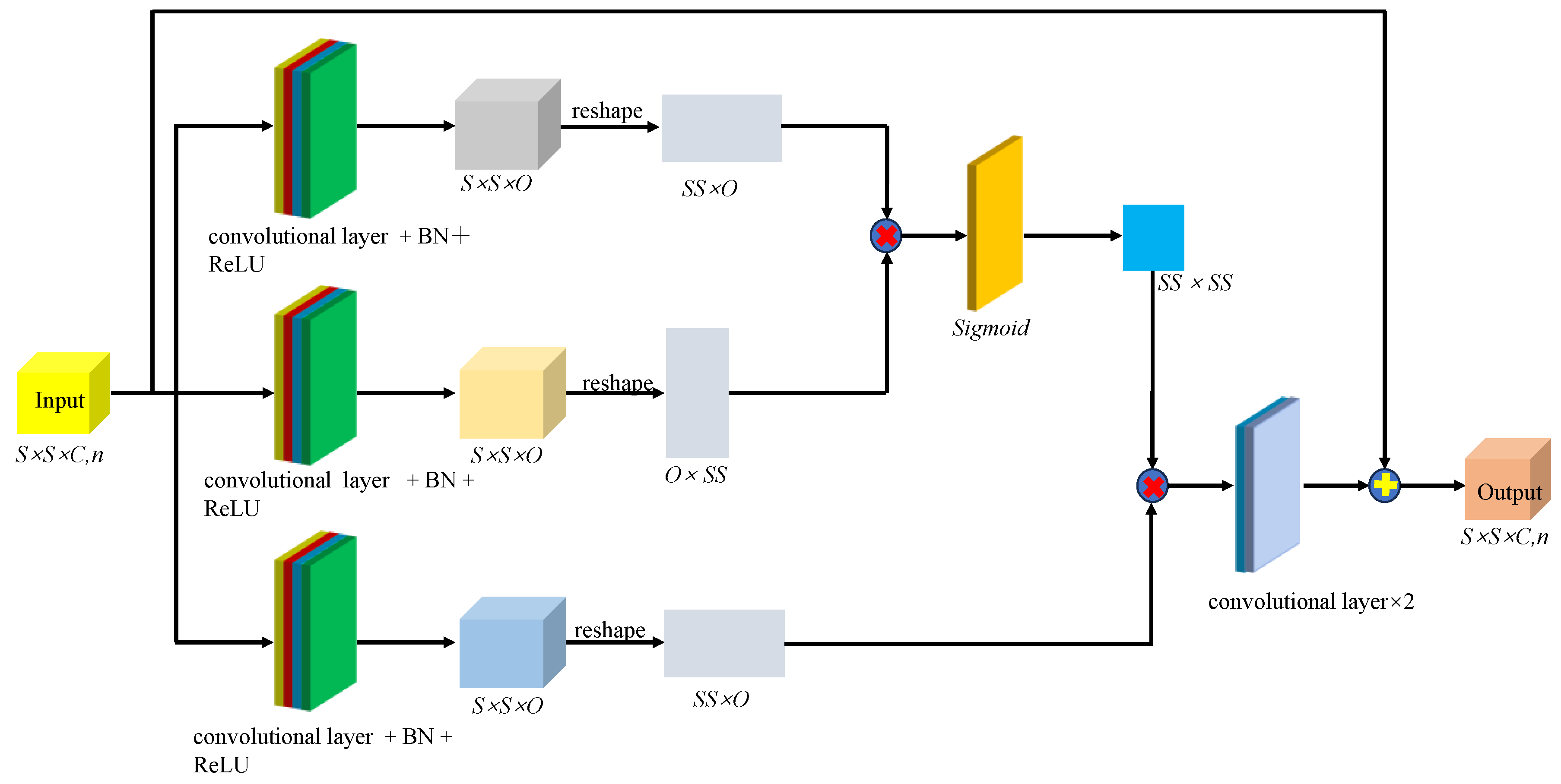
31] relied on hyperspectral images of gastric juice to classify Helicobacter pylori infection and used a ResNet model to learn from two-dimensional data, achieving 91.48% classification accuracy. We improved SMLME-Resnet by introducing a SE module after the pretreatment process, such as PCA, and used it to learn the relationship between channels by assigning corresponding weights to each channel, so as to realize the effective fusion of features between channels.
Figure 7 shows the adopted implementation process, which can effectively reduce redundant information and improve the feature extraction ability of the network.
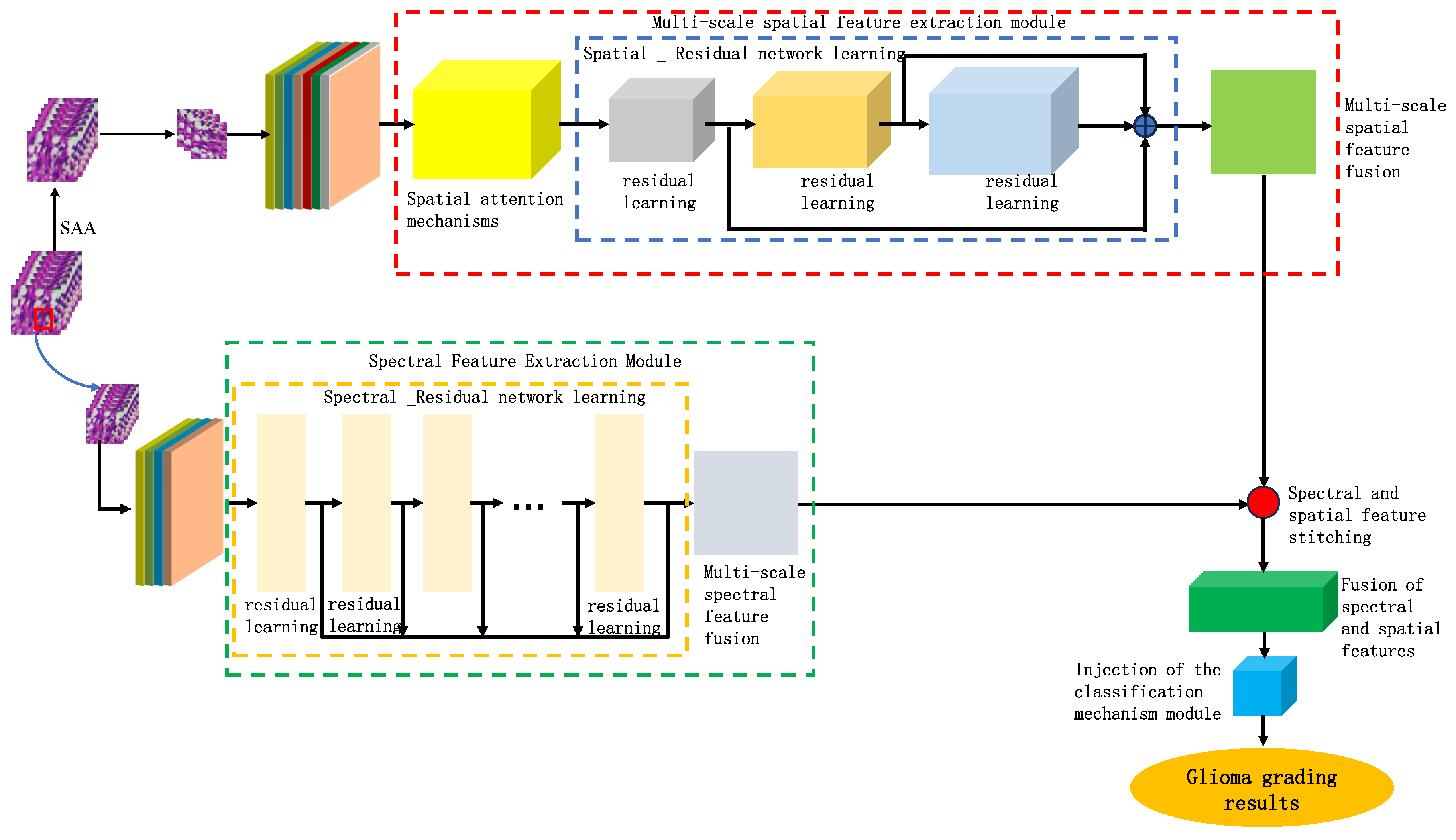
Based on the findings of this study, we proposed a model for identifying the malignancy level of glioma pathological tissues by integrating the basic framework of SENet and ResNet residual network models. This model, named SMLMERNet (Neural Network Modeling of Multimodal Multiscale Hierarchical Spectral-Spatial Fusion Features), combines spectral-spatial multiscale features. The overall network architecture comprises four main modules: spectral feature extraction, spatial feature extraction, higher-order spatial-spectral fusion features, and a classifier module.
Figure 8 shows the overall framework of the designed model, while
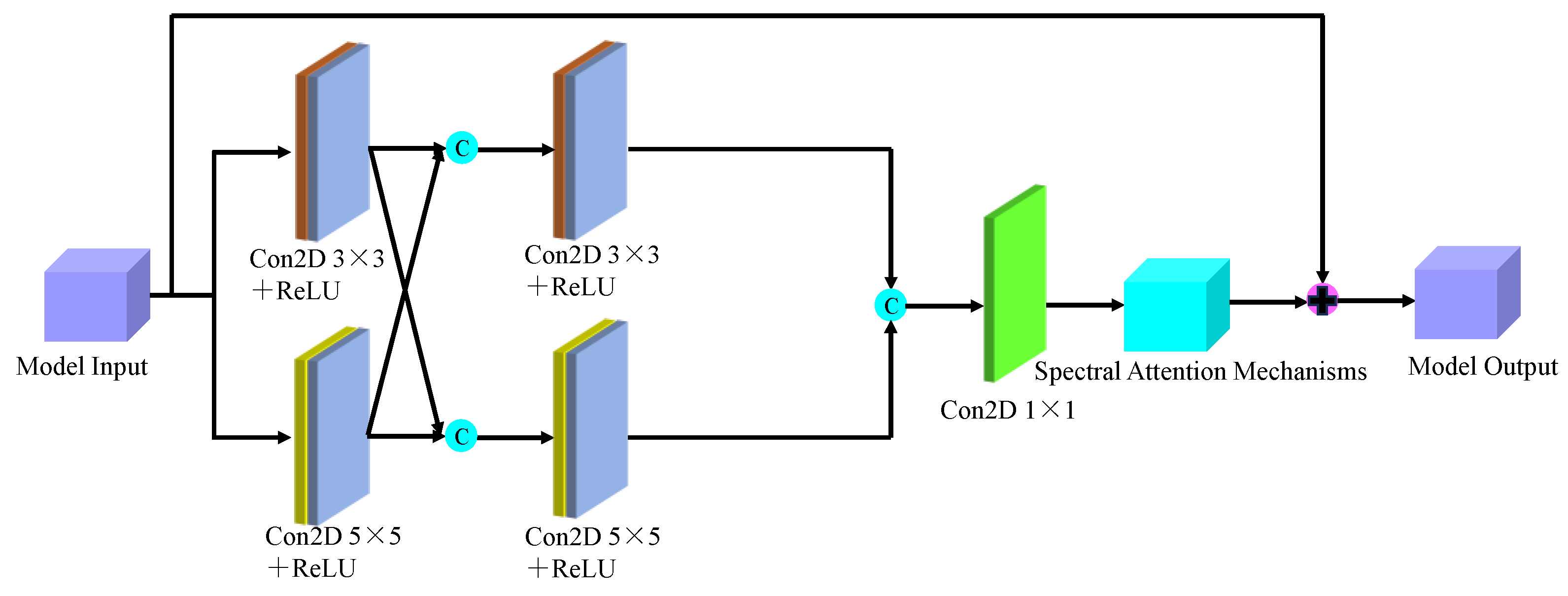
Figure 9 depicts the multi-scale spectral feature extraction framework.In this study, we inputted an image block of size 7×7×200 into the spectral feature extraction module. Initially, an initialization process was conducted, consisting of a convolutional layer with 64 filters of size 3×3, a BN layer, and a ReLU activation function. Following the initialization module, the size of the original 3D data cube was changed to 7×7×64. Subsequently, the downscaled transformed data cube was further inputted into the multiscale spectral feature extraction module to capture multiscale spectral features and eliminate redundant information interference. By integrating six combined SE modules with the residual learning network, we achieved multi-scale spectral feature extraction, resulting in six local spectral features of size 7×7×64. Finally, global spectral features were obtained through spectral feature fusion. The multiscale spectral feature fusion module employed a combined processing strategy involving cascade operation and bottleneck layer, with the bottleneck layer consisting of 64 2D convolution kernels of size 1×1. Consequently, a multiscale spectral feature matrix of dimensions 7×7×64 was obtained on the spectral feature extraction branch.
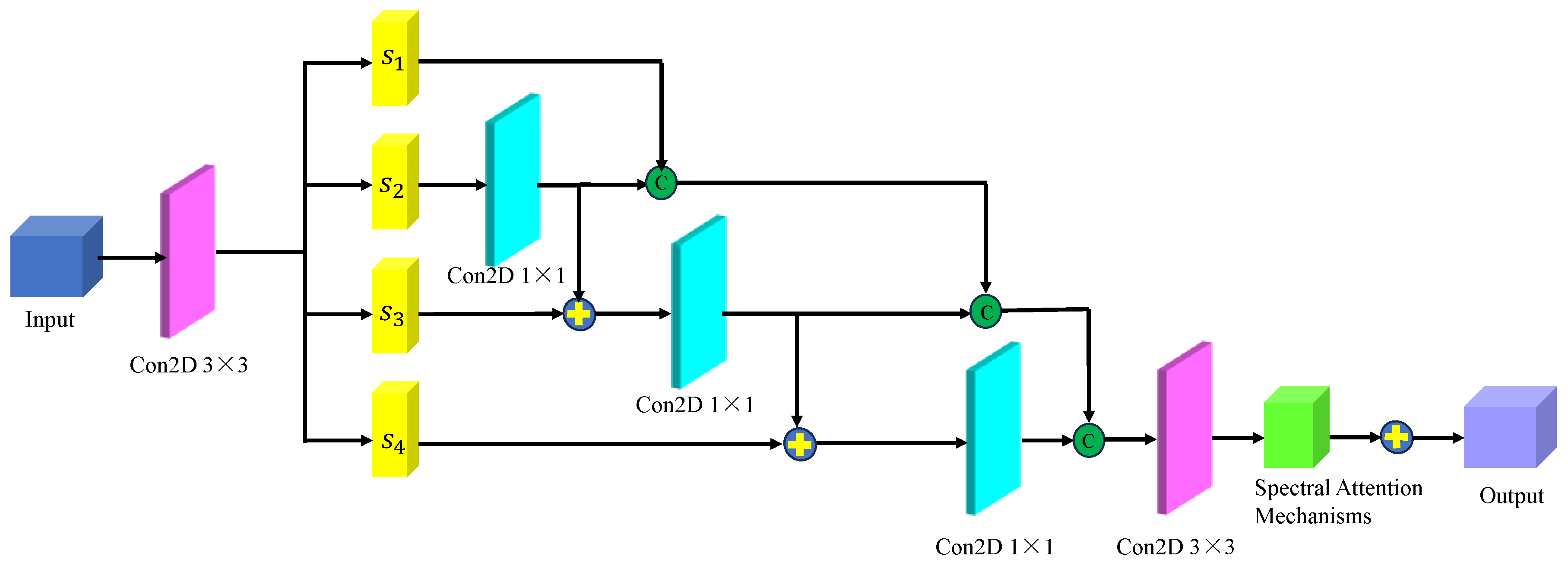
Figure 10 shows the multi-scale hierarchical spatial feature extraction module. In this module, the original micro-hyperspectral data of glioma pathology were initially downscaled using PCA, and image blocks of size 27×27×30 were inputted. Similar to the spectral feature extraction process, an initialization process was performed, comprising a 3D convolutional layer, a BN layer, and an activation function containing 16 filters of size 5×5×30 and 16 filters of size 3×3×1, with ReLU as the activation function. After the initialization module, the original input data was transformed into a four-dimensional vector, and the size of the dimensions 23×23×1×16 was calculated. The transformed data was then inputted into the spatial feature extraction module with a spatial attention mechanism to achieve multilevel spatial feature extraction. The multi-scale spatial feature extraction module included a spatial attention mechanism and a multi-stage spatial feature fusion module. After multiscale spatial feature extraction, the spatial feature size was changed to 23×23×16×16, followed by spatial feature fusion. To be spliced with spectral features from another branch, the spatial features were transformed into 7×7×8 of multilevel spatial features.
Figure 11 shows the spatial-spectral fusion feature extraction module. This module effectively captured the close connection between spatial and spectral features, integrating multiscale spectral features, multilevel spatial features, and higher-order fusion features for further refinement and extraction. After processing, more comprehensive and integrated multiscale features were obtained for analyzing and identifying the malignancy level of glioma pathology sections. Finally, to prevent overfitting of the model after deep-level feature extraction, we applied layer-specific regularization and normalization to the classifier, resulting in adaptive adjustment of the weights of the fused features. Given that the glioma pathology slice dataset contained 5 categories, the model outputs a one-dimensional vector of dimension 5.
3. Results
3.1. Model Parameter Analysis
3.1.1. Image Block Size Analysis
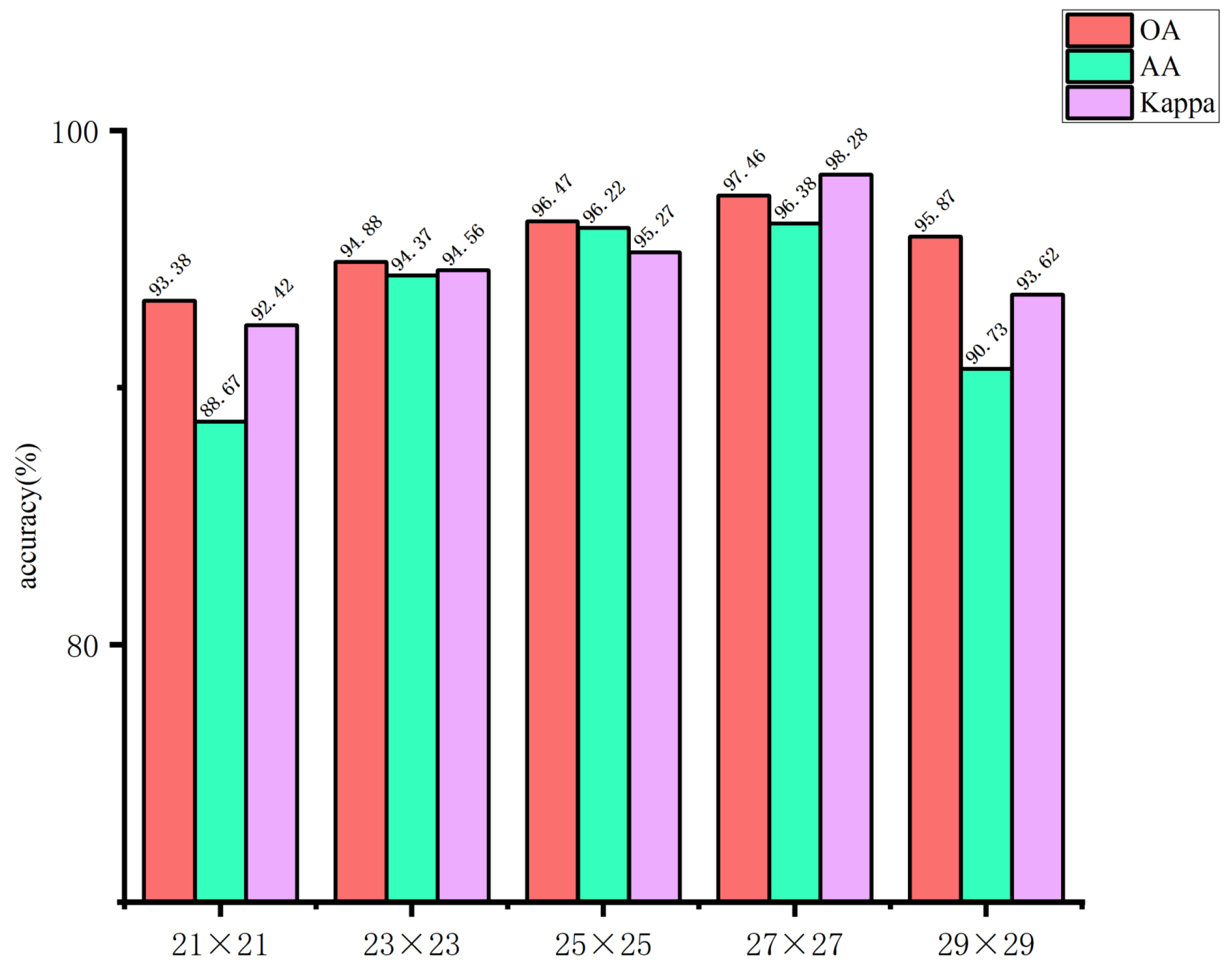
The use of larger image blocks can facilitate the capture of more complex features by the network. However, if the image block is too large, it may introduce irrelevant information, which could affect the extraction of spatial features for small targets, thereby hindering the network’s ability to learn features useful for classification. Conversely, if the image block size is too small, it may not be possible to extract sufficient spatial information, which could result in a decline in the overall classification accuracy of the model. Accordingly, in this study, the input image block sizes of the spectral feature extraction module were set to 5×5, 7×7, 9×9, 11×11, and 13×13, respectively. The input image block sizes of the spatial feature extraction module are set as 21×21, 23×23, 25×25, 27×27, and 29×29. The impact of the spatial dimensions of the model input image blocks on the classification outcomes of the glioma pathology sample dataset is illustrated in
Figure 12 and
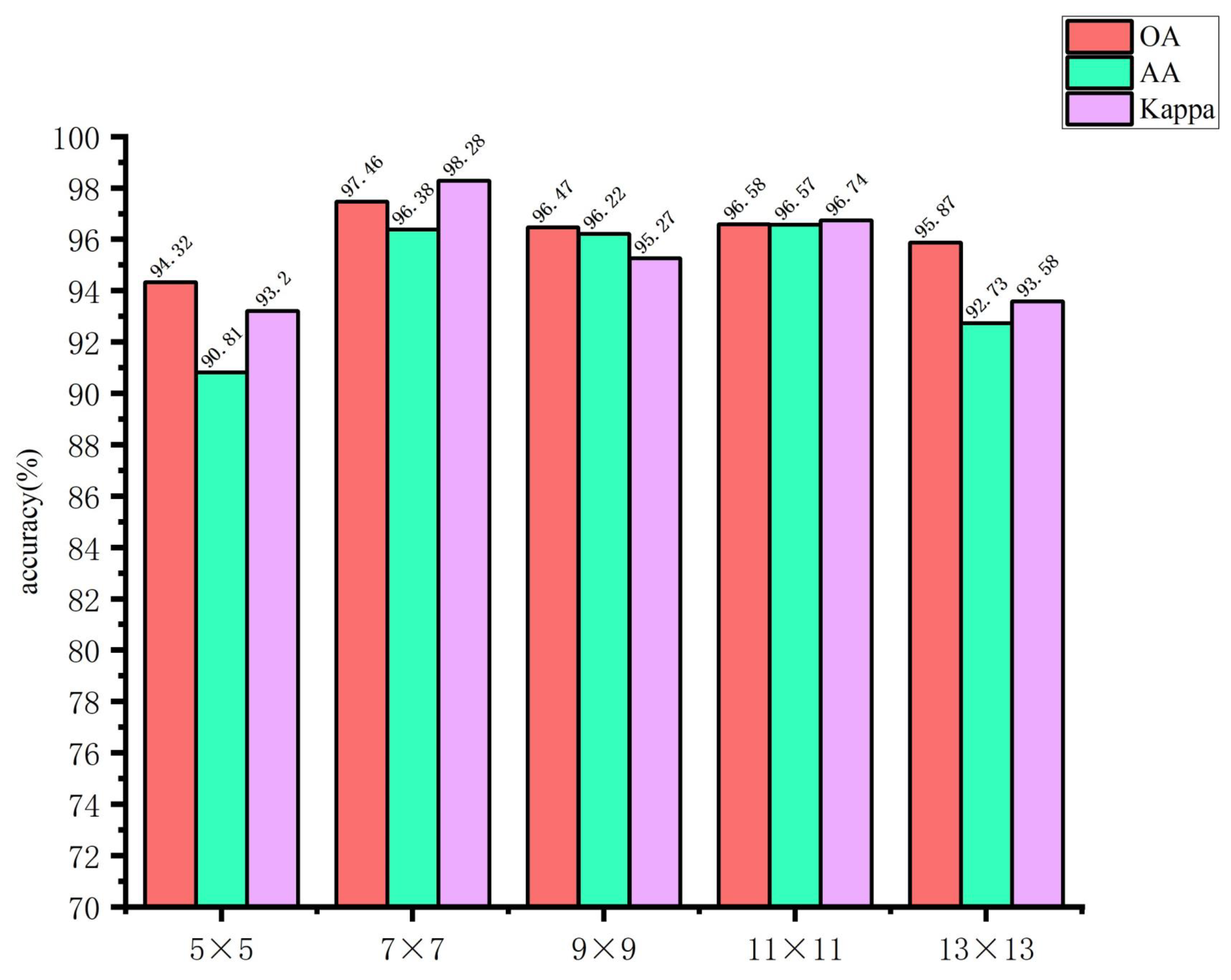
Figure 13. These figures demonstrate that when the spectral feature extraction module inputs an image block with a size of 7 × 7, while the spatial feature extraction module inputs an image block with a size of 27 × 27, the SMLMERNet model proposed in this study exhibits superior performance metrics on the glioma pathology sample dataset. The input image block size for the spatial feature extraction module is 27 × 27, while the input image block size for the spectral feature extraction module is 7 × 7. The SMLMERNet model proposed in this study exhibits superior performance metrics on the glioma pathology sample dataset. Consequently, the optimal input image block sizes for the SMLMERNet network model were determined to be 7 × 7 (the input image block size for the spectral feature extraction module) and 27 × 27 (the input image block size for the spatial feature extraction module), respectively. This was done by considering the classification accuracy and the amount of network computation.
3.1.2. Model Learning Rate Analysis
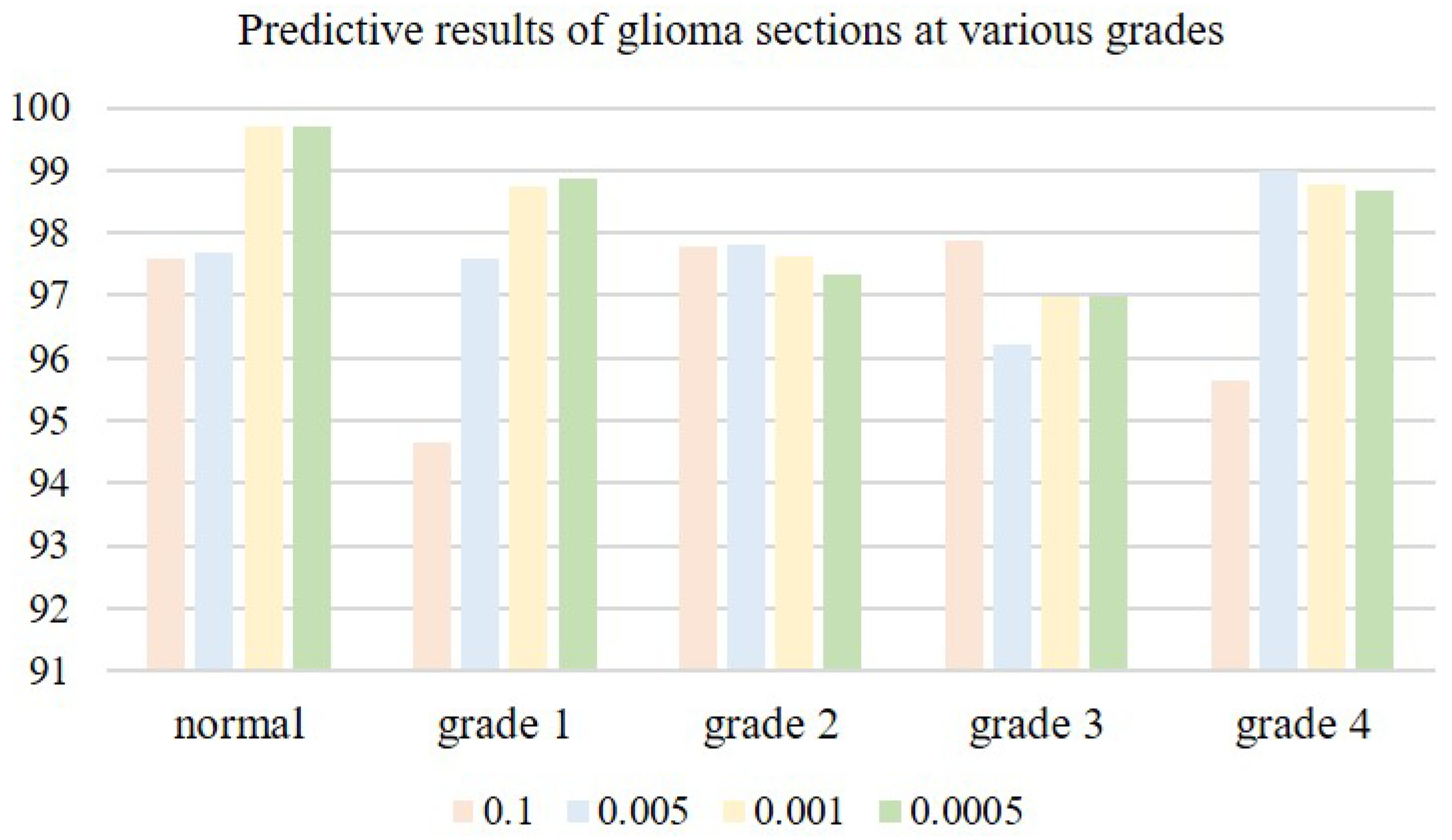
The choice of an appropriate learning rate can help the network model deliver better training results. When the learning rate is very small, the convergence speed is slow, and overfitting can easily occur; when the learning rate is high, the gradient may oscillate back and forth around the minimum value, and may even fail to converge. We tested four learning rates on microscopic hyperspectral data from pathological sections of brain glioma: 0.01, 0.005, 0.001, and 0.0005. Classification results associated with different learning rates are shown in
Table 2. As the learning rate is decreased, classification accuracy gradually improves. When the learning rate is too small, the convergence speed of the model slows down, and classification accuracy decreases. When the learning rate is 0.001, the model reaches its peak classification accuracyin Figure, as is shown in
Figure 14. We therefore settled on a learning rate of 0.001.
3.2. Comparison between the Improved Model and the Traditional Model
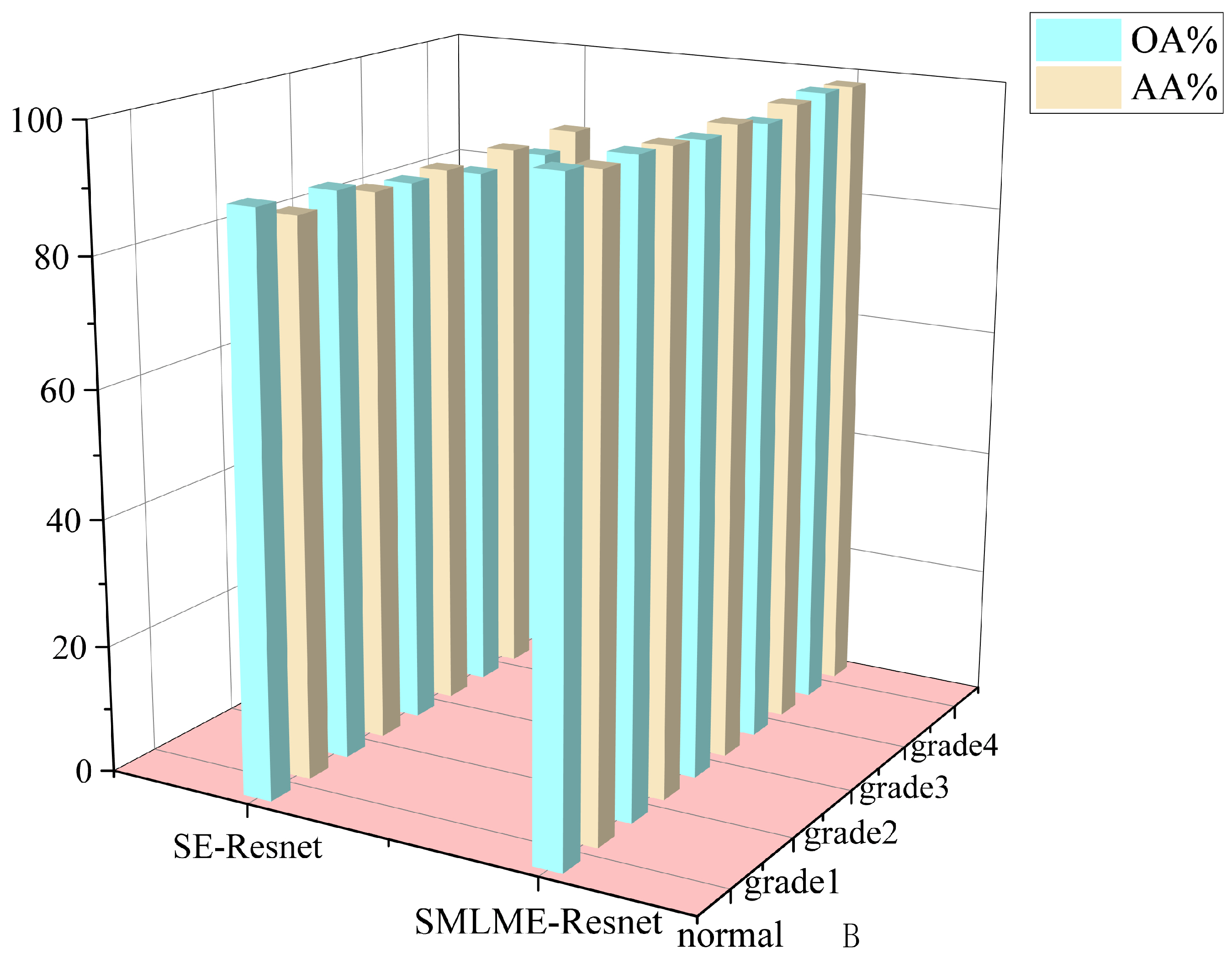
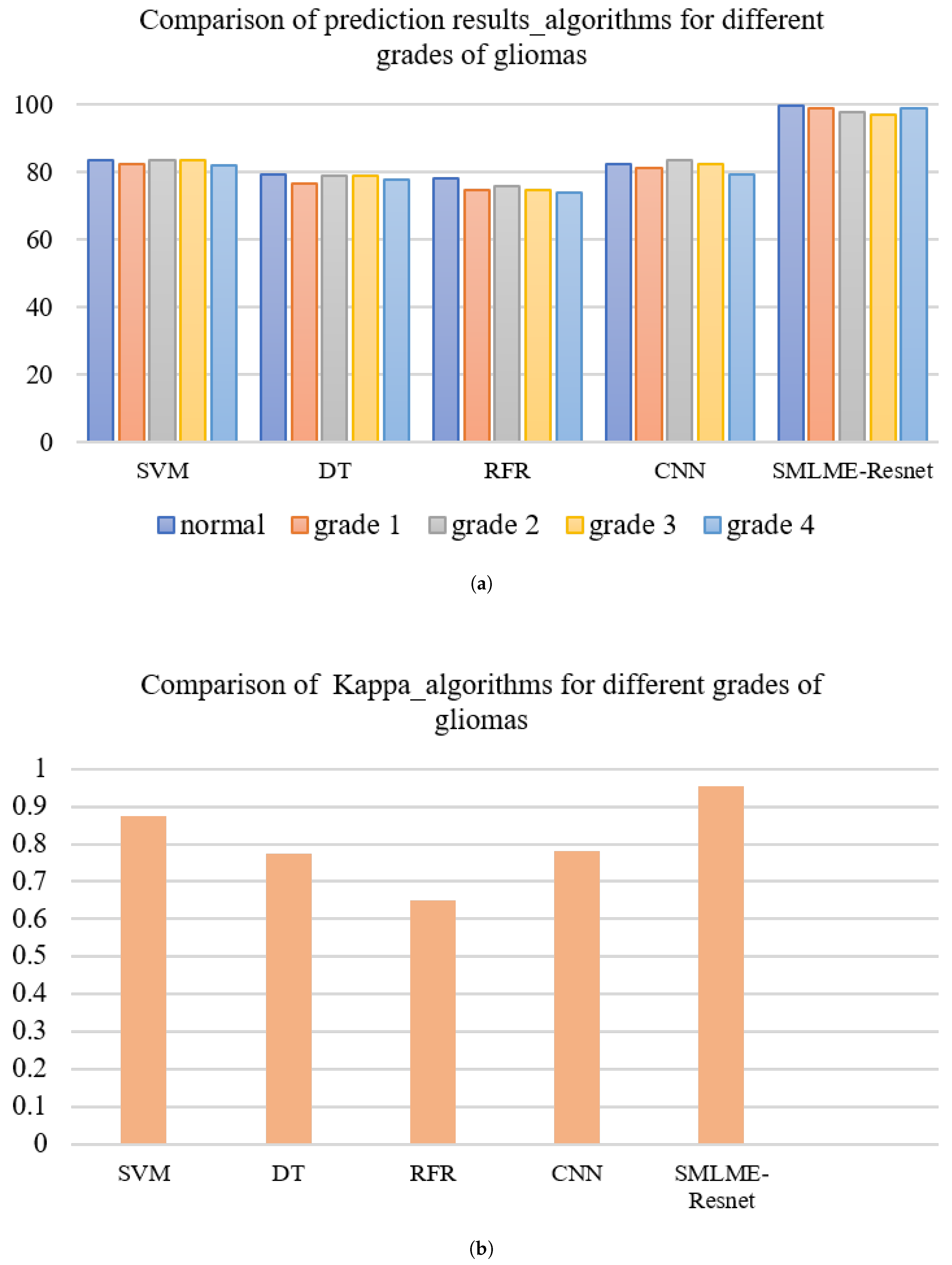
In order to verify the improvement in model performance, we compared the traditional SE-Resnet algorithm with the SMLME-Resnet algorithm proposed in this study for their ability in classifying pathological sections of brain gliomas at different levels. We used the same dataset and preprocessing steps for the two models. The classification results are shown in Table , and recognition results for different algorithms are shown in
Figure 15 and
Figure 16. Overall accuracy (OA), average accuracy (AA), and Kappa coefficient of SMLME-Resnet are superior to those produced by SE-Resnet and are much higher than those produced by other machine learning algorithms. These results demonstrate that the improvements introduced by our study can extract spatial-spectral features from brain gliomas pathological slices of different levels more effectively. SMLME-Resnet has slightly more tunable parameters (126540) than SE-Resnet (123780), but accuracy has been greatly improved, as is shown in
Table 3, demonstrating that the SMLME-Resnet method proposed in this study can retain the lightweight nature of the original model while achieving better accuracy in identifying malignant grades in
Figure 16.
4. Discussion
In recent years, machine learning methods, especially deep learning algorithms, have become an emerging technology to assist automated diagnosis in pathology. It can increase the speed of diagnosis and reduce inter-pathology variability [
32,
33,
34,
35,
36]. Previous studies have mainly used larger RGB image blocks and low magnification to include sufficient histologic features. Relatively complex network architectures are generally required to effectively classify these image blocks.HSI technology can capture small spectral differences caused by molecular changes, providing more useful information than traditional three-channel RGB tissue color information. As a result, HSI technology can benefit in many different microscopy applications, such as cancer detection in tissue sections.
This study focuses on the identification of malignant grade from pathological sections of glioma using microscopic hyperspectral images. We combine hyperspectral images with neural network algorithm properties at a deep level to accomplish the identification and classification of pathological sections of different subtypes of gliomas.And We introduce a multimodal and multiscale hyperspectral image feature extraction model, SMLME-Resnet, which integrates a channel attention mechanism. This model comprises a multiscale spectral feature extraction module, a hierarchical spatial feature extraction module, and a null-spectral fusion feature extraction module. These modules are designed for the classification of microscopic hyperspectral images of glioma pathology slices. The SMLME-Resnet module first assigns weights to the features of each channel to learn feature correlations between different channels, thus realizing the fusion of features across channels. The original convolution layer is replaced by a maximum pooling layer and by an up-sampling layer to extract salient features of the image, and features at different scales in the channel dimension are extracted by a parallel feature fusion method. The 1×1 convolutional layer is added at the level of the jump connection to compensate for the local information lost within the pooling layer. Concurrently, we extracted both shallow and deep semantic features of glioma pathology tissue sections, thereby laying the foundation for improving model accuracy. We first selected the best data dimensionality reduction method (SAA) through machine learning. In that process we found that the accuracy of subtype identification in pathological sections of gliomas in the brain was no more than 90% when different dimensionality reduction methods were combined with classification modeling methods, and that the SAA-SVM, SAA-DT, and SAA-RFR models outperformed the SPA-SVM, SPA-DT, and SPA-RFR models in terms of both accuracy and kappa. And then designed our experiments to identify optimal parameters of the SMLME-Resnet model. We set the image block size to 7×7 and 27×27. We set the learning rate to 0.001.The number of parameters in the improved SMLME-Resnet model of 126540 is higher than the number of parameters in the SE-Resnet model of 123,780, but the recognition accuracy is improved from 88.1% to 99.4%, which we consider acceptable By comparing several commonly used identification algorithms, we efficiently and accurately classified pathological sections of diffuse glioma in compliance with the classification method introduced by the WHO in 2016. Average classification accuracy values were 97.32% and the Kappa coefficient was 0.954. Compared with other deep learning methods, the SMLME-Resnet module proposed in this paper can effectively extract spatial features and channel features from micro-hyperspectral images, achieving better classification results with less computational cost and increased robustness. Compared with traditional machine learning methods, the model used in this paper can automatically learn relevant features from hyperspectral images and continuously optimize network parameters through backpropagation, which is more suitable for accurate classification of microscopic hyperspectral images.
5. Conclusion
The SMLME-Resnet algorithm proposed in this paper introduces the channel attention mechanism SE module before the traditional residual connection module. Building on this design, the algorithm introduces a parallel multi-scale image feature method to further extract relevant features from three-dimensional hyperspectral images. Our design improves the convolutional layer within the residual connection by converting it into multi-branch modules with different image sizes, which effectively reduces redundant information and improves the feature extraction ability of the network. This study effectively improves the accuracy of subtype identification in glioma pathology sections compared to conventional algorithms. Using this method, we are able to reach deep into the atlas information of glioma pathological sections, and identify diffuse glioma pathological sections efficiently and accurately according in compliance with the classification criteria introduced by the WHO in 2016. Our approach solves the problems associated with the poorly efficient, time-consuming, and laborious classification of malignant grades that limit existing auxiliary detection methods. Our study provides an effective tool for auxiliary diagnosis that can be effectively used by clinical pathologists.
However our work has some limitations. We obtained a small sample size for this study. In the future, we will increase our sample size and make full use of the available clinical resources at the Third Hospital of Jilin University to further validate our model, with the goal of promoting the adoption of our hyperspectral image analyzer within the clinical community. Another of our future endeavors is to explore which particular hyperspectral image bands or pathologic features in hyperspectral images that improve the classification results and work with hospitals to complete studies of medical mechanisms and improve the interpretability of the test.
Author Contributions
Conceptualization, jiaqi chen.and jinyu wang.; methodology, ci sun.; software,mingjia wang.; validation, jiaqi chen., jinyu wang., Yinyan Wang and zitong zhao.; formal analysis, zitong zhao.; investigation, jinyu wang.; resources, jin yang.; data curation,ci sun.; writing—original draft preparation, jiaqi chen and Jinnan Zhang.; writing—review and editing, jiaqi chen.; visualization, shulong feng.; supervision, jin yang.; project administration,nan song.; funding acquisition, jiaqi chen. All authors have read and agreed to the published version of the manuscript.
Funding
National Natural Science Foundation of China (62375265); Jilin Province and the Chinese Academy of Sciences Science and Technology Cooperation in High-tech Industrialisation Special Funds Project (2022SYHZ0025); Jilin Province Science & Technology Development Program Project in China(20210204216YY); Jilin Province Science & Technology Development Program Project in China(20210204146YY).
Data Availability Statement
Data underlying the results presented in this paper are not publicly available at this time but may be obtained from the authors upon reasonable request.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Weller, M.; Wick, W.; Aldape, K.; Brada, M.; Berger, M.; Pfister, S.M.; Nishikawa, R.; Rosenthal, M.; Wen, P.Y.; Stupp, R.; et al. Glioma. Nature reviews Disease primers 2015, 1, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Macyszyn, L.; Akbari, H.; Pisapia, J.M.; Da, X.; Attiah, M.; Pigrish, V.; Bi, Y.; Pal, S.; Davuluri, R.V.; Roccograndi, L.; et al. Imaging patterns predict patient survival and molecular subtype in glioblastoma via machine learning techniques. Neuro-oncology 2015, 18, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Kros, J.M.; Gorlia, T.; Kouwenhoven, M.C.; Zheng, P.P.; Collins, V.P.; Figarella-Branger, D.; Giangaspero, F.; Giannini, C.; Mokhtari, K.; Mørk, S.J.; et al. Panel review of anaplastic oligodendroglioma from European Organization For Research and Treatment of Cancer Trial 26951: assessment of consensus in diagnosis, influence of 1p/19q loss, and correlations with outcome. Journal of Neuropathology & Experimental Neurology 2007, 66, 545–551. [Google Scholar]
- Janowczyk, A.; Madabhushi, A. Deep learning for digital pathology image analysis: A comprehensive tutorial with selected use cases. Journal of pathology informatics 2016, 7, 29. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Yang, D.M.; Rong, R.; Zhan, X.; Fujimoto, J.; Liu, H.; Minna, J.; Wistuba, I.I.; Xie, Y.; Xiao, G. Artificial intelligence in lung cancer pathology image analysis. Cancers 2019, 11, 1673. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, R.S.; Graham, A.R.; Richter, L.C.; Barker, G.P.; Krupinski, E.A.; Lopez, A.M.; Erps, K.A.; Bhattacharyya, A.K.; Yagi, Y.; Gilbertson, J.R. Overview of telepathology, virtual microscopy, and whole slide imaging: prospects for the future. Human pathology 2009, 40, 1057–1069. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Xu, R.; Mei, H.; Zhang, L.; Yu, Q.; Liu, R.; Fan, B.; et al. Application of enhanced T1WI of MRI Radiomics in Glioma grading. International journal of clinical practice 2022, 2022. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, C.; Cheng, Y.; Teng, Y.; Guo, W.; Xu, H.; Ou, X.; Wang, J.; Li, H.; Ma, X.; et al. Ability of radiomics in differentiation of anaplastic oligodendroglioma from atypical low-grade oligodendroglioma using machine-learning approach. Frontiers in oncology 2019, 9, 1371. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.; Huang, S.; Pan, X.; Liao, X.; Yang, R.; Liu, J. Machine learning-based radiomics predicting tumor grades and expression of multiple pathologic biomarkers in gliomas. Frontiers in Oncology 2020, 10, 1676. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Reifenberger, G.; Von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta neuropathologica 2016, 131, 803–820. [Google Scholar] [CrossRef]
- Bjorgan, A.; Denstedt, M.; Milanič, M.; Paluchowski, L.A.; Randeberg, L.L. Vessel contrast enhancement in hyperspectral images. Optical Biopsy XIII: Toward Real-Time Spectroscopic Imaging and Diagnosis. SPIE, 2015, Vol. 9318, pp. 52–61.
- Akbari, H.; Kosugi, Y.; Kojima, K.; Tanaka, N. Blood vessel detection and artery-vein differentiation using hyperspectral imaging. 2009 Annual International Conference of the IEEE Engineering in Medicine and Biology Society. IEEE, 2009, pp. 1461–1464.
- Akbari, H.; Kosugi, Y.; Kojima, K.; Tanaka, N. Detection and analysis of the intestinal ischemia using visible and invisible hyperspectral imaging. IEEE Transactions on Biomedical Engineering 2010, 57, 2011–2017. [Google Scholar] [CrossRef] [PubMed]
- Mordant, D.; Al-Abboud, I.; Muyo, G.; Gorman, A.; Sallam, A.; Ritchie, P.; Harvey, A.; McNaught, A. Spectral imaging of the retina. Eye 2011, 25, 309–320. [Google Scholar] [CrossRef] [PubMed]
- Milanic, M.; Bjorgan, A.; Larsson, M.; Strömberg, T.; Randeberg, L.L. Detection of hypercholesterolemia using hyperspectral imaging of human skin. European Conference on Biomedical Optics. Optica Publishing Group, 2015, p. 95370C.
- Fabelo, H.; Ortega, S.; Kabwama, S.; Callico, G.M.; Bulters, D.; Szolna, A.; Pineiro, J.F.; Sarmiento, R. HELICoiD project: A new use of hyperspectral imaging for brain cancer detection in real-time during neurosurgical operations. Hyperspectral Imaging Sensors: Innovative Applications and Sensor Standards 2016. SPIE, 2016, Vol. 9860, p. 986002.
- Ogihara, H.; Hamamoto, Y.; Fujita, Y.; Goto, A.; Nishikawa, J.; Sakaida, I.; et al. Development of a gastric cancer diagnostic support system with a pattern recognition method using a hyperspectral camera. Journal of Sensors 2016, 2016. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, H.; Li, Q. Tongue tumor detection in medical hyperspectral images. Sensors 2011, 12, 162–174. [Google Scholar] [CrossRef] [PubMed]
- Akbari, H.; Halig, L.V.; Schuster, D.M.; Osunkoya, A.; Master, V.; Nieh, P.T.; Chen, G.Z.; Fei, B. Hyperspectral imaging and quantitative analysis for prostate cancer detection. Journal of biomedical optics 2012, 17, 076005–076005. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, T.; Zheng, Y.; Yin, X. Recognition of liver tumors by predicted hyperspectral features based on patient’s Computed Tomography radiomics features. Photodiagnosis and Photodynamic Therapy 2023, 103638. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, B.; Wang, Y.; Zhou, C.; Zou, D.; Vonsky, M.S.; Mitrofanova, L.B.; Li, Q. Dual-modality image feature fusion network for gastric precancerous lesions classification. Biomedical Signal Processing and Control 2024, 87, 105516. [Google Scholar] [CrossRef]
- Du, J.; Tao, C.; Xue, S.; Zhang, Z. Joint Diagnostic Method of Tumor Tissue Based on Hyperspectral Spectral-Spatial Transfer Features. Diagnostics 2023, 13, 2002. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Qiu, S.; Li, Q. Hyperspectral image segmentation of cholangiocarcinoma based on Fourier transform channel attention network. Journal of Image and Graphics 2021, 26, 1836–1846. [Google Scholar]
- Zhang, L.; Ye, N.; Ma, L. Hyperspectral Band Selection Based on Improved Particle Swarm Optimization. Spectroscopy and Spectral Analysis 2021, 41, 3194–3199. [Google Scholar]
- Mantripragada, K.; Dao, P.D.; He, Y.; Qureshi, F.Z. The effects of spectral dimensionality reduction on hyperspectral pixel classification: A case study. PLoS One 2022, 17, e0269174. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Luo, T.; Nie, J.; Chu, Y. Blood cancer diagnosis using hyperspectral imaging combined with the forward searching method and machine learning. Analytical Methods 2023, 15, 3885–3892. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Wang, Q.; Song, G. Selecting informative bands for partial least squares regressions improves their goodness-of-fits to estimate leaf photosynthetic parameters from hyperspectral data. Photosynthesis Research 2022, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Bian, X.H.; Li, S.J.; Fan, M.R.; Guo, Y.G.; Chang, N.; Wang, J.J. Spectral quantitative analysis of complex samples based on the extreme learning machine. Analytical Methods 2016, 8, 4674–4679. [Google Scholar] [CrossRef]
- Kifle, N.; Teti, S.; Ning, B.; Donoho, D.A.; Katz, I.; Keating, R.; Cha, R.J. Pediatric Brain Tissue Segmentation Using a Snapshot Hyperspectral Imaging (sHSI) Camera and Machine Learning Classifier. Bioengineering 2023, 10, 1190. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Hu, B.; Zhang, Z. Study on the classification method of gastric cancer tissues based on convolutional neural network and micro-hyperspectral. Acta Optica 2018, 38, 7. [Google Scholar]
- Tian, C.; Hao, D.; Ma, M.; Zhuang, J.; Mu, Y.; Zhang, Z.; Zhao, X.; Lu, Y.; Zuo, X.; Li, W. Graded diagnosis of Helicobacter pylori infection using hyperspectral images of gastric juice. Journal of Biophotonics 2023, e202300254. [Google Scholar] [CrossRef]
- Halicek, M.; Shahedi, M.; Little, J.V.; Chen, A.Y.; Myers, L.L.; Sumer, B.D.; Fei, B. Detection of Squamous Cell Carcinoma in Digitized Histological Images from the Head and Neck Using Convolutional Neural Networks. Proceedings of SPIE 2019, 109560K. [Google Scholar]
- Iizuka, O.; Kanavati, F.; Kato, K.; Rambeau, M.; Arihiro, K.; Tsuneki, M. Deep Learning Models for Histopathological Classification of Gastric and Colonic Epithelial Tumours. SCIENTIFIC REPORTS 2019. [Google Scholar] [CrossRef]
- Geijs, D.J.; Pinckaers, H.; Amir, A.L.; Litjens, G.J.S. End-to-end classification on basal-cell carcinoma histopathology whole-slides images. SCIENTIFIC REPORTS, 2021; 1160307. [Google Scholar]
- Wang, K.S.; Yu, G.; Xu, C.; Meng, X.H.; Zhou, J.; Zheng, C.; Deng, Z.; Shang, L.; Liu, R.; Su, S.; Zhou, X.; Li, Q.; Li, J.; Wang, J.; Ma, K.; Qi, J.; Hu, Z.; Tang, P.; Deng, J.; Qiu, X.; Li, B.Y.; Shen, W.D.; Quan, R.P.; Yang, J.T.; Huang, L.Y.; Xiao, Y.; Yang, Z.C.; Li, Z.; Wang, S.C.; Ren, H.; Liang, C.; Guo, W.; Li, Y.; Xiao, H.; Gu, Y.; Yun, J.P.; Huang, D.; Song, Z.; Fan, X.; Chen, L.; Yan, X.; Li, Z.; Huang, Z.C.; Huang, J.; Luttrell, J.; Zhang, C.Y.; Zhou, W.; Zhang, K.; Yi, C.; Wu, C.; Shen, H.; Wang, Y.P.; Xiao, H.M.; Deng, H.W. Accurate diagnosis of colorectal cancer based on histopathology images using artificial intelligence. BMC MEDICINE 2021, 19. [Google Scholar] [CrossRef]
- Liu, Y.; Yin, M.; Sun, S. DetexNet: Accurately Diagnosing Frequent and Challenging Pediatric Malignant Tumors. IEEE TRANSACTIONS ON MEDICAL IMAGING 2021, 40, 395–404. [Google Scholar] [CrossRef]
Figure 1.
Schematic diagram of glioma pathological chip array.
Figure 1.
Schematic diagram of glioma pathological chip array.
Figure 2.
brain glioma processing flow.
Figure 2.
brain glioma processing flow.
Figure 3.
Schematic diagram of microscopic hyperspectral acquisition system
Figure 3.
Schematic diagram of microscopic hyperspectral acquisition system
Figure 4.
Schematic diagram of hyperspectral data images from pathological sections of brain gliomas of different grades.
Figure 4.
Schematic diagram of hyperspectral data images from pathological sections of brain gliomas of different grades.
Figure 5.
Example spectral reflectance curve obtained from pathological section of brain glioma.
Figure 5.
Example spectral reflectance curve obtained from pathological section of brain glioma.
Figure 6.
Feature extraction process of hyperspectral images of pathological sections of gliomas of differentgrades. (a) AA%; (b) Kappa
Figure 6.
Feature extraction process of hyperspectral images of pathological sections of gliomas of differentgrades. (a) AA%; (b) Kappa
Figure 7.
Procedure adopted to implement the attention mechanism in different spectral channels.
Figure 7.
Procedure adopted to implement the attention mechanism in different spectral channels.
Figure 8.
The overall framework of the designed model.
Figure 8.
The overall framework of the designed model.
Figure 9.
The multi-scale spectral feature extraction framework.
Figure 9.
The multi-scale spectral feature extraction framework.
Figure 10.
The multi-scale hierarchical spatial feature extraction module.
Figure 10.
The multi-scale hierarchical spatial feature extraction module.
Figure 11.
The multi-scale spectral feature extraction framework.
Figure 11.
The multi-scale spectral feature extraction framework.
Figure 12.
The input image block sizes of the spectral feature extraction module.
Figure 12.
The input image block sizes of the spectral feature extraction module.
Figure 13.
The input image block sizes of the spatial feature extraction module.
Figure 13.
The input image block sizes of the spatial feature extraction module.
Figure 14.
Predictive results of glioma sections at various grades.
Figure 14.
Predictive results of glioma sections at various grades.
Figure 15.
Comparison of results before and after model improvement.
Figure 15.
Comparison of results before and after model improvement.
Figure 16.
Feature extraction process of hyperspectral images of pathological-sections of gliomas of different grades. (a) OA%, (b) Kappa
Figure 16.
Feature extraction process of hyperspectral images of pathological-sections of gliomas of different grades. (a) OA%, (b) Kappa
Table 1.
Comparison of results for different dimensionality reduction methods
Table 1.
Comparison of results for different dimensionality reduction methods
| |
|
|
|
OA% |
|
|
AA% |
Kappa |
| |
|
normal |
grade 1 |
grade 2 |
grade 3 |
grade 4 |
|
|
| Simulated |
SVM |
83.381 |
82.186 |
83.292 |
83.347 |
81.911 |
82.823 |
0.875 |
| annealing |
DT |
79.291 |
76.322 |
78.676 |
78.747 |
77.452 |
78.097 |
0.776 |
| algorithm (SAA) |
RFR |
77.852 |
74.667 |
75.781 |
74.575 |
73.866 |
75.348 |
0.648 |
| Successive |
SVM |
83.182 |
83.823 |
81.456 |
82.902 |
81.824 |
82.637 |
0.68 |
| Projections |
DT |
77.442 |
76.714 |
77.185 |
77.943 |
78.538 |
77.564 |
0.702 |
| Algorithm (SPA) |
RFR |
97.297 |
78.383 |
75.267 |
71.332 |
68.989 |
78.253 |
0.3803 |
Table 2.
Comparison of evaluation indicators for different learning rates
Table 2.
Comparison of evaluation indicators for different learning rates
| Learning rate |
|
|
OA% |
|
|
AA% |
| |
normal |
grade 1 |
grade 2 |
grade 3 |
grade 4 |
|
| 0.1 |
97.602 |
94.633 |
97.792 |
97.863 |
95.628 |
96.704 |
| 0.005 |
97.681 |
97.587 |
97.796 |
96.216 |
98.982 |
97.652 |
| 0.001 |
99.705 |
98.746 |
97.633 |
96.992 |
98.785 |
97.32 |
| 0.0005 |
99.701 |
98.874 |
97.342 |
96.99 |
98.689 |
98.319 |
Table 3.
Comparison of model results before and after improvement
Table 3.
Comparison of model results before and after improvement
Hierarchical
model |
|
|
OA/% |
|
|
AA/% |
Kappa |
Parameter
quantity |
| |
normal |
grade 1 |
grade 2 |
grade 3 |
grade 4 |
|
|
|
| SE-Resnet |
89.687 |
88.663 |
86.383 |
84.562 |
84.353 |
86.729 |
0.881 |
123780 |
| SMLME-Resnet |
99.705 |
98.746 |
97.633 |
96.992 |
98.785 |
97.32 |
0.954 |
126540 |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).